A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency
Figures
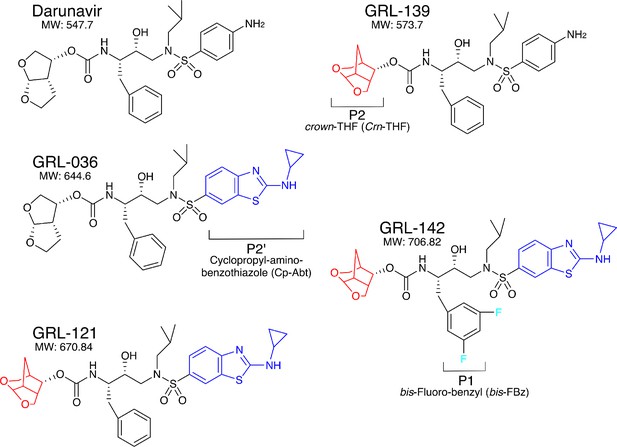
Chemical structure of compounds.
Structures of DRV, GRL-139, GRL-036, GRL-121, and GRL-142. P2-Crn-THF and P2’-Cp-Abt moieties are shown in red and blue, respectively.
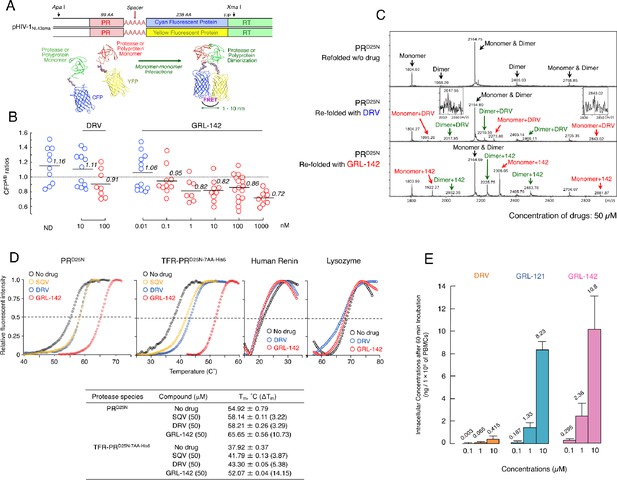
GRL-142 has significantly more potent HIV-1 protease dimerization inhibition activity, much greater thermal stability, and much higher intracellular concentration than DRV.
(A) The FRET-based HIV-1 expression assay system detecting HIV-1 PR dimerization and its disruption by DRV or GRL-142. In an attempt to elucidate the dynamics of HIV-1 PR dimerization and the mechanisms of the emergence of HIV-1 resistance against certain PR inhibitors (PIs), we developed an intermolecular FRET-based HIV-1 expression assay system using CYP- and YFP-tagged PR monomers. Using this FRET-based HIV-1 expression assay system, we have previously identified nonpeptidyl small-molecule inhibitors of HIV-1 PR dimerization, including DRV(Koh et al., 2007; Aoki et al., 2012). Various plasmids encoding full-length molecular infectious HIV-1 (HIV-1NL4-3) clones producing CFP- or YFP-tagged PR using the PCR-mediated recombination method were prepared. A linker consisting of five alanines was inserted between PR and fluorescent protein. A phenylalanine-proline site (F/P) that HIV-1 PR cleaves was also introduced between the fluorescent protein and reverse transcriptase. (Lower) Structural representations of PR monomers and dimer in association with the linker atoms and fluorescent proteins. FRET occurs only when the fluorescent proteins are 1–10 nm apart. (B) GRL-142 more potently inhibits PRWT dimerization by a factor of ~1000 than DRV. COS7 cells were exposed to various concentrations (0.01 to 1000 nM) of GRL-142 or DRV and were subsequently co-transfected with two plasmids, pHIV-PRWT-CFP and pHIV-PRWT-YFP, respectively. After 72 hr, cultured cells were examined in the FRET-based HIV-1 expression assay and the CFPA/B ratios (Y axis) were determined. The arithmetic mean values of the ratios obtained are shown as horizontal bars. A CFPA/B ratio that is greater than one signifies that protease dimerization occurred, whereas a ratio that is less than one signifies that the disruption of protease dimerization occurred (Koh et