Developmental adaptations of trypanosome motility to the tsetse fly host environments unravel a multifaceted in vivo microswimmer system
Figures
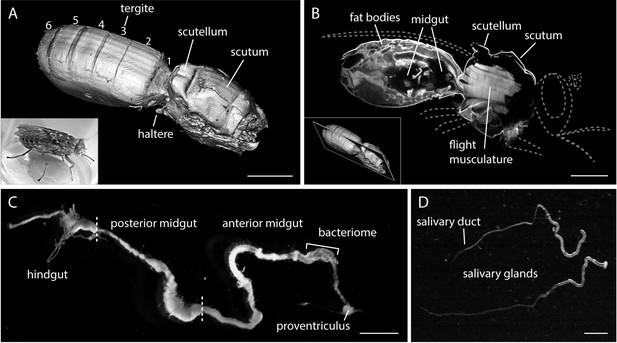
The alimentary tract is the trypanosome habitat within the tsetse fly.
(A) A surface rendering model of an intact female tsetse fly thorax and abdomen, visualised by multicolour LSFM. Head, extremities and a part of the back shield (scutellum) were removed. The fly was fixed, bleached and cleared to enable the autofluorescence recording of the complete abdomen and thorax. The inset shows a living tsetse fly. Scale bar: 1 mm. (B) Mid-section of the three-dimensional data set (Video 1) showing internal anatomical details. The grey dotted lines indicate removed body parts (not to scale). Inside the thorax the flight musculature is strongly autofluorescent, whereas mainly fat bodies are visible in the abdomen. The abdominal midgut is detected as a void volume of low to negligible autofluorescence. The inset indicates the position in the image stack. Scale bar: 1 mm. (C) Surgically removed, intact alimentary tract of a teneral fly. The midgut was sprawled out and freed from remaining tissue. The midgut is divided into a posterior part, where the blood meal is digested, and a thinner anterior part, which includes the bacteriome and ends in the proventriculus. Scale bar: 3 mm. (D) Two salivary glands with the thinner salivary ducts. Scale bar: 1 mm.
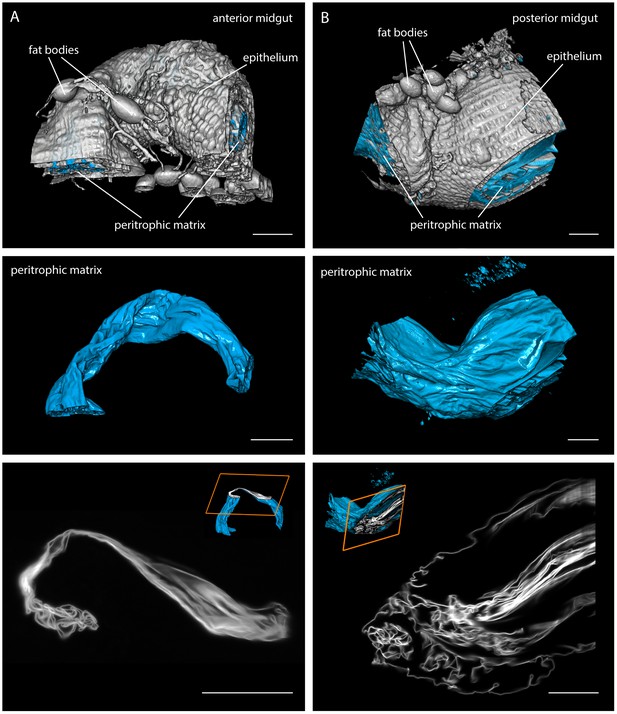
Multicolour light sheet fluorescence microscopy details the highly convoluted peritrophic matrix within the tsetse midgut.
