Activation of the NRF2 antioxidant program generates an imbalance in central carbon metabolism in cancer
Figures
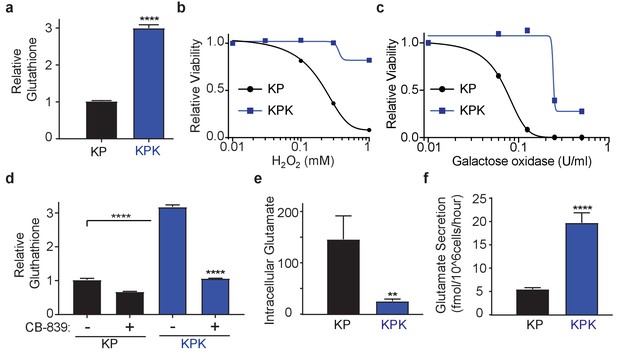
Keap1 mutations cause increased dependency on exogenous glutamine.
(a) Measurement of whole cell glutathione levels in wild type (KP) and Keap1 mutant (KPK) in isogenic clones derived from mouse lung tumors (n = 3, triplicate wells). (b) Relative viability assayed with cell-titer glo (relative luminescent units) in KP and KPK cells in RPMI after treatment with H2O2 for 12 hr. All data points are relative to vehicle treated controls (n = 8/data point from replicate wells) (c) Relative viability assayed with cell-titer glo (relative luminescent units) in KP and KPK cells in RPMI after treatment with galactose oxidase for 72 hr. All data points are relative to vehicle treated controls (n = 4/data point from replicate wells). d) Measurement of whole cell glutathione levels in KP and KPK cells after 24 hr of CB-839 treatment where indicated (n = 3, triplicate wells). (e) Intracellular glutamate levels per cell (n = 6, two independent experiments with triplicate wells). (f) Glutamate secretion in KP and KPK cells (n = 6, two independent experiments with triplicate wells). All error bars depict s.e.m. **p<0.01, ****p<0.0001.
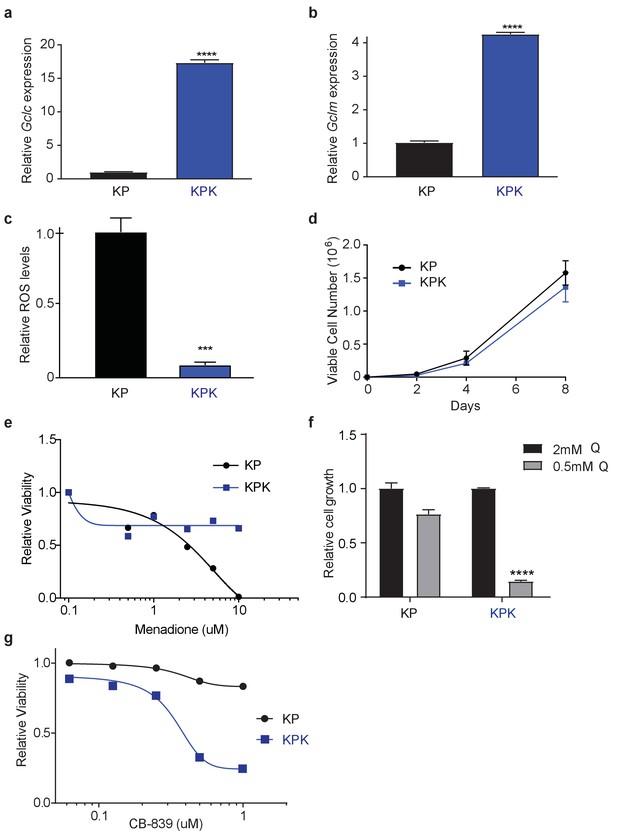
Keap1 mutation increases cellular antioxidant capacity and sensitizes KPK cells to glutamine deprivation or glutaminase inhibition.