al., 2007; Aoki et al., 2012). All the experiments were conducted in a blind fashion. The P values were determined using the Wilcoxon rank-sum test (JMP software, SAS, Cary, NC) and were 0.6721 for the CFPA/B ratio in the absence of drug (CFPA/BNo-Drug) versus the CFPA/B ratio in the presence of 10 nM DRV (CFPA/B10-DRV), 0.0262 for CFPA/BNo-Drug versus CFPA/B100-DRV, 0.2483 for CFPA/BNo-Drug versus CFPA/B0.01- GRL-142, 0.0585 for CFPA/BNo-Drug versus CFPA/B0.1-GRL-142, 0.0145 for CFPA/BNo-Drug versus CFPA/B1-GRL-142, 0.0042 for CFPA/BNo-Drug versus CFPA/B10-GRL-142, 0.0056 for CFPA/BNo-Drug versus CFPA/B100-GRL-142, and 0.0019 for CFPA/BNo-Drug versus CFPA/B1000-GRL-142. (C) The ESI-MS spectrum of PRD25N in the absence or presence of 50 µM of DRV or GRL-142. Addition of DRV yielded two DRV-bound monomers and two DRV-bound dimers, while three GRL-142-bound monomers and three GRL-142-bound dimers were seen. Note that GRL-142 more tightly binds to monomers and dimers than DRV (by 6.78- and 3.13-fold; see the text), explaining at least in part the reason GRL-142 much more strongly blocks PR dimerization than DRV. (D) Thermal stability of PRD25N and TFR-PRD25N-7AA-His6 in the absence or presence of SQV, DRV, or GRL-142 was determined using the differential scanning fluorimetry. Tm (50% melting temperature) values were determined as the temperature at which the relative fluorescent intensity became 50%. Note that the thermal stability curves with GRL-142 significantly shifted to the higher temperature (to the right) than those with no agent, SQV, or DRV. The Tm values with GRL-142 were much higher than those with no agent, SQV, or DRV. The thermal stability curves of human renin and lysozyme did not shift at all with GRL-142 as compared with those with no agent, indicating highly specific binding of GRL-142 to PRD25N and TFR-PRD25N-7AA-His6. (E) GRL-142 achieves markedly higher intracellular concentration compared with DRV. PBMCs were incubated with 0.1, 1, and 10 μM of DRV, GRL-121, or GRL-142 for 60 min and vigorously washed with PBS and intra-cellular concentrations of each compound were determined using LC/MS. Arithmetic mean values shown are from the data derived from three independent experiments.
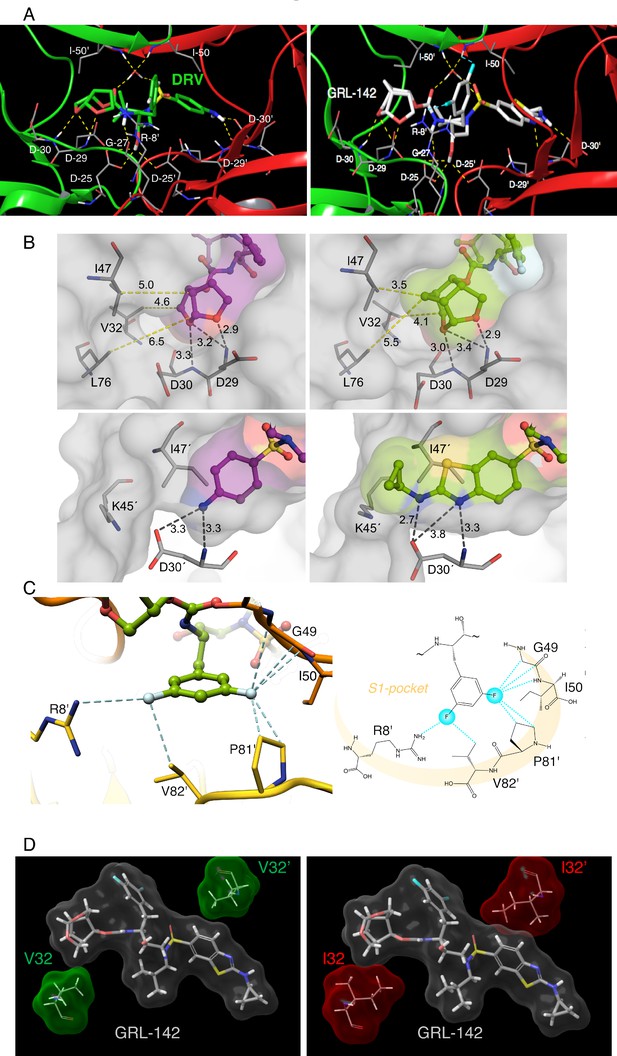
X-ray crystal structure analysis of GRL-142 with HIV-1 protease.
