A novel role for Ets4 in axis specification and cell migration in the spider Parasteatoda tepidariorum
Figures
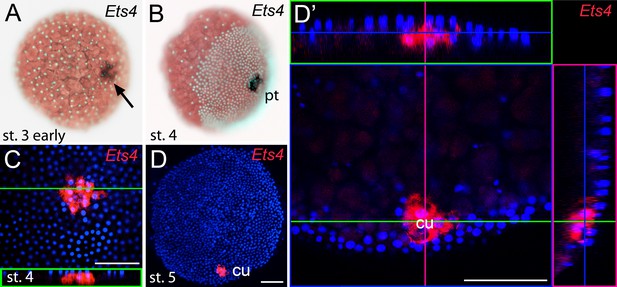
Early embryonic expression of Pt-Ets4.
(A and B) Pt-Ets4 is expressed within the cluster of cells (arrow in A) that will develop into the central primary thickening (pt) (B). (C–D’) Confocal scans (single optical slices (C and D’); maximum intensity projection (D)) of embryos stained for Pt-Ets4 (FastRed stain; red) and nuclei (DAPI; blue). Pt-Ets4 is expressed within the primary thickening and in the migrating cumulus (cu), which are covered by the surface epithelium of the germ-disc. Orthogonal views are boxed in green and magenta. The same embryo is depicted in D and D’. Scale bar is 100 µm.
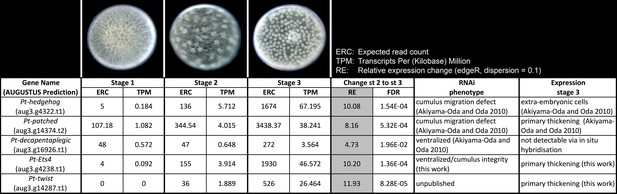
Expression profile analysis.
Expression profile in embryonic stages 1–3, RNAi phenotypes and expression of the genes Pt-hh, Pt-ptc, Pt-dpp, Pt-Ets4 and Pt-twi.
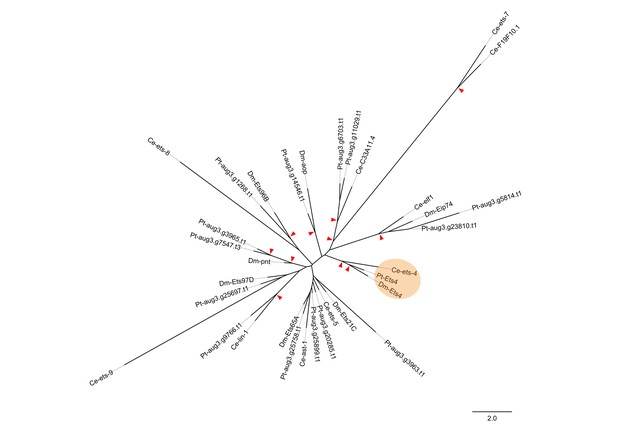
Phylogenetic analysis.
Maximum likelihood phylogeny of all ETS-family genes from D. melanogaster (Dm), C. elegans (Ce) and P. tepidariorum (Pt). Red arrowheads mark nodes with bootstrap support greater than 80. Scale is substitutions per site.
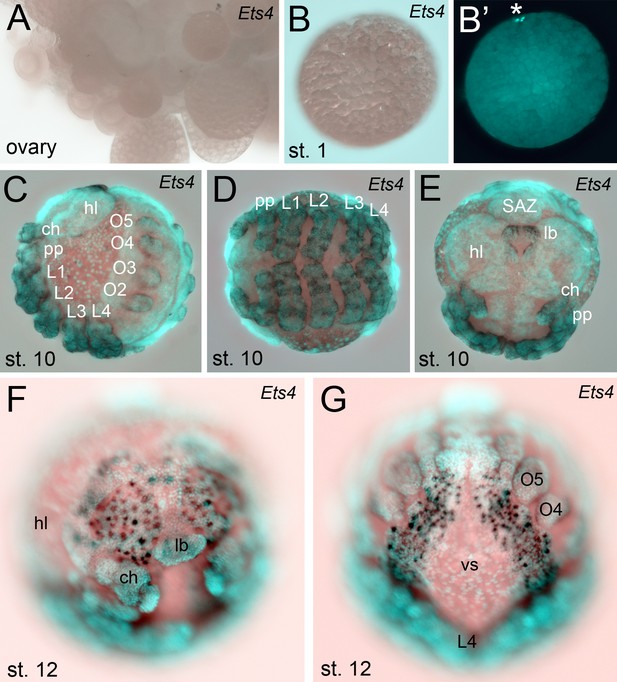
Extended expression analysis of Pt-Ets4.