Real-time quantitative PCR of (a) Gclc and (b) Gclm in KP and KPK cells. Data is presented as relative to KP cells (n = 4, technical replicates). (c) Relative ROS levels in KP and KPK cells. Data is presented relative to KP cells (n = 4, replicate wells). (d) Proliferation of KP and KPK cells in RPMI (n = 4, replicate wells). (e) Relative viability assayed with cell-titer glo (relative luminescent units) in KP and KPK cells in RPMI after 72 hr of treatment with menadione. All data is presented relative to vehicle treated controls. (n = 4/data points, replicate wells) (f) Cell proliferation in RPMI media containing either 2 mM or 0.5 mM glutamine (Q). Data presented relative to KP cell proliferation in 2 mM condition (n = 6, two independent cell lines with triplicate wells). (g) Relative viability assayed with cell-titer glo (relative luminescent units) in KP and KPK cells in RPMI after CB-839 treatment for 4 days. All data points are relative to vehicle treated controls (n = 8/data point from replicate wells). All error bars depict s.e.m. ****p<0.0001.
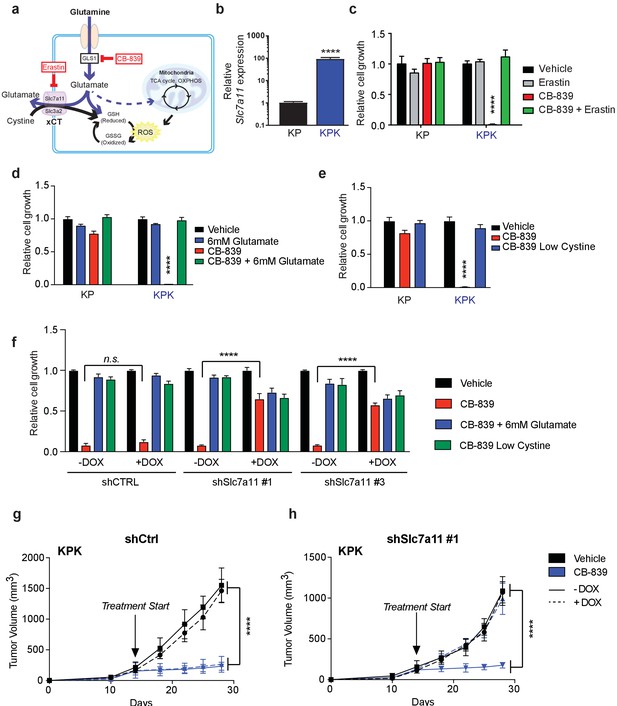
xCT/Slc7a11-dependent glutamate secretion in Keap1 mutant cells causes glutamine dependency.
(a) Schematic depicting glutaminolysis and subsequent glutamate export by the xCT/Slc7a11 antiporter. Treatment with the small molecule CB-839 inhibits Gls1, blocking the conversion of glutamine to glutamate while the small molecule Erastin inhibits Slc7a11 preventing glutamate export. (b) Quantitative real-time PCR of mRNA expression of Slc7a11 in KP and KPK cells (n = 3, technical replicates). Data presented as relative to Slc7a11 expression in KP cells. (c) Proliferation of KP and KPK cells after overnight pretreatment with 500 nM Erastin followed by 250 nM CB-839 treatment for 5 days (n = 3, triplicate wells). Data presented as relative to vehicle only treated condition for each cell line. (d) Proliferation of KP and KPK cells after addition of 6 mM glutamate followed by 250 nM CB-839 treatment for 5 days (n = 3, triplicate wells). Data presented as relative to vehicle only treated condition for each cell line. (e) Proliferation of KP and KPK cells in media containing low cystine (20 μM, 10X reduction from RPMI which contains 208 μM cystine) followed by 250 nM CB-839 treatment for 5 days (n = 3, triplicate wells). Data presented as relative to vehicle only treated condition for each cell line. (f) Proliferation of KP and KPK expressing either a doxycycline inducible control shRNA (shCTRL) or an shRNA targeted against Slc7a11 (shSlc7a11). Cells were cultured in RPMI with or without doxycycline that was supplemented with 6 mM glutamate or RPMI containing low cystine (20 μM, 10X reduction from RPMI which contains 208 μM cystine) followed by 250 nM CB-839 treatment for 5 days (n = 3, triplicate wells). (g) Subcutaneous tumor volumes of KPK tumors expressing a doxycycline inducible control shRNA (shCtrl). Animals were treated with vehicle (black) or CB-839 (blue) and received normal (solid line) or doxycycline (dashed line) feed starting from day 13 (arrow indicating treatment start, n = 6 tumors) (h) Subcutaneous tumor volumes of KPK tumors expressing a doxycycline inducible shRNA targeted against Slc7a11 (shSlc7a11). Animals were treated with vehicle (black) or CB839 (blue) and received normal (solid line) or doxycycline (dashed line) feed starting from day 13 (arrow indicating treatment start, n = 6 tumors). All error bars depict s.e.m. ****p<0.0001.