(A) X-ray crystal structure of wild-type HIV-1 protease (PRWT) in complex with DRV or GRL-142. The crystal structure of GRL-142 in complex PRWT was solved (PDB ID: 5TYS). The polar interactions of GRL-142 with protease residues in the active site are shown, and the interactions of DRV with PRWT (PDB ID: 4HLA) are shown for comparison. Cross-section of protease backbone is shown in green and red ribbons. The carbon atoms of GRL-142 and DRV are shown in off-white and green respectively. Nitrogen, oxygen, sulfur, fluorine, and hydrogen atoms are shown in blue, red, yellow, cyan, and white, respectively. Hydrogen bond interactions are shown by yellow dashed lines and polar interactions from fluorine are shown by cyan dashed lines. (B) Focus on the P2 and P2´ site interactions of DRV vs GRL-142. On the top panel, side by side comparison of bis-THF group of DRV (left panel) with crn-THF group of GRL-142 (right panel) in complex with HIV-1 protease. The crn-THF is larger with two two extra carbon atoms that contribute additional van der Walls interactions, particularly with three residues, I47, V32 and L76 in close proximity. On the lower panel, benzothioazole moiety of GRL-142 (right panel) with formation of extra ring effectively fills up the S2´subpocket. While sulfur atom forms close contact with I47’, cyclopropyl moiety protrudes outside the binding pocket and forms close contact with K45´. (C) Zoomed-in feature of the fluorine-mediated interactions inside the S1 pocket of dimerized PRWT. The distances between the fluorine atoms and the interacting atoms less than 3.2 Å are shown as dashed cyan lines. While one fluorine of the bis-meta-fluorophenyl group is heavily involved in halogen bonding with G49, I50 and P81’, the other fluorine atom forms halogen bonds with the positively charged guanidinium group of R8’ as well as the side chain of V82’. The right panel scheme shows the halogen bonding within the S1 pocket as highlighted in light orange crescent shade. (D) GRL-142 has greater vdW contacts with I32 than with V32. The vdW interactions of GRL-142 with V32 and V32' (green surfaces) in PRWT and with I32 and I32' (red surfaces) in PRV32I mutant protease are shown. GRL-142 is shown in grey surface.
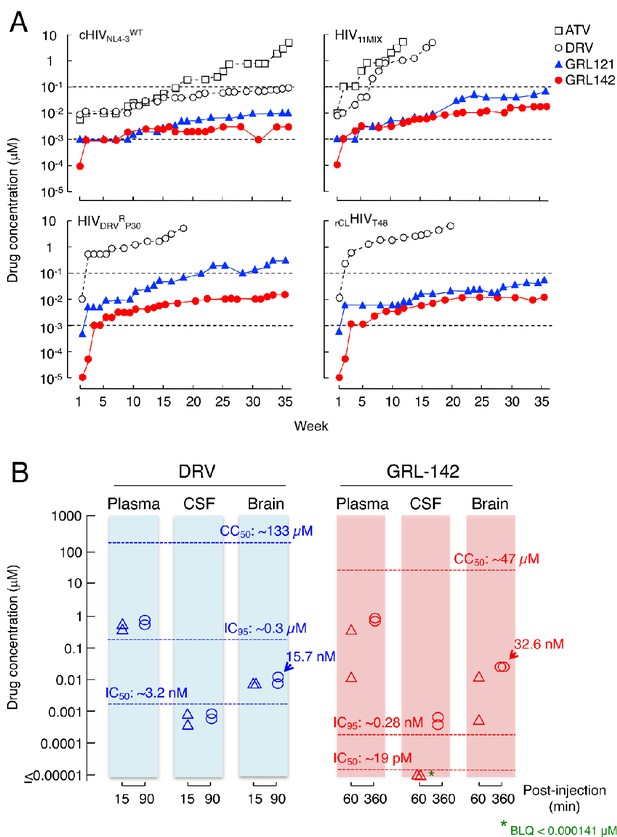
GRL-142 has an extremely high genetic barrier to the emergence of HIV-1 variants resistant to GRL-142 in vitro and effective penetration into brain in rats.