Fly midguts were surgically removed 1–2 days after the last meal. The intestinal tissue exhibits a strong autofluorescence signal, whereas the PM (cyan) is visualised with rhodamine-labelled WGA. The grey surfaces in the top panel show the epithelial tissue surrounding the PM and attached residual fat bodies. The outer surfaces of the PMs are depicted in the middle panel. The bottom panel shows single image slices of 3D stacks, illustrating the complex membrane folding. The position within the volume stack is shown as orange box in the cutaway model (inset). (A) Representative part of the anterior midgut region. (B) The posterior midgut region has a larger diameter and contains a more convoluted PM, as well as large void sections of the endoperitrophic gut lumen. Scale bars: 100 µm. Video 2 contains animated versions of the 3D-PM models and the entire LSFM stacks.
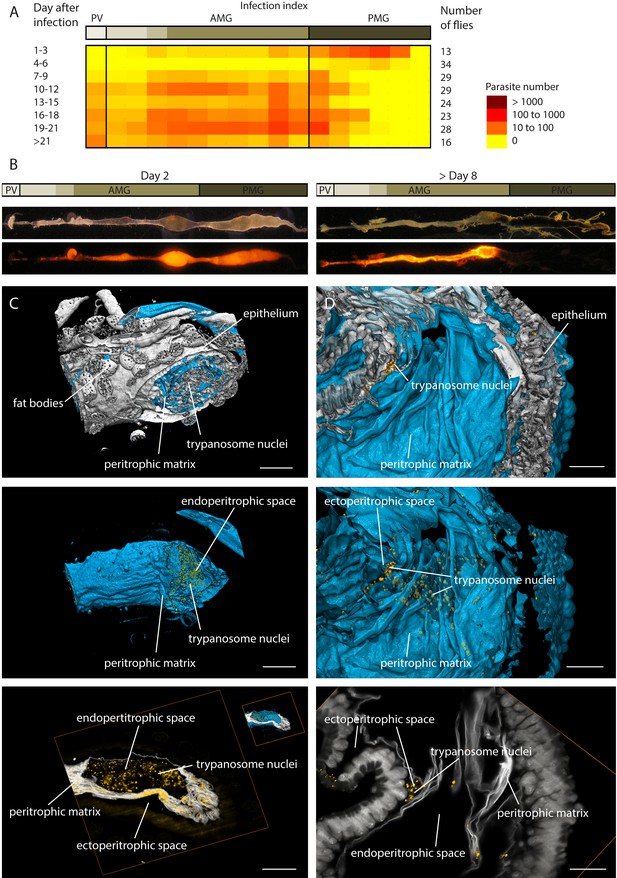
Trypanosome midgut infections can be monitored dynamically and with single parasite resolution.
(A) Heat map of the early infection process, analysed by the amount of fluorescent trypanosomes in different areas: proventriculus (PV), anterior midgut (AMG) and posterior midgut (PMG). Trypanosomes are located in the posterior midgut between 1–3 days after the infective blood meal. The flies´ immune system clears a large part of the parasite population between days 4–6 (Aksoy et al., 2003; Van Den Abbeele et al., 1999). In flies that were not able to eliminate all parasites, the population expands again, while the trypanosomes concentrate in anterior regions. The midgut stays infected for the remaining life-span (Gibson and Bailey, 2003). (B) The distribution of trypanosomes at day two after the infective meal is shown on the left. The typical infection pattern after a stable infection (>8 days) is shown on the right, where the trypanosomes have invaded the ectoperitrophic space. (C) and (D) show surface rendering models of isolated infected fly guts. The intestinal tissue is visualised by autofluorescence (grey). The PM is stained with rhodamine-labelled WGA (cyan) and the trypanosome nucleus with a GFP-reporter (yellow). (C) Dissected part of the midgut 2 days post-infection. The PM is shown isolated in the middle panel, together with the fluorescent trypanosome nuclei, which are located exclusively inside the internal midgut lumen (animated in Video 3). In the bottom panel a single plane shows the localisation of the trypanosomes within the endoperitrophic space (animation of full stack in Video 3). Scale bars: 100 µm. (D) Dissected midgut >day 8 post-infection. The top view is onto the inside surface of the PM (view point in the gut lumen), with underlying epithelial tissue and trypanosomes between folds of the PM belonging to the ectoperitrophic space. The middle panel allows the same view, albeit with the PM rendered transparent, in order to visualise the trypanosomes concentrated around the outer surface of the PM. The single slice in the bottom panel resolves groups of nuclei in the ectoperitrophic space whereas the endoperitrophic space is void (animation of full stack in Video 3). Scale bars: 50 µm. Figure 3—Videos 1 and 2. Infective metacyclic cells expressing nuclear GFP (Figure 3—Video 1) or PFR-GFP (Figure 3—Video 2) are imaged immediately after release from the salivary glands, showing that the transgenic cell lines successfully complete the developmental cycle in the tsetse fly. Figure 3—Video 3. Multicolour light sheet fluorescence microscopy details the peritrophic matrix within the tsetse midgut and allows identification of trypanosomes with single cell accuracy (Figure 3—Video 3).