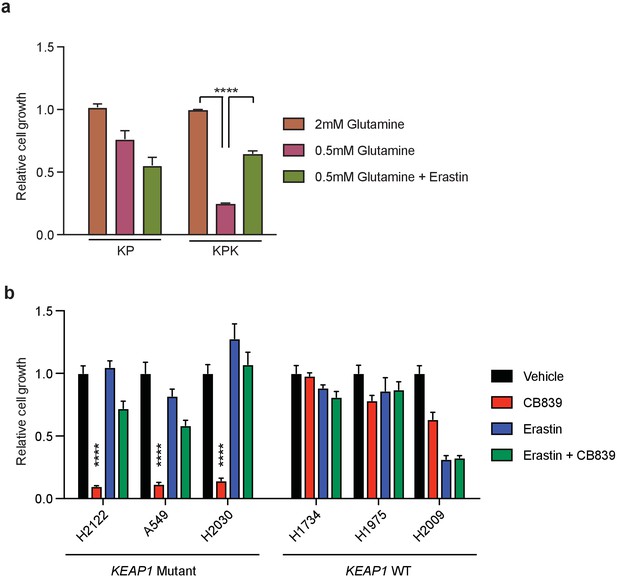
Inhibition of xCT/Slc7a11 rescues glutamine dependency in Keap1 mutant cells.
(a) Proliferation of KP and KPK cells in RPMI media containing either 2 mM or 0.5 mM glutamine (Q). Where indicated cells were treated with 500 nM of Erastin (n = 3, triplicate wells). Data presented relative to proliferation in 2 mM glutamine condition. (b) Proliferation of wild type and KEAP1 mutant human LUAD cell lines. Cells were pretreated with 500 nM erastin followed by treatment with 250 μM CB-839 treatment for 5 days (n = 3, triplicate wells). All error bars depict s.e.m. ****p<0.0001.
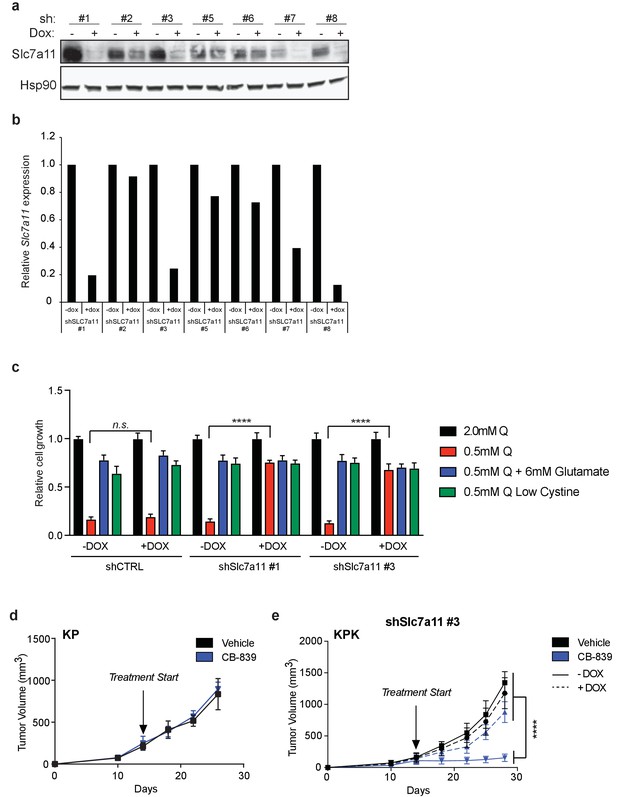
Reduction in xCT/Slc7a11 expression rescues glutamine dependency both in vitro and in vivo.