Pt-Ets4 is not expressed in ovaries (A) and early stage 1 embryos (B and B’). Asterisk in B’ marks the polar bodies. (C–E) Expression of Pt-Ets4 at stage 10 (lateral view (C), ventral view (D), anterior view (E)). Pt-Ets4 is expressed in all developing appendages of the prosoma (ch-L4) and of the opisthosoma (O2–O5). Within the prosomal appendages Pt-Ets4 is expressed in rings (D). Pt-Ets4 is strongly expressed within the labrum (E) and in lateral domains surrounding the stomodeum. (F and G) Additional expression of Pt-Ets4 in the nervous system is detectable at embryonic stage 12 (anterior view (F); ventral view on the anterior part of the opisthosoma (G)). Abbreviations: ch: chelicera; pp: pedipalpus; L1-L4: walking legs 1–4, O2-O5: opisthosomal limb buds 2–5 (O2 develops into the book lung, O3 develops into trachea, O4 and O5 develop into the spinnerets); hl: head lobes; lb: labrum; vs: ventral sulcus; SAZ: segment addition zone.
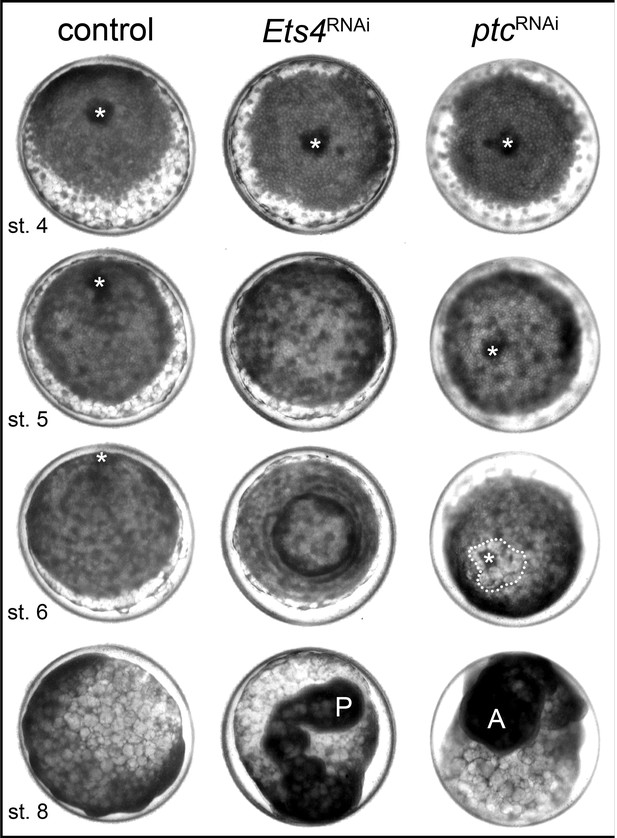
Pt-Ets4 is required for cumulus integrity.
Stills from the embryos shown in Video 1. The cumulus (asterisk) migrates in the control, disappears in the Pt-Ets4 RNAi and stays in the center of the germ-disc in Pt-ptc RNAi embryo. Ectopic, central opening (induction of the dorsal field) of the germ-disc is depicted via the dotted line (Pt-ptc RNAi st. 6). Posterior (P) and anterior (A) tube formation in Pt-Ets4 and Pt-ptc knockdown embryos is also indicated.