Metacyclic cells expressing nuclear GFP successfully complete the developmental cycle in the tsetse fly.
https://doi.org/10.7554/eLife.27656.007Metacyclic cells expressing PFR-GFP successfully complete the developmental cycle in the tsetse fly.
https://doi.org/10.7554/eLife.27656.008Multicolour light sheet fluorescence microscopy details the peritrophic matrix within the tsetse midgut and allows identification of trypanosomes with single cell accuracy.
https://doi.org/10.7554/eLife.27656.009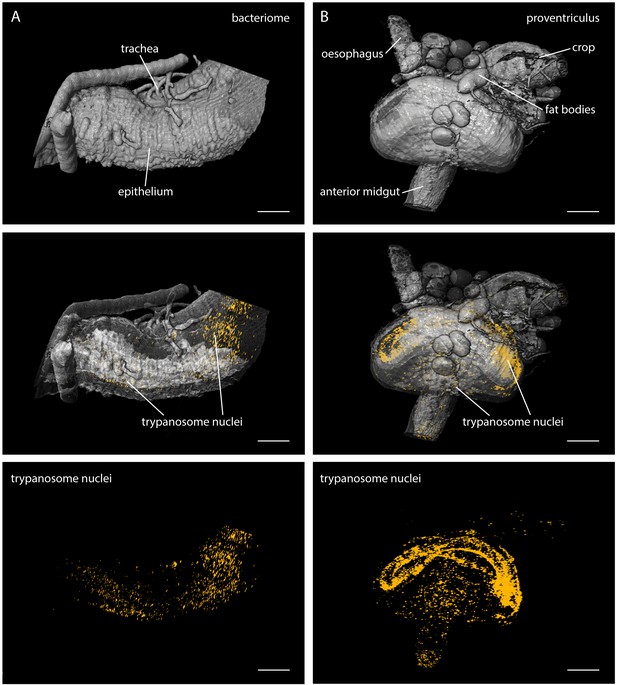
LSFM quantifies trypanosome populations in complex tsetse organs.
Infected tsetse organs were surgically removed and rendered using LSFM autofluorescence data in the top panel (grey). Nuclear GFP reveals the distribution of trypanosomes in the corresponding volume in the lower panels (yellow). The middle panel shows the merged 3D-localisations with semi-transparent organ models. (A) The bacteriome is located in the anterior midgut region and harbours endosymbiotic bacteria. It has a characteristic three-dimensional structure which is discernible with the appropriate transparency settings, due to higher autofluorescence levels (middle panel). The trypanosomes are excluded from this organ and located in the ectoperitrophoic space around the juxtaposed PM (not labelled in this specimen). (B) The proventriculus connects the anterior midgut, the crop and the salivary duct. The PM (not labelled) is produced here by a ring of specialised cells. Trypanosomes accumulate in partially high cell densities around this toroid structure. Scale bars: 100 µm. Video 4 contains animations of the proventriculus model and of the corresponding LSFM data stack.