(A) High genetic barrier of GRL-142 against the emergence of GRL-142-resistant variants. cHIVNL4-3WT (top left panel), a mixture of 11 multi-PI-resistant HIV-1 isolates (HIV11MIX)(top right panel), a DRV-resistant HIV-1 variant obtained from in vitro passage 30 with DRV (HIVDRVRP30)(bottom left panel), and an infectious molecular HIV-1 clone derived from a heavily treated ART-experienced HIV-1-infected patient (rCLHIVT48)(bottom right panel) were propagated in the presence of increasing concentrations of each compound in MT-4 cells in a cell-free manner over 36 weeks. When HIV11MIX, HIVDRVRP30 and rCLHIVT48 were employed as a starting HIV-1 population, the virus quickly became highly resistant to ATV and DRV; however, all the starting virus populations failed to propagate in the presence of low concentrations of GRL-121 and GRL-142. GRL-142 did not allow the virus to acquire resistance and propagate more persistently than GRL-121. (B) Favorable penetration of GRL-142 into brain in rats. When DRV and GRL-142 were perorally administered to rats (n = 2) at a dose of 5 mg/kg plus RTV (8.33 mg/kg), the Cmax was achieved around 90 and 360 min after the administration, respectively. Thus, the concentrations of DRV and GRL-142 in plasma, CSF, and brain were determined in 15 and 90 min for DRV and 60 and 360 min for GRL-142 after the administration. The concentrations of GRL-142 in brain were 7.24 ± 9.65 and 32.6 ± 1.4 nM in 60 and 360 min, respectively. The latter concentration (32.6 nM) represents ~1,882 fold greater than the IC50 value and ~114 fold greater than the IC95 value of GRL-142. The concentrations of each compound were determined using LC-MS/MS and the lowest detection limit was 0.1 ng/ml (GRL-142: 14.1pM, DRV: 18.2 pM). These data strongly suggest that GRL-142 would potently block the infection and replication of HIV-1 in the brain.
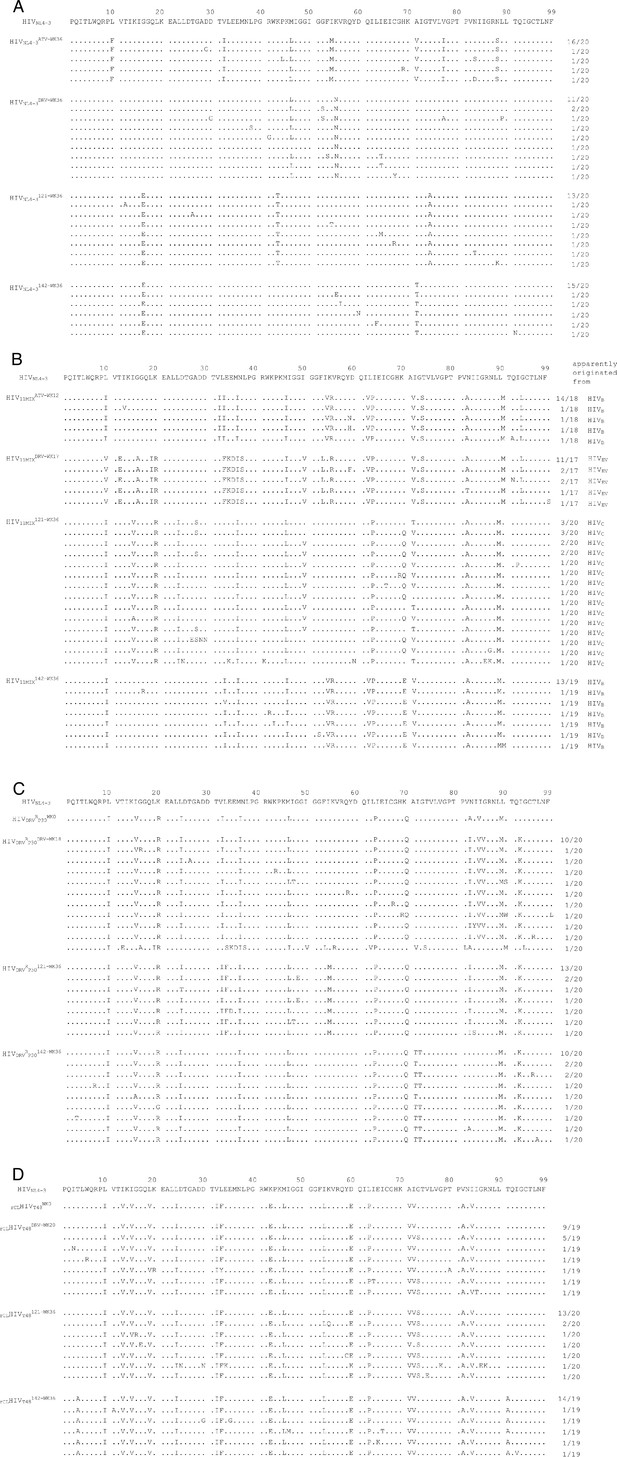
Amino acid sequences of the protease-encoding regions of cHIVNL4-3WT, HIV11MIX, HIVDRVRP30, and rCLHIVT48in vitro selected with various agents.