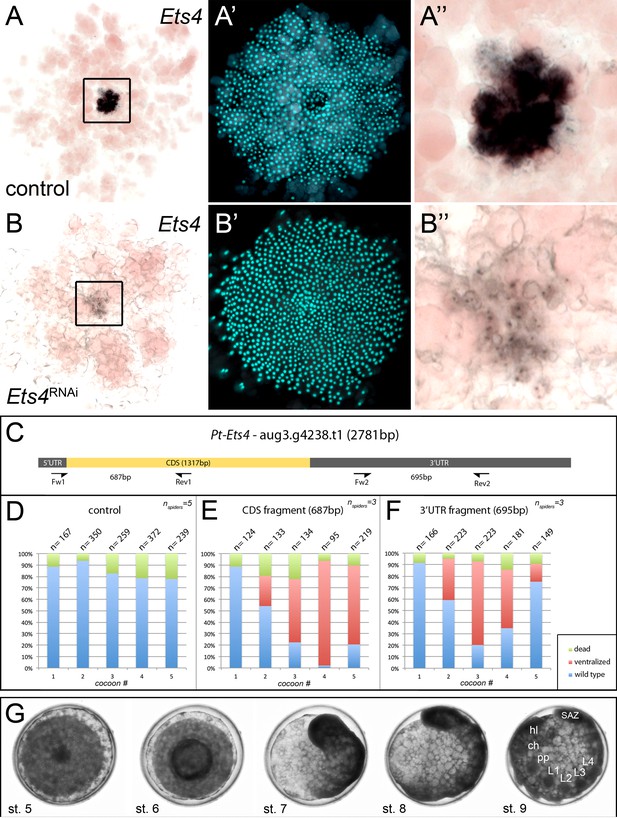
Knockdown efficiency after RNAi with Pt-Ets4.
In comparison to the control embryo (A–A’’) the expression of Pt-Ets4 is strongly silenced after Pt-Ets4 RNAi (B–B’’). Nascent Pt-Ets4 transcripts are still detectable in the nuclei of the primary thickening. The boxed region in A and B is magnified in A’’ and B’’. (C) Schematic representation of the AUGUSTUS prediction for Pt-Ets4 including the location of the untranslated regions (5’ and 3’ UTR), the coding sequence (CDS) and the primers (T7-Pt-CDS-Ets4-Fw (Fw1), T7-Pt-CDS-Ets4-Rev (Rev1), T7-Pt-3’Ets4-Fw (Fw2), T7-Pt-3’Ets4-Rev (Rev2), see Material and methods) that were used to generate two non-overlapping fragments of Pt-Ets4 that were used in the RNAi experiments (see Material and methods). (D–F) Statistical analysis of the knockdown efficiency after pRNAi with the CDS fragment and the 3’UTR fragment of Pt-Ets4. As many embryos were able to completely recover from the Pt-Ets4 knockdown at later stages of development (see embryo shown in G) the embryos were analyzed for their phenotypes at embryonic stages 6 and 7. (G) Stills from a time-lapse imaging experiment showing a strongly affected Pt-Ets4 RNAi embryo at developmental stages 6 and 7. The tube-like structure elongates from the posterior, folds back onto the yolk and re-establishes a DV axis during stages 8 and 9. The embryo has fully recovered from the early DV phenotype at stage 9 of development. This demonstrates the regulatory capacities of spider embryos.
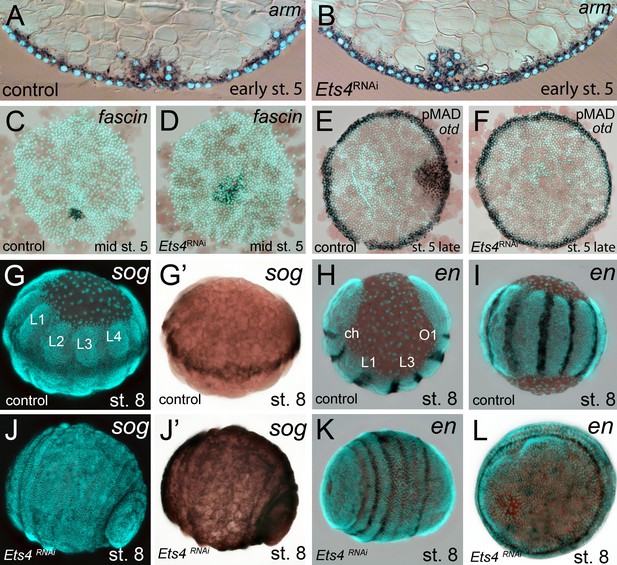
Cumulus integrity and signaling is affected in Pt-Ets4 knockdown embryos.