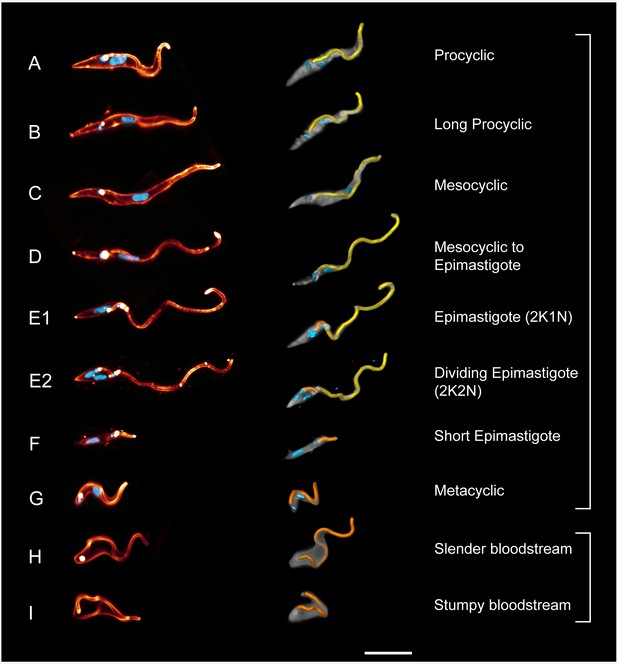
High-resolution 3D-morphometry of all trypanosome life cycle stages.
Trypanosomes were isolated from infected tsetse flies (A–G) or mice (H–I). The cell surface was fluorescently labelled with a sulfo-NHS dye (red) and the cell nuclei and kinetoplasts were labelled with DAPI (cyan). In the left panel, representative 3D-volume models of surface-labelled parasites are shown. The right panel presents the corresponding 3D surface models, with the cell body in grey and the attached flagellum in yellow or orange. Scale bar: 10 µm. Procyclic cells (A) exhibit a 180° right hand turn of the flagellum around the cell body. Long procyclic cells (B) are larger than normal procyclics, but reveal the same characteristic flagellar attachment. Mesocyclic cells (C) show a more elongated cell body and a straighter flagellar attachment, also fulfilling a 180° flagellar turn, but lack a free flagellar tip. During the transformation from mesocyclic to epimastigote cells (D), the nucleus elongates and moves posterior, and the cytoplasm at the anterior tip retracts. After repositioning, the epimastigote cell begins cell division, with the kinetoplast duplicating first (E1, 2K1N configuration). The orange flagellum represents the new flagellum of the adolescent short epimastigote daughter cell. Shortly before cytokinesis, the dividing epimastigote cell (E2, 2K2N configuration) shows an almost sperm-like appearance. The short epimastigote cell (F) resulting from this division has a thin, straight cell body with an extremely reduced flagellum protruding from the anterior end. Infective metacyclic cells (G) in contrast, have a curly appearance with a 180° turn of the flagellum, which originates from the posterior end. Metacyclic cells thus re-establish the trypomastigote configuration, which is maintained in the slender (H) and stumpy bloodstream forms (I). (H, I, adapted from [Bargul et al., 2016]). (Video 5 contains animations of the 3D-surface models)
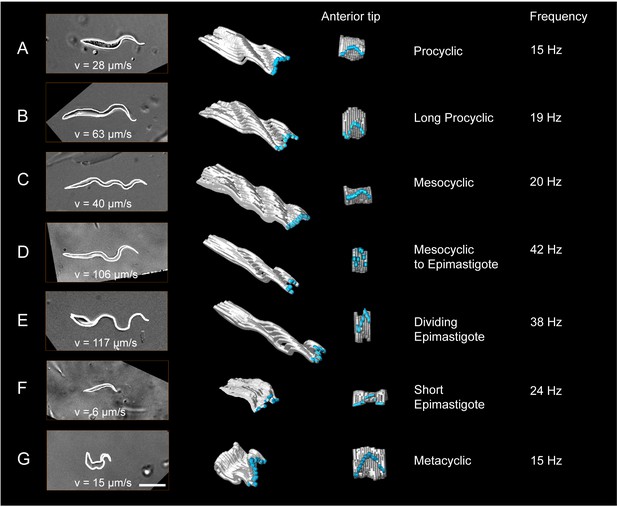
Measurement of the dynamic cellular waveforms of different trypanosome morphotypes.