Shown are the amino acid sequences deduced from the nucleotide sequences of the protease-encoding region of proviral DNA isolated from cHIVNL4-3WT at week 36 with ATV, DRV, GRL-121, or −142. (A) HIV11MIX at week 12 with ATV, week 17 with DRV, and week 36 with GRL-121, or −142. (B) HIVDRVRP30 at week 0 and week 18 with DRV, and week 36 with GRL-121 or −142. (C) and rCLHIVT48 at week 0 and week 20 with DRV and week 36 with GRL-121 or −142. (D) The consensus sequence of HIVNL4-3 is illustrated at the top as a reference. Identity with sequence at individual amino acid positions is indicated by dots. The fractions of the virus, which each clone is presumed to have originated from, over the number of clones examined are shown on the right.
Tables
Antiviral activity of novel four compounds against highly DRV-resistant HIV-1 variants.
https://doi.org/10.7554/eLife.28020.003| Mean IC50 in nM ± SD (fold-change) | |||||||
|---|---|---|---|---|---|---|---|
| LPV | ATV | DRV | GRL-139 | GRL-036 | GRL-121 | GRL-142 | |
| cHIVNL4-3WT | 13 ± 2 | 4.0 ± 2.3 | 3.2 ± 0.7 | 2.8 ± 0.8 | 1.9 ± 0.2 | 0.26 ± 0.05 | 0.019 ± 0.017 |
| HIVDRVRP20 | >1000 (>77) | 450 ± 20 (113) | 51 ± 3 (16) | 36 ± 8 (13) | 5.9 ± 6 (3) | 0.075 ± 0.058 (0.3) | 0.0024 ± 0.002 (0.1) |
| HIVDRVRP30 | >1000 (>77) | >1000 (>250) | 220 ± 40 (79) | 350 ± 10 (125) | 28 ± 2 (15) | 1.9 ± 0.1 (7) | 0.023 ± 0.018 (1) |
| HIVDRVRP51 | >1000 (>77) | >1000 (>250) | 2500 ± 100 (781) | >1000 (>357) | 530 ± 70 (279) | 32 ± 4 (123) | 1.2 ± 2 (63) |
-
Numbers in parentheses represent fold changes in IC50s for each isolate compared to the IC50s for wild-type cHIVNL4-3WT. All assays were conducted in triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments.
Antiviral activity of GRL-121 and −142 against cHIVNL4-3WT and two HIV-2 strains and their cytotoxicities in vitro.
https://doi.org/10.7554/eLife.28020.004| Drug | Mean IC50(nM) ± SD | CC50 (μM) | Selectivity index* | ||
|---|---|---|---|---|---|
| cHIVNL4-3WT | HIV-2ROD | HIV-2EHO | |||
| SQV | 12 ± 3 | 9.0 ± 5.0 | 8.8 ± 2.1 | 33 | 2750 |
| IDV | 18 ± 5 | 31 ± 3 | 55 ± 25 | 75 | 4167 |
| NFV | 23 ± 5 | 27 ± 0.4 | 84 ± 20 | 32 | 1391 |
| RTV | 34 ± 10 | 136 ± 165 | 278 ± 88 | 35 | 1.029 |
| TPV | 330 ± 13 | 293 ± 45 | 313 ± 48 | 34 | 103 |
| APV | 26 ± 8 | 170 ± 82 | 305 ± 78 | >150 | >4167 |
| LPV | 13 ± 8 | 40 ± 28 | 11 ± 2 | 33 | 2538 |
| ATV | 4.0 ± 2.3 | 28 ± 6 | 10 ± 8 | 80 | 20,000 |
| DRV | 3.2 ± 0.7 | 8.5 ± 0.7 | 6.2 ± 0.7 | 133 | 41,562 |
| GRL-121 | 0.26 ± 0.05 | 0.020 ± 0.014 | 0.071 ± 0.071 | 34 | 130,769 |
| GRL-142 | 0.019 ± 0.017 | 0.00032 ± 0.00015 | 0.000059 ± 0.000025 | 47 | 2,473,684 |
-
*Each selectivity index denotes a ratio of CC50 to IC50 against cHIVNL4-3WT.