(A and B) Cross-section through the central cumulus of ubiquitously stained (via Pt-arm RNA in situ hybridization) control (A) and Pt-Ets4 RNAi (B) embryos. (C and D) control and Pt-Ets4 RNAi embryos stained for the cumulus marker Pt-fascin. Cells of the cumulus are dispersing in the Pt-Ets4 knockdown embryo (D). (E and F) Single color double stain of anterior Pt-otd expression (anterior ring) and nuclear localized pMAD in the cells overlaying the cumulus. pMAD signal is absent in Pt-Ets4 RNAi embryos (F). In situ hybridization for the ventral fate marker Pt-sog (G, G’, J and J’) or the segmental marker Pt-en (H, I, K and L) in control (G–I) and Pt-Ets4 knockdown embryos (J–L). The same embryos in fluorescence vs. bright field channel are shown in G and G’ as well as in J and J’. Nuclear stain (DAPI)/bright field overlay is shown in A-F, H, I, K and L. Flat mounted embryos in C-F. Lateral-ventral view (G, G’, J–K), lateral view (H), ventral view (I). Abbreviations: ch: cheliceral segment; L1-L4: walking leg bearing segments 1–4; O1: opisthosomal segment 1.
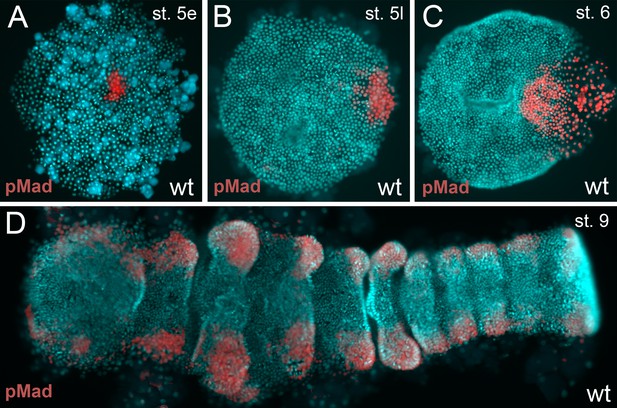
BMP pathway activity in wt embryos.
(A–D) The BMP signaling pathway (visualized via pMad antibody staining; false color images) is active from early stage 5 onwards. However, the germ-disc does not open up before late stage 5, after the cumulus has reached the rim of the disc.
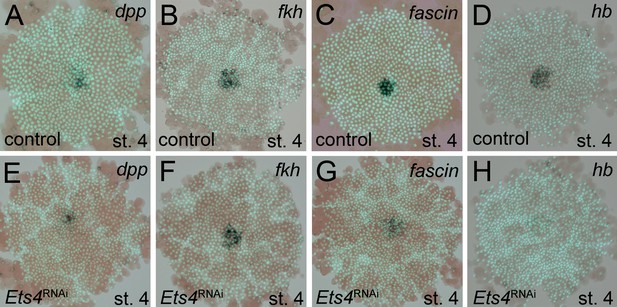
Analysis of ‘cumulus marker’ genes.
Flat mount preparation of in situ stained stage 4 embryos. While Pt-dpp and Pt-fkh expression is unaffected (compare A to E and B to F), Pt-fascin expression is slightly (compare C to G) and Pt-hb expression is strongly down regulated in Pt-Ets4 RNAi embryos (compare D to H).
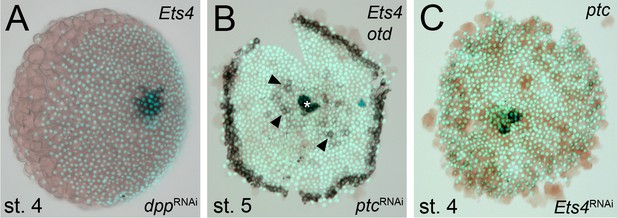
Regulation of Pt-Ets4 and Pt-ptc.
(A) Pt-dpp does not regulate the expression of Pt-Ets4. (B) Single color double staining detecting cumulus-specific Pt-Ets4 and anterior Pt-otd transcripts in a Pt-ptc RNAi embryo. Strong Pt-Ets4 expression (asterisk) is detectable in a Pt-ptc RNAi embryo. As already described (Akiyama-Oda and Oda, 2010), Pt-otd is ectopically activated in the center of the germ-disc (arrowheads). (C) Pt-Ets4 does not regulate the expression of Pt-ptc.