(a) Western blot depicting expression of eight different doxycycline inducible shRNAs against Slc7a11 (shSlc7a11) in KPK cell line. The shSlc7a11 was induced for 72 hr using doxycycline. First panel depicts an Slc7a11 blot. The second panel depicts an Hsp90 loading control. (b) Real-time quantitative PCR of Slc7a11 in KPK cells expressing inducible shSLC7a11, as depicted in (a). Data is presented as relative to untreated control for each shRNA (n = 3, technical replicates). (c) Proliferation of KPK cells expressing either a doxycycline inducible control shRNA (shCT) or an shRNA targeted against Slc7a11 (shSlc7a11). Cells were cultured in RPMI containing 2 mM or 0.5 mM glutamine (Q). Cells were treated doxycycline where indicated. Where indicated, media was supplement with 6 mM glutamate or media with reduced cystine (20 μM, 10X reduction from RPMI which contains 208 μM cystine) was supplied. Data is represent as relative to growth with 2 mM glutamine (n = 3, triplicate wells). (d) Subcutaneous tumor volumes of KP tumors. Animals were treated with vehicle (black) or CB-839 (blue) starting from day 13 (arrow indicating treatment start, n = 6 tumors). (e) Subcutaneous tumor volumes of KPK tumors expressing a second independent doxycycline inducible shRNA targeted against Slc7a11 (shSlc7a11). Animals were treated with vehicle (black) or CB-839 (blue) and received normal (solid line) or doxycycline (dashed line) feed starting from day 13 (arrow indicating treatment start, n = 6 tumors). All error bars depict s.e.m. ****p<0.0001.
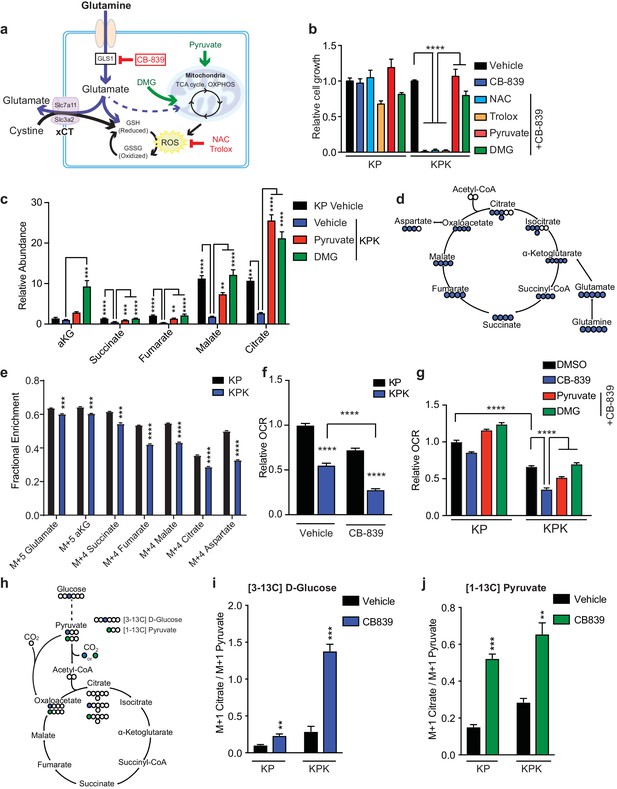
Keap1 mutants have defects in glutamine anaplerosis.
(a) Schematic depicting glutaminolysis and subsequent glutamate export by the xCT/Slc7a11 antiporter. Treatment with the small molecule CB-839 inhibits Gls1, blocking the conversion of glutamine to glutamate. Treatment with the antioxidants N-acetylcysteine (NAC) or trolox scavenge cellular reactive oxygen species (ROS) while treatment with either pyruvate or dimethyl 2-oxoglutarate (DMG) provide carbons to the TCA cycle (b) Proliferation of KP and KPK cells after overnight pretreatment with either 500 nM NAC, 500 nM Trolox, 2 mM pyruvate or 2 mM DMG followed by 250 nM CB-839 treatment for 5 days (n = 3, triplicate wells). Data presented as relative to vehicle only treated condition for each cell line. (c) Relative abundance of TCA cycle metabolites in KP and KPK cells supplemented with 2 mM pyruvate or DMG where indicated (n = 3, triplicate wells). Data is normalized by cell counts for each cell line in each condition. (d) Schematic depicting the TCA cycle. Filled blue circles represent 