The data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments.
Antiviral activity of GRL-121 and −142 against highly PI-resistant HIV-1 variants.
https://doi.org/10.7554/eLife.28020.005| Mean IC50 in nM ± SD (fold-change) | ||||||
|---|---|---|---|---|---|---|
| Virus species | LPV | ATV | DRV | GRL-121 | GRL-142 | |
| Wild-type | cHIVNL4-3WT | 13 ± 2 | 4.0 ± 2.3 | 3.2 ± 0.7 | 0.26 ± 0.05 | 0.019 ± 0.017 |
| invitroHIVPIR* | HIVSQV-5μM | >1000 (>77) | 430 ± 20 (108) | 17 ± 7 (5) | 0.026 ± 0.01 (0.1) | 0.00018 ± 0.00003 (0.009) |
| HIVAPV-5μM | 280 ± 15 (22) | 3.0 ± 1.0 (1) | 39 ± 16 (12) | 0.13 ± 0.08 (0.5) | 0.0000085 ± 0.000008 (0.0004) | |
| HIVLPV-5μM | >1000 (>77) | 46 ± 10 (12) | 280 ± 50 (86) | 0.0018 ± 0.0006 (0.007) | 0.0000019 ± 0.0000014 (0.0001) | |
| HIVIDV-5μM | 250 ± 15 (19) | 56 ± 9 (14) | 37 ± 8 (12) | 0.0092 ± 0.0163 (0.04) | 0.00018 ± 0.00028 (0.009) | |
| HIVNFV-5μM | 37 ± 3 (3) | 12 ± 2 (3) | 7.7 ± 3 (2) | 0.048 ± 0.018 (0.2) | 0.00024 ± 0.00026 (0.01) | |
| HIVATV-5μM | 310 ± 20 (24) | >1000 (>250) | 25 ± 1 (8) | 0.092 ± 0.097 (0.4) | 0.015 ± 0.004 (0.8) | |
| HIVTPV-15μM | >1000 (>77) | >1000 (>250) | 40 ± 3 (13) | 0.063 ± 0.016 (0.2) | 0.00024 ± 0.00007 (0.01) | |
| rCLHIV** | rCLHIVF16 | >1000 (>77) | 193 ± 23 (48) | 3357 ± 600 (1,049) | 2.5 ± 0.9 (10) | 0.016 ± 0.016 (0.8) |
| rCLHIVF39 | >1000 (>77) | 374 ± 23 (94) | 313 ± 230 (98) | 0.028 ± 0.02 (0.1) | 0.0061 ± 0.002 (0.3) | |
| rCLHIVV42 | >1000 (>77) | 270 ± 20 (68) | 343 ± 28 (107) | 3.1 ± 1.9 (12) | 0.026 ± 0.023 (1) | |
| rCLHIVT44 | >1000 (>77) | >1000 (>250) | 2487 ± 210 (777) | 12 ± 6 (46) | 0.69 ± 0.66 (36) | |
| rCLHIVM45 | >1000 (>77) | >1000 (>250) | 1924 ± 1570 (601) | 3.8 ± 0.5 (15) | 0.094 ± 0.113 (5) | |
| rCLHIVT48 | >1000 (>77) | 440 ± 26 (110) | 315 ± 61 (98) | 1.1 ± 1.1 (4) | 0.0052 ± 0.0017 (0.3) | |
-
*invitroHIVPIR, in vitro PI-selected HIV-1 variants; **rCLHIV, recombinant clinical HIV-1 variants.
Numbers in parentheses represent fold changes in IC50s for each isolate compared to the IC50s for wild-type cHIVNL4-3WT. All assays were conducted in triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of at least three independent experiments.