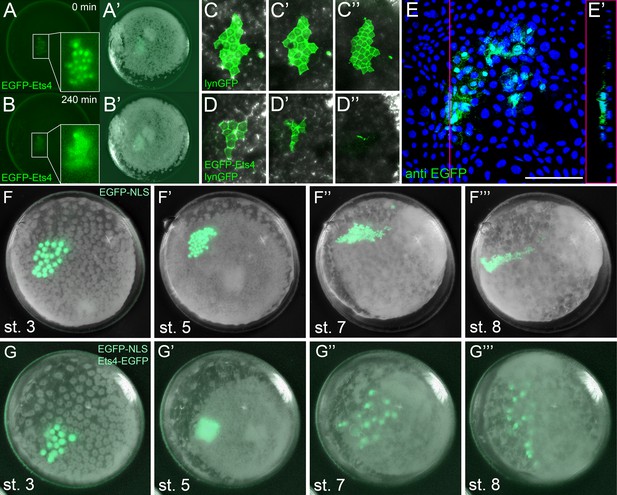
Ectopic expression of Pt-Ets4 causes the delamination and migration of cells.
(A and A’) A stage 4 embryo in which a cell clone has been marked via the ectopic expression of an EGFP-Pt-Ets4 fusion construct. The fusion protein localizes to the nuclei. (B and B’) The cell clone has delaminated four hours later. As the overlaying epithelium is highly light-scattering, the nuclear EGFP signal is no longer visible. The inset shows the magnification of the boxed region in A and B. (C and D) Stills from Video 2A and D (magnifications of the lynGFP positive regions). Cells expressing lynGFP alone (C–C’’) stay at the surface epithelium of the germ-disc. Cells expressing lynGFP/EGFP-Pt-Ets4 (D–D’’) constrict and delaminate. (E) EGFP antibody staining (maximum intensity projection is shown in E and the orthogonal view is shown in E’) of a fixed embryo ectopically expressing EGFP-Pt-Ets4. (F–F’’’) Stills from Video 3A (control). A cell clone marked via the ectopic expression of nuclear localized EGFP (EGFP-NLS) marks the ectoderm during germ-band formation. (G–G’’’) Stills from Video 3B (ectopic expression of Pt-Ets4). A cell clone ectopically expressing EGFP-Pt-Ets4 in combination with EGFP-NLS delaminates after germ-disc formation (G’). The cells of this cell clone start to disperse during later stages of development (G’’ and G’’’). Scale bar is 100 µm in E.
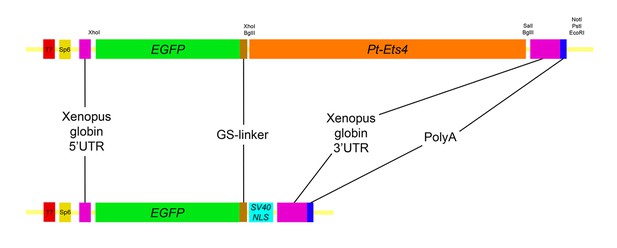
Constructs.
Schematic representation of the constructs that were used for the production of capped mRNA. The EGFP-Pt-Ets4 fusion construct was synthesized and cloned into the pUC57 vector by Eurofins Genomics. This construct was modified (see Material and methods) to generate the nuclear localized form of EGFP (via insertion of the SV40-NLS sequence). Location of used restriction sites (used for modification and linearization) is indicated above the EGFP-Pt-Ets4 fusion construct. The PolyA tail consists of 25 residues.