13C atoms derived from [U-13C]-L glutamine. (e) Mass isotopomer analysis of TCA cycle metabolites in KP and KPK cells cultured for 8 hr with [U-13C]-L glutamine (n = 3, triplicate wells). (f) Relative mitochondrial respiration measured by oxygen consumption rate (OCR, pmol O2/min) in KP and KPK cells treated either with vehicle or 250 nM CB-839 for 4 hr (n = 5, replicate wells). Data is presented as relative to vehicle treated KP cells. (g) Relative mitochondrial respiration measured by oxygen consumption rate (OCR, pmol O2/min) in KP and KPK cells pretreated either 2 mM pyruvate or 2 mM DMG followed by treatment with vehicle or 250 nM CB-839 for 4 hr (n = 3, triplicate wells). Data is presented as relative to vehicle treated KP cells. (h) Schematic depicting the entry of glucose derived carbon into the TCA cycle via pyruvate carboxylase activity. Filled circles represent 13C atoms derived from [313C]-D glucose (blue) or [113C]-pyruvate (green). Carbon flux through pyruvate carboxylase results in M + 1 labeled citrate while flux through pyruvate dehydrogenase results in unlabeled citrate. (i) Mass isotopermer analysis of citrate in KP and KPK cells cultured for 8 hr with [313C]-D glucose and 250 nM CB-839 where indicated (n = 3, triplicate wells). Data is normalized with respect to M + 1 pyruvate to account for differences in basal rates of glycolysis. (j) Mass isotopermer analysis of citrate in KP and KPK cells pretreated with [113C]-pyruvate overnight and then treated 250 nM CB-839 for 8 hr where indicated (n = 3, triplicate wells). Data is normalized with respect to M + 1 pyruvate to account for differences in basal rates of glycolysis. All error bars depict s.e.m. *p<0.05, **p<0.01, ****p<0.0001.
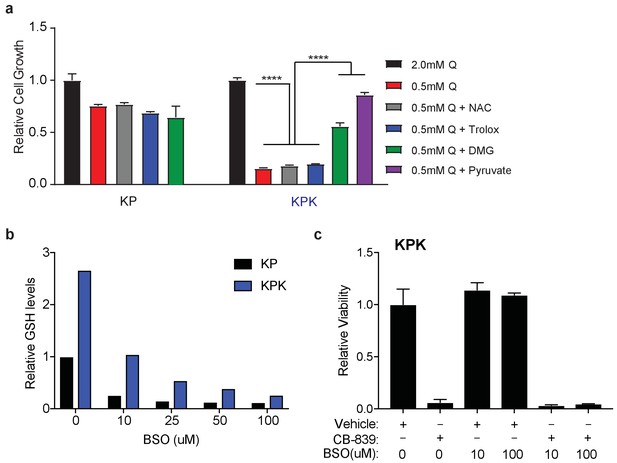
Antioxidants do not rescue glutamine dependency in Keap1 mutant cells.
(a) Proliferation of KP and KPK cells in RPMI containing 2 mM or 0.5 mM glutamine (Q). Cells were treated with 500 nM N-acetyl-cystine (NAC), 500 nM trolox, 2 mM DMG, or 2 mM pyruvate where indicated. All data is presented as relative to 2 mM glutamine condition for each cell line (n = 3, triplicate wells). (b) Relative glutathione levels in KP and KPK cells treated with BSO. Data is presented relative to vehicle treated KP cells. (n = 2, replicate wells). (c) Relative viability assayed with cell-titer glo (relative luminescent units) in KP and KPK cells in RPMI. Cells were treated with the indicated concentration of BSO and 250 nM CB-839 for 72 hr where indicated. Data is presented as relative to vehicle treated control (n = 4, replicate wells). All error bars depict s.e.m. ****p<0.0001.