Antiviral activity of GRL-121 and −142 against HIV-1 variants carrying single amino acid substitution in PR region.
https://doi.org/10.7554/eLife.28020.006| Mean IC50 ± SD (nM) | ||||
|---|---|---|---|---|
| Infectious clone | Amino acid substitution in PR | DRV | GRL-121 | GRL-142 |
| cHIVNL4-3WT | none | 3.2 ± 0.7 | (2.6 ± 0.5)×10−1 | (1.9 ± 1.7)×10−2 |
| cHIVNL4-3L10F | L10F | 3.3 ± 1.0 | (3.4 ± 1)×10−2 | (2.9 ± 1.2)×10−3 |
| cHIVNL4-3L24I | L24I | 3.1 ± 0.6 | (1.2 ± 1.1)×10−4 | (2.9 ± 1.6)×10−5 |
| cHIVNL4-3D30N | D30N | 4.7 ± 1.0 | (2.0 ± 3.0)×10−2 | (4.7 ± 2.0)×10−3 |
| cHIVNL4-3V32I | V32I | (3.0 ± 1.0)×10−1 | (8.0 ± 10.5)×10−5 | (1.2 ± 1.6)×10−8 |
| cHIVNL4-3L33F | L33F | 2.8 ± 1.1 | (3.5 ± 2.5)×10−1 | (1.8 ± 1.0)×10−2 |
| cHIVNL4-3M46I | M46I | 3.3 ± 0.2 | (2.2 ± 0.8)×10−2 | (2.2 ± 1.5)×10−3 |
| cHIVNL4-3I47V | I47V | 3.0 ± 0.6 | (3.2 ± 0.7)×10−2 | (1.2 ± 0.9)×10−3 |
| cHIVNL4-3G48V | G48V | (2.9 ± 0.6)×10−1 | (6.0 ± 1.1)×10−5 | (3.6 ± 6.0)×10−8 |
| cHIVNL4-3I50V | I50V | 2.7 ± 1.0 | (1.5 ± 2.4)×10−5 | (9.3 ± 15.1)×10−8 |
| cHIVNL4-3I54M | I54M | 3.2 ± 0.8 | (3.1 ± 3.4)×10−3 | (1.9 ± 2.3)×10−4 |
| cHIVNL4-3I54L | I54L | 3.3 ± 0.2 | (3.2 ± 0.3)×10−1 | (3.1 ± 2.6)×10−3 |
| cHIVNL4-3I54V | I54V | 3.0 ± 0.4 | (3.2 ± 1)×10−4 | (2.7 ± 1.1)×10−5 |
| cHIVNL4-3L63P | L63P | 2.3 ± 0.7 | (2.5 ± 2.6)×10−2 | (5.9 ± 5.8)×10−3 |
| cHIVNL4-3V82A | V82A | 2.9 ± 0.1 | (5.0 ± 2.0)×10−3 | (3.6 ± 10)×10−4 |
| cHIVNL4-3V82I | V82I | 4.0 ± 1.1 | (1.8 ± 1.8)×10−1 | (2.0 ± 0.5)×10−2 |
| cHIVNL4-3V82T | V82T | (5.7 ± 2.0)×10−1 | (1.5 ± 1.0)×10−5 | (3.1 ± 4.0)×10−6 |
| cHIVNL4-3I84V | I84V | 2.7 ± 1.1 | (1.7 ± 0.9)×10−3 | (3.9 ± 1.6)×10−5 |
| cHIVNL4-3L90M | L90M | 4.2 ± 0.5 | (3.2 ± 2.7)×10−2 | (5.8 ± 0.9)×10−4 |
-
All assays were conducted in triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of at least three independent experiments.
van der Waals interaction energies between GRL-142 or DRV with the protease dimer and selected active site residues.
https://doi.org/10.7554/eLife.28020.009| GRL-142 | DRV | |
|---|---|---|
| (kcal/mol) | (kcal/mol) | |
| Protease dimer | −67.5 | −57.8 |
| Asp29 & Asp29’ | −7.4 | −4.2 |
| Asp30 & Asp30’ | −5.4 | −2.9 |
| Val82 & Val82’ | −2.2 | −2.3 |
| Ile84 & Ile84’ | −4.3 | −4.6 |
| Ile47 & Ile47’ | −6.5 | −3.6 |
| Gly48 & Gly48’ | −3.3 | −2.8 |
| Gly49 & Gly49’ | −3.7 | −3.3 |
| Ile50 & Ile50’ | −8.7 | −9.2 |
| Pro81 & Pro81’ | −1.6 | −1.9 |
| Arg8 & Arg8’ | −2.2 | −2 |
-
Average van der Waals energies were calculated by analyzing the trajectories from a 1.2 ns molecular dynamics simulation using Desmond molecular dynamics system (D.E. Shaw Research, New York, NY 2017).