Images in the left panel are stills from videos, captured with a frame rate of 250 fps. The speed (v in µm/s) represent the highest velocity reached during the video sequence analysed. The panel in the middle is a model of the outline for one single flagellar beat analysed frame by frame. The frames were stacked along the time axis in a three-dimensional surface representation, which allows the visualisation of the cellular waveform produced by the flagellum and the cell body in two different views. The flagellar tip was highlighted in blue. The model shows the travelling waves running along the cell body in a top-diagonal view and one wavelength of the flagellar beat in the view of the anterior tip. The frequency (Hz) of the analysed flagellar beat is shown on the right. Procyclic cells (A) and long procyclic cells (B) show similar waveform patterns, although the long procyclic cells generally swim faster. Mesocyclic cells (C) show a characteristic waveform due to their small amplitude during flagellar beating. When they start differentiating from mesocyclic to epimastigote cells (D), the amplitude increases again with a higher frequency and cells gain more speed. Dividing epimastigote cells (2K2N) (E) have proven to be the fastest swimmers of tsetse fly stages. Short epimastigote cells (F) are weak swimmers, despite beat frequencies similar to procyclic cells, due to their lack of a free flagellum. Infective metacyclic (G) cells show an increase in amplitude and a characteristic curly waveform, while reaching medium beat frequencies and swimming speeds. Video 6 contains all original video sequences selected for waveform analysis. The videos of trypanosome stages are consecutively played to show the position of each traced waveform along the time-axis in the 3D-models.
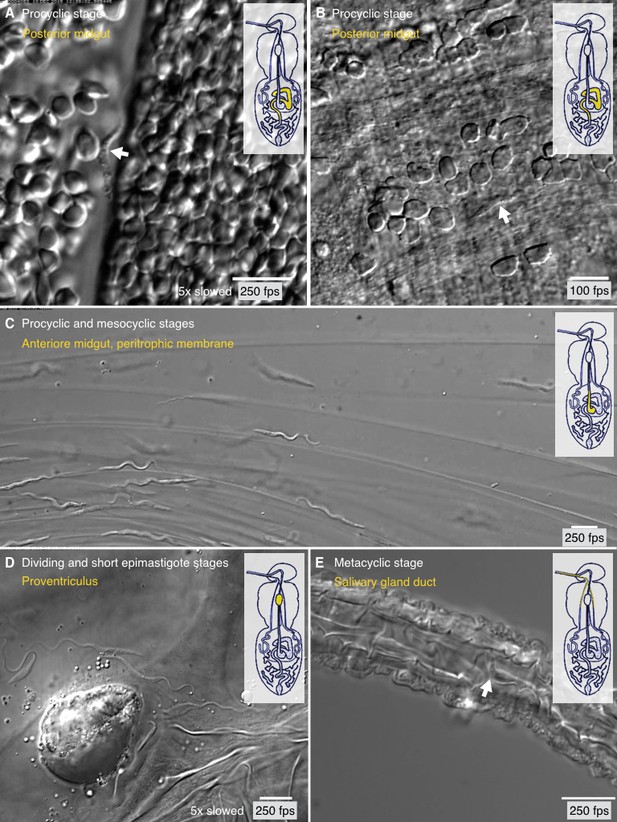
Trypanosome life cycle stages as solitary microswimmers in varying tsetse environments.
Infected tsetse flies were dissected in PBS and various regions, marked in the inset cartoon fly, were analysed by high speed microscopy (100 to 250 fps). Images are stills of the corresponding videos (Video 7), showing trypanosomes (white arrows) of various developmental stages. Scale bars: 10 µm. (A) Procyclic trypanosome swimming between blood cells in the posterior midgut lumen shortly after feeding. (B) A procyclic cell in tissue of the posterior midgut, confined by gut epithelium. (C) Procyclic to mesocyclic transition stages swimming along sheets of dissected PM in the anterior midgut. Trypanosomes experience different degrees of confinement and display characteristic straight trajectories and U-turns. (D) Various epimastigote cells inside the proventriculus, confined to a limited fluid-filled cavern. (E) Single metacyclic cells inside the thin salivary gland duct. The cells are motile, but mainly tumble around one position, as they await the tsetse fly´s next blood meal. Video 7 plays the original videos simultaneously with the annotated speeds.