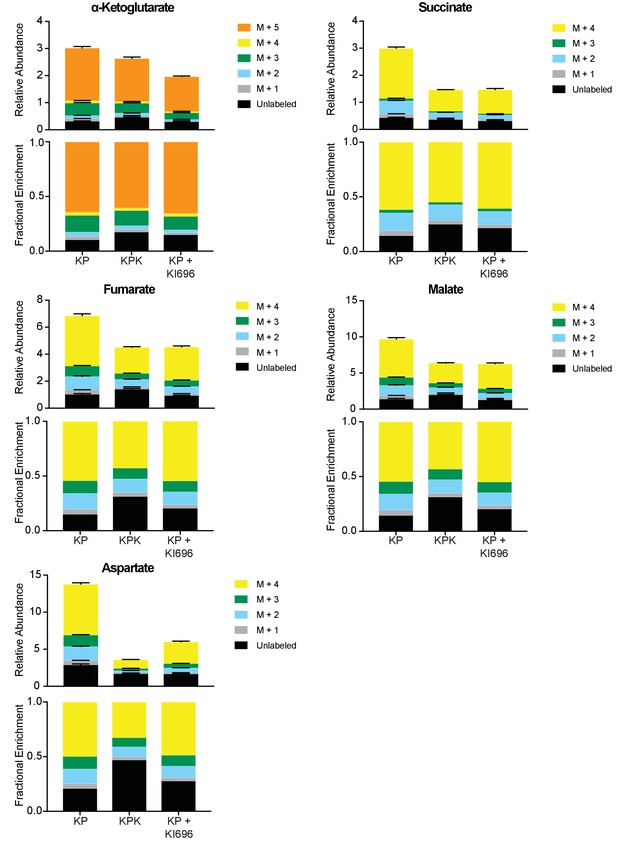
Fractional labeling of glutamine derived carbon in TCA cycle intermediates.
Mass isotopomer analysis of TCA cycle intermediates in KP, KPK, and KP cells pretreated with KI696. Cells were cultured for 8 hr with [U-13C]-L-glutamine in the presence or absence of KI696 (1 μM). Metabolite pool sizes (top) and fraction of isotopomers derived from labeled glutamine (bottom) for each metabolite indicated are presented in separate graphs. Relative pool sizes are normalized to cell counts for each condition (n = 3, triplicate wells). All error bars depict s.e.m.
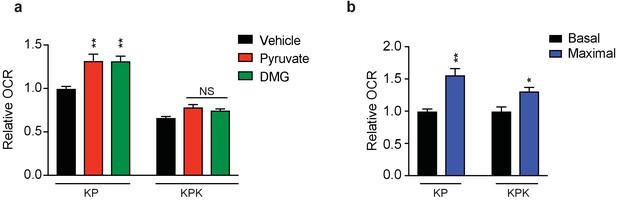
Basal respiration in KP and KPK cells.
(a) Relative mitochondrial respiration measured by oxygen consumption rate (OCR, pmol O2/min) in KP and KPK cells pretreated with either 2 mM pyruvate or DMG where indicated (n = 3, triplicate wells). Data is presented as relative to vehicle treated KP cells. (b) Relative basal and maximal mitochondrial respiration measured by oxygen consumption rate (OCR, pmol O2/min) in KP and KPK cells (n = 3, triplicate wells). Basal respiration is OCR of cells alone and maximal respiration is OCR in cells with mitochondria uncoupled by FCCP. Data is presented as relative to basal respiration for each cell line. All error bars depict s.e.m. *p<0.05, **p<0.01.
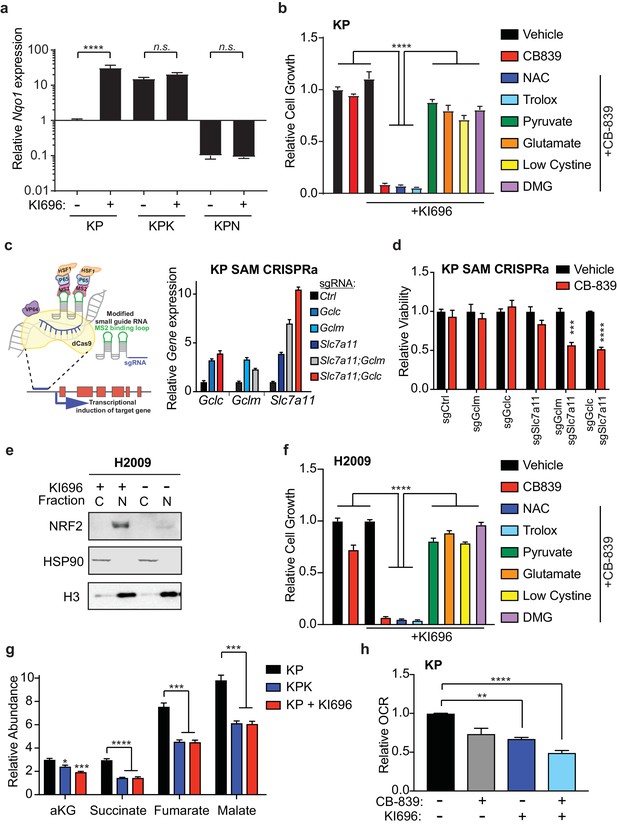
Nrf2 activation is sufficient to sensitize cells to glutaminase inhibition.