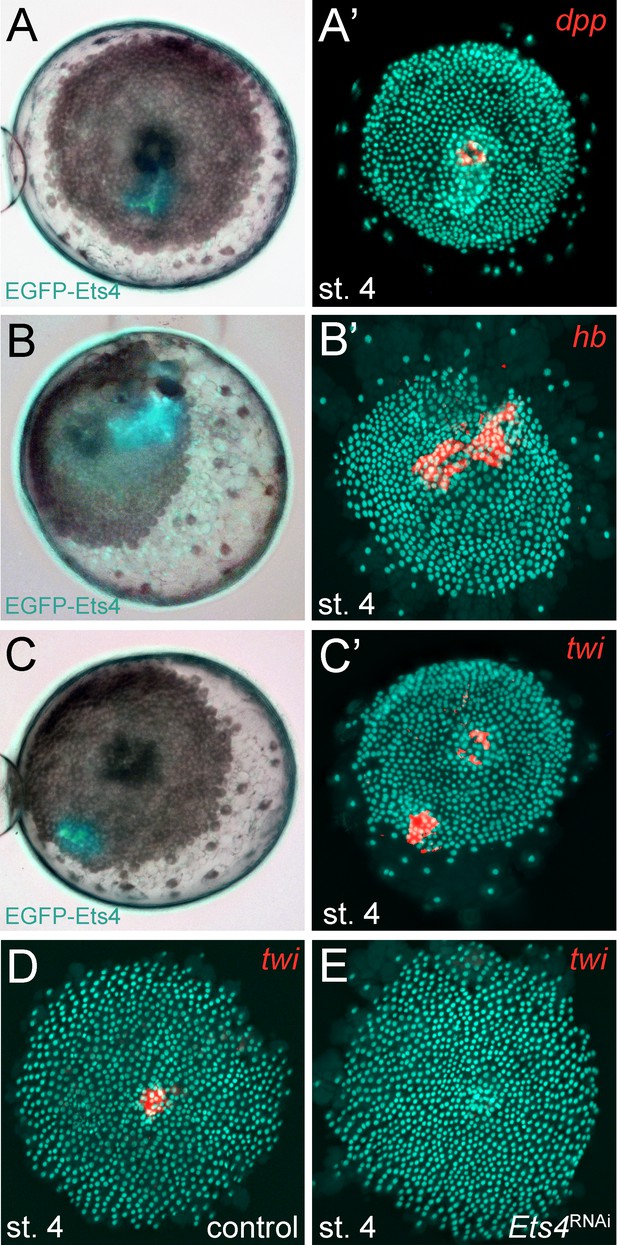
Pt-Ets4 regulates Pt-hb and Pt-twi expression within the primary thickening.
Live stage 4 embryos in which a cell clone is marked via the ectopic-expression of EGFP-Ets4 are depicted in A-C. The same embryos have been fixed and analyzed for their expression of Pt-dpp (A’), Pt-hb (B’) and Pt-twi (C’), respectively. Expression of Pt-twi in a control (D) and a Pt-Ets4 knockdown embryo (E). A’, B’, C’, D and E are false-color overlays of in situ hybridization images.
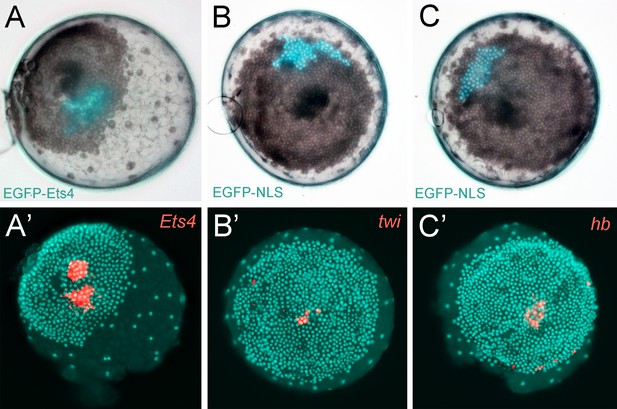
Controls for the ectopic expression of EGFP-Ets4.
(A–C) Live stage 4 control embryos in which a cell clone is marked via the ectopic expression of EGFP-Ets4 (A) or EGFP-NLS (B and C). The same embryos have been fixed and analyzed for their expression of Pt-Ets4 (A’), Pt-twi (B’) and Pt-hb (C’), respectively (false-color overlays of in situ hybridization images). Injected EGFP-Ets4 mRNA is detectable within the marked cell clone (A’). The cell clones ectopically expressing EGFP-NLS are negative for Pt-twi and Pt-hb transcripts (B’ and C’).