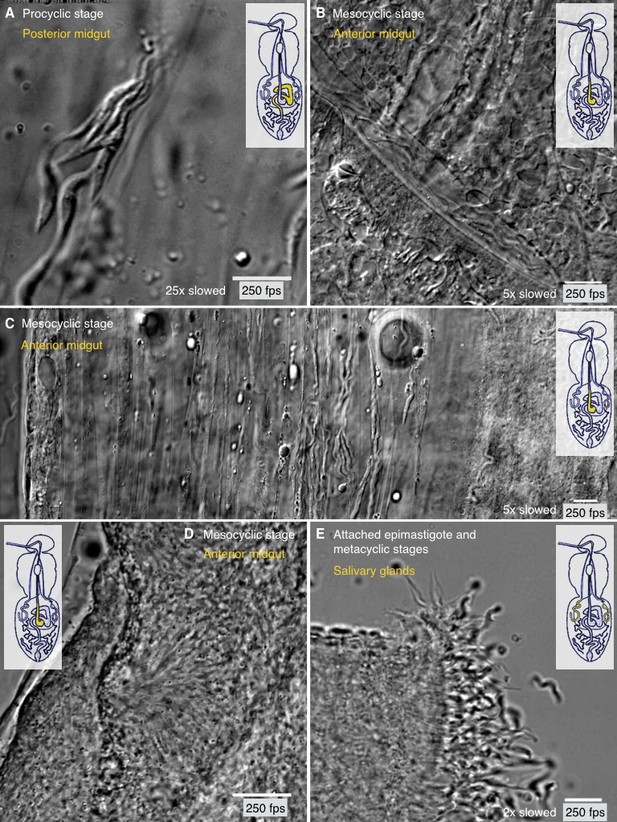
Different degrees of trypanosome crowding and environmental confinement can be found throughout the fly.
Infected tsetse flies were dissected in PBS and various regions, marked in the inset cartoon fly, were analysed by high speed microscopy (250 fps). Images are stills of the corresponding videos (Video 8). Scale bars: 10 µm. (A) Procyclic cells at day two after infection in the posterior midgut show the ability to form clusters and synchronise their flagellar oscillations. (B) Long procyclic to mesocyclic transition stage cells packed within a channel in the anterior midgut tissue of a late stage infected tsetse. (C) Mesocyclic cells in anterior midgut tissues and encased in folds of the PM. Depending on the degree of confinement, partly synchronised clusters of cells are visible. Strongly confined single cells display significant bending of the cell body and are able to perform sharp U-turns in the limited space. (D) High density swarms of mesocyclic cells inside the midgut ectoperitrophic space create superordinate wave patterns and generate tissue deforming force. (E) Sliced salivary gland with epithelium-attached epimastigote cells and free pre-metacyclic cells floating in the surrounding medium. The intact tissue was too dense to allow imaging of salivary gland stages by light microscopy, therefore the organ was dissected to show the free posterior ends of attached epimastigote trypanosomes in a limited region. Video 8 plays the original videos simultaneously with the annotated speeds.
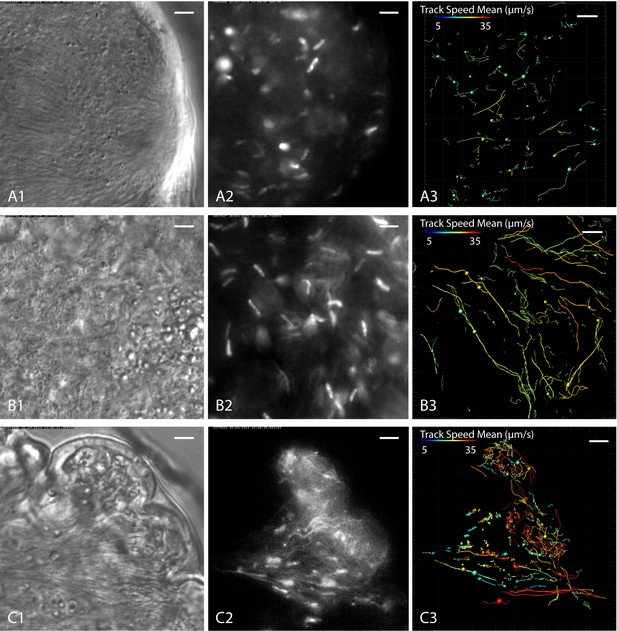
Cell tracking details single trypanosome motion behaviour in parasite clusters.