Antiviral activity of GRL-142 against viruses obtained after in vitro selection.
https://doi.org/10.7554/eLife.28020.012| Virus before selection | Mean IC50 in nM ± SD | Virus after selection | Mean IC50 in nM ± SD | Fold-change |
|---|---|---|---|---|
| cHIVNL4-3WT | 0.019 ± 0.017 | HIVNL4-3142-WK36* | 0.29 ± 0.06 | 15 |
| rCLHIVT48 | 0.0052 ± 0.0017 | rCLHIVT48142-WK36* | 4.0 ± 0.5 | 769 |
-
*Viruses were obtained at 36 week of the in vitro GRL-142 selection.
All assays were conducted in triplicate, and the data shown represent mean values (±1 standard deviation) derived from the results of three independent experiments.
X-ray diffraction data processing details for PRWT in complex with GRL-121 or −142.
https://doi.org/10.7554/eLife.28020.013| PRWT + GRL-121 | PRWT + GRL142 | |
|---|---|---|
| PDB entry | 5TYR | 5TYS |
| Resolution range (Å) | 50.0–1.7 | 50.0–2.0 |
| Unit cell - a (Å) | 62.56 | 63.13 |
| b (Å) | 62.56 | 63.13 |
| c (Å) | 82.66 | 82.23 |
| α () | 90 | 90 |
| β () | 90 | 90 |
| γ () | 120 | 120 |
| Space group | P61 | P61 |
| Solvent content (%) | 54.22 | 54.8 |
| No. of unique reflections | 20,171 (994)* | 12,475 (624) |
| Mean (I/σ(I)) | 29.05 (3.3) | 26.6 (4.8) |
| †Rmerge | 0.09 (0.55) | 0.10 (0.46) |
| Data redundancy | 10 (10.1) | 10.2 (9.7) |
| Completeness (%) | 100 (100) | 99.9 (100) |
-
*Values in parentheses are for the highest resolution shell
†Rmerge = Σ |I - < I > | / Σ I
Refinement statistics for structure solutions of PRWT in complex with GRL-121 or −142.
https://doi.org/10.7554/eLife.28020.014| PRWT + GRL-121 | PRWT + GRL-142 | |
|---|---|---|
| PDB entry | 5TYR | 5TYS |
| Resolution range (Å) | 45.32–1.8 | 32.8–2.01 |
| No. of reflections used | 17,043 | 12,421 |
| *Rcryst | 0.191 | 0.1949 |
| Rfree | 0.232 | 0.2366 |
| No. of protease dimers per †AU | 1 | 1 |
| No. of protein atoms per AU | 1516 | 1516 |
| No. of ligand molecules per AU | 2 | 2 |
| No. of ligand atoms per AU | 92 | 96 |
| No. of water molecules | 148 | 90 |
| Mean temperature factors: | ||
| Protein (Å2) | 24.21 | 28.36 |
| Main chains (Å2) | 22.02 | 25.84 |
| Side chains (Å2) | 26.6 | 31.11 |
| Ligand (Å2) | 16.55 | 20.05 |
| Waters (Å2) | 34.24 | 34.9 |
| RMSD bond lengths (Å) | 0.007 | 0.008 |
| RMSD bond angles (Å) | 1.015 | 1.022 |
| Ramachandran plot: | ||
| Most favored (%) | 99.48 | 97.94 |
| Additional allowed (%) | 0.52 | 2.06 |
| Generously allowed (%) | 0 | 0 |
| Disallowed (%) | 0 | 0 |
-
*Rcryst = Σ ||Fobs| - |Fcalc|| / Σ|Fobs|
†AU - Asymmetric unit
Additional files
-
Supplementary file 1
Antiviral activity of five PIs and four NRTIs against laboratory-selected PI-resistant HIV-1 variants.
- https://doi.org/10.7554/eLife.28020.015
-
Supplementary file 2
Representative dose-response profiles of DRV, GRL-121, and GRL-142 against cHIVNL4-3WT, cHIVNL4-3V32I, cHIVNL4-3G48V, cHIVNL4-3I50V, and cHIVNL4-3V82T are shown.
- https://doi.org/10.7554/eLife.28020.016
-
Supplementary file 3
List of 37 HIVs used in antiviral assay and the amino acid sequences of the protease-encoding region of HIVs used in this study.
- https://doi.org/10.7554/eLife.28020.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28020.018