(a) Quantitative real-time PCR of mRNA expression of Nqo1 in KP, KPK, and KPN (Nrf2 null) cells after pretreatment with 1 μM KI696 for 36 hr (n = 3, technical replicates). Data presented as relative to Nqo1 expression in untreated KP cells. (b) Proliferation of KP cells after pretreatment with the small molecule activator of Nrf2, 1 μM KI696, followed by treatment with either 500 nM NAC, 500 nM trolox, 2 mM pyruvate, 2 mM DMG, supplementation with 6 mM glutamate, or media containing low cystine (20 μM, 10X reduction from RPMI which contains 208 μM cystine) followed by 250 nM CB-839 treatment for 5 days (n = 5, replicate wells). Data is presented as relative to vehicle treated. (c) Left: Schematic depicting CRISPR synergistic activation mediated (SAM) system. A modified sgRNA recruits catalytically dead Cas9 that is fused to VP64 to promoter regions up stream of the transcription start site of target genes and recruits additional transcriptional activators to induced endogenous transcription of target genes. Right: Quantitative real-time PCR of mRNA expression of Gclc, Gclm, and Slc7a11 in KP SAM CRISPRa cell lines with sgRNAs targeting each gene of interest (n = 3, technical replicates). Data presented as relative to expression of each gene in sgCtrl cells. (d) Proliferation of KP SAM CRISPRa cells expressing sgRNAs against Gclc, Gclm, and Slc7a11. Cells were seeded in media lacking glutamate and after 24 hr were treated with 100 nM of CB-839 for 5 days (n = 3, replicate wells). Data is presented as relative to vehicle treated control for each cell line. (e) Western blot depicting Nrf2 expression in KEAP1 wild type human lung adenocarcinoma line (H2009) after treatment with KI696. Cells were treated with 1 μM KI696 for 36 hr and then nuclear (N) and cytoplasmic (C) fractions were collected. First panel depicts an Nrf2 blot, second panel depicts an Hsp90 loading control for cytoplasmic fraction, and third panel depicts an H3 loading control for nuclear fraction. f) Proliferation of KEAP1 wild type human lung cancer cell line H2009 after pretreatment with the small molecule activator of NRF2, 1 μM KI696, followed by treatment with either 500 nM NAC, 500 nM trolox, 2 mM pyruvate, 2 mM DMG, supplementation with 6 mM glutamate, or media with reduced cystine (20 uM) followed by 250 nM CB-839 treatment for 5 days (n = 5, replicate wells). Data is presented as relative to vehicle treated. (g) Relative abundance of TCA cycle metabolites in KP, KPK, and KP cells treated with 1 μM KI696 (n = 3, triplicate wells). Data is normalized by cell counts for each cell line. (h) Relative mitochondrial respiration measured by oxygen consumption rate (OCR, pmol O2/min) in KP cells treated with either vehicle or 1 μM KI696 followed by treatment with vehicle or 250 nM CB-839 for 4 hr (n = 3, triplicate wells). Data is presented as relative to vehicle treated KP cells. All error bars depict s.e.m. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
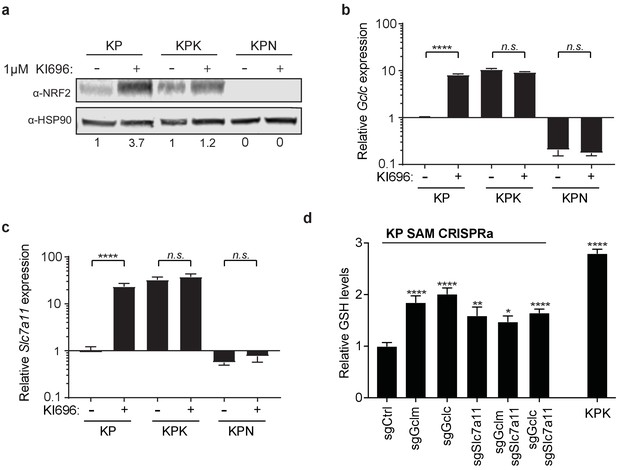
Small molecule KI696 induces Nrf2 activation in KP cells.