Infected tsetse flies were dissected in PBS and specific areas containing high trypanosome concentrations selected. Fluorescent trypanosomes express GFP signal in the nucleus (A and B) or additionally in the flagellum (C). The images are stills of the corresponding videos captured with 250 fps (Video 9). The left panel shows transmitted light images, the middle panel shows fluorescence images of the same region and the right panel the corresponding tracking analysis. The mean track speed is colour coded and shows a range of 5–35 µm/s. Scale bars: 5 µm. (A) Accumulation of mesocyclic trypanosomes inside the ectoperitrophic space in anterior midgut tissue. The trypanosomes show less persistent swimming in this area, due to dense synchronised clusters of parasites. (B) Mesocyclic cells and mesocyclic to epimastigote transition stages with elongated cell nuclei inside the proventriculus. There are more persistent swimmers in this region, probably due to lesser cell crowding and topographical structures effectively producing microswimmer channels. (C) Trypanosomes labelled with a nuclear and/or a flagellar marker, in anterior midgut tissue, experiencing different levels of confinement in close proximity. In the left region a high degree of clustering and synchronisation is obvious, whereas to the right, fast single parasites are tracked swimming in fluid-filled cavernous regions. Single parasites are tracked swimming into the cluster at the left, synchronise their oscillations temporarily and eventually reverse swimming direction and leave the swarm. Video 9 shows the original videos and the synchronous animated tracking data in original speed.
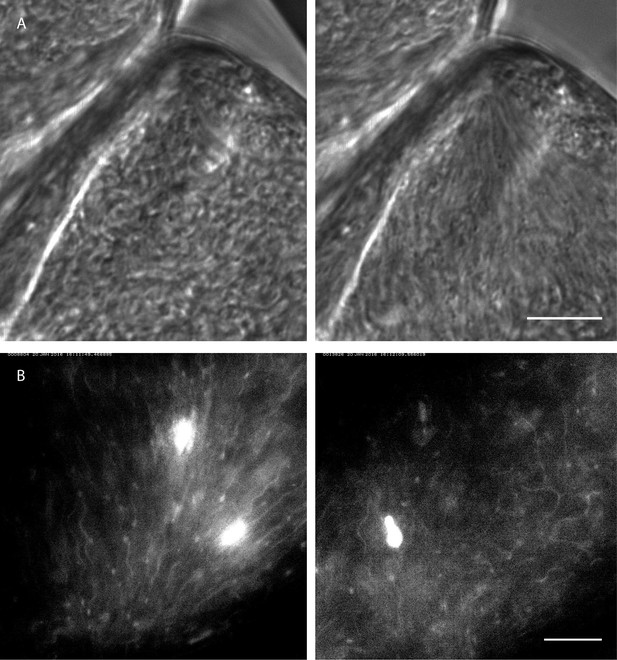
Rapid switching between synchronised and chaotic motion inside the ectoperitrophic space of the tsetse fly midgut.
Infected tsetse fly midguts were dissected and analysed in PBS. Swarms of trypanosomes in the long procyclic to mesocyclic transition stages were recorded with 250 fps in the ectoperitrophic space of the anterior midgut (Video 10). Scale bars: 10 µm. (A) Switch from chaotic (left panel) to synchronised motion (right panel). (B) Synchronised motion (left panel) of cells and transition to chaotic movement (right panel) within a few seconds. Fluorescent trypanosomes express GFP in the nucleus and/or the flagellum.