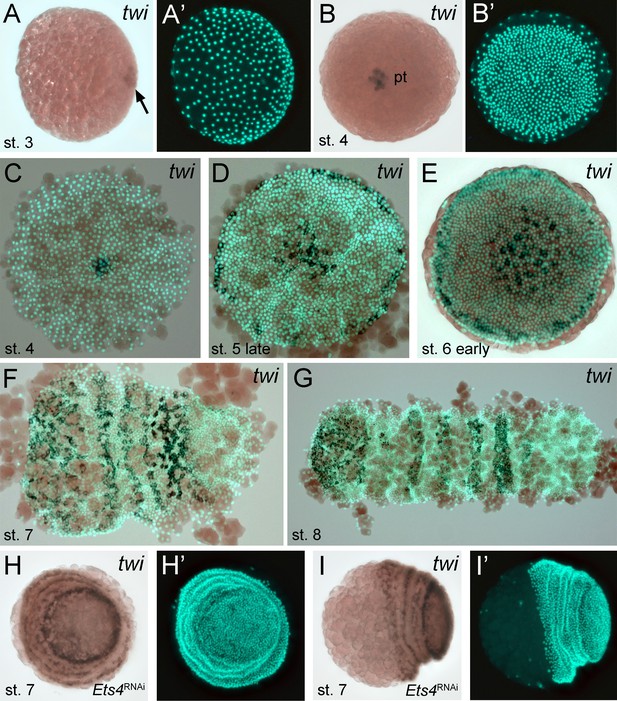
Expression of Pt-twi in wt and Pt-Ets4 RNAi embryos.
Newly discovered expression of Pt-twi in the central cell cluster of the forming germ-disc (arrow in A) and the primary thickening (pt in B) is depicted in (A–C). The already published expression of Pt-twist (Yamazaki et al., 2005) is depicted in (D–G). The expression of Pt-twi in stage 7 Pt-Ets4 RNAi embryos is not affected (H and I). Anterior is to the left (where possible).
Videos
Knockdown of Pt-Ets4 and Pt-ptc.
Live imaging of a control (A), a Pt-Ets4 RNAi (B) and a Pt-ptc RNAi (C) embryo under transmitted light conditions. The video starts at stage 3 and ends at stage 9 of embryonic development. Cumulus migration and normal germ-band formation is visible in the control embryo (A). The cells of the cumulus disperse in the Pt-Ets4 knockdown embryo (B). The ventralized Pt-Ets4 RNAi embryo stays radially symmetric and posterior tube formation is initiated (30 hr onwards). The cumulus of the Pt-ptc RNAi embryo does not migrate (C). The germ-disc opens up at the central position and the radially symmetric embryo overgrows the yolk and anterior tube formation is initiated (48 hr onwards).
Ectopic expression of Pt-Ets4 causes the delamination of cells.
Ectopic expression of lynGFP (A) and lynGFP in combination with EGFP-Ets4 (B-D). The lynGFP positive cell clone (A) stays in the ectodermal cell layer of the germ-disc. Regardless of the shape of the cell clones, Pt-Ets4 positive cells apically constrict and delaminate (B-D).
Ectopic expression of Pt-Ets4 causes the delamination and migration of cells.
Ectopic expression of EGFP-NLS (A) and EGFP-NLS in combination with EGFP-Ets4 (B). The cell clone positive for EGFP-NLS (A) stays in the ectodermal cell layer of the germ-disc. Cells further divide and form a long and thin cell clone (via convergent extension [Kanayama et al., 2011]) at stage 8 of embryonic development. The Pt-Ets4 positive cell clone delaminates at stage 4 (B). Cells stop dividing and start to disperse as soon as the cells of the cumulus start to migrate (st. 5).
Ectopic expression of EGFP-NLS and EGFP-Pt-Ets4 in multiple embryos.
Ectopic expression of EGFP-NLS (A-D) and EGFP-NLS in combination with EGFP-Ets4 (E-H). EGFP-NLS (A-D) and ectopic clones expressing Pt-Ets4 (E-H) have equal positions within the germ-disc. While all of the control cell clones (A-D) form long and thin stretched cell clones at stage 8 of embryonic development, the Pt-Ets4 positive cells clones (E-H) delaminate and the cells disperse from stage 5 onwards. Only the EGFP channel is shown for all embryos.
Additional files
-
Supplementary file 1
Full sequence of the EGFP-Pt-Ets4-PolyA fusion construct
- https://doi.org/10.7554/eLife.27590.022