(a) Western blot depicting Nrf2 expression in KP, KPK, and KPN (Nrf2 null) cells after treatment with KI696. Cells were treated with 1 μM KI696 for 36 hr and then whole cell lysates were collected. First panel depicts an Nrf2 blot and the second panel depicts an Hsp90 loading control. Nrf2 protein normalized with respect to Hsp90 and presented as relative to untreated controls. Real-time quantitative PCR of Gclc (b) and Slc7a11 (c) in KP, KPK, and KPN cells after treatment with 1 μM KI696 for 36 hr where indicated. Data is presented as relative to untreated KP cells for each gene (n = 3, technical replicates). (d) Measurement of whole cell glutathione levels in KP SAM CRISPRa cells with sgRNAs against Gclc, Gclm, and Slc7a11 and KPK cells (n = 6, replicate wells). Data is presented relative to sgCtrl. All error bars depict s.e.m. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
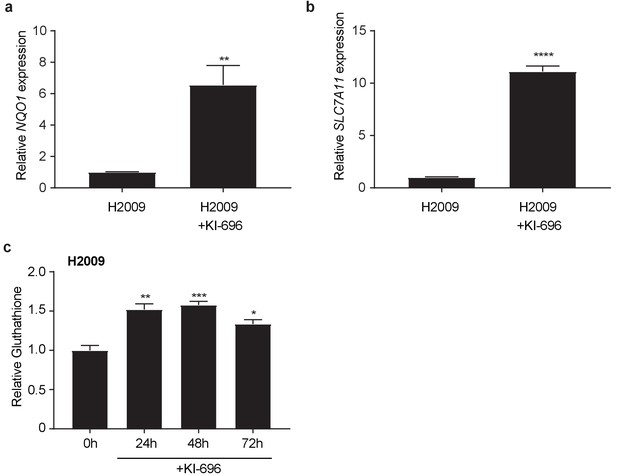
Small molecule KI696 induces Nrf2 activation in KEAP1 wild type human lung cancer cells.
Real-time quantitative PCR of NQO1 1 (a) and SLC7A1 (b) in H2009 cells after treatment with 1 μM KI696 for 36 hr where indicated. Data is presented as relative to untreated H2009 cells for each gene (n = 3, technical replicates). (c) Glutathione levels in H2009 after KI696 treatment for indicated time points (n = 3, replicate wells). All error bars depict s.e.m. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
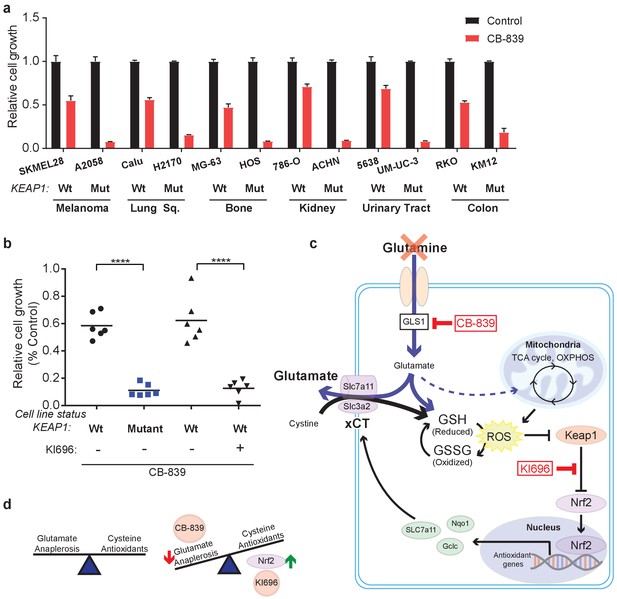
KEAP1 mutations predict sensitivity to glutaminase inhibitors across multiple cancer subtypes.
(a) Proliferation of a panel of human cancer cell lines of various tissues of origin treated with vehicle or 250 nM CB-839 for 4 days. One wild type and one KEAP1 mutant cell line assessed for each tissue (n = 3, triplicate wells). Data is presented as relative to vehicle treated. (b) Proliferation of cell lines shown in (a) along with pretreatment of 1 μM KI696 in KEAP1 wild type cells followed by 250 nM CB-839 treatment for 4 days. Each data point represents an independent cell line (n = 3, triplicate wells). Data is presented as relative to vehicle treated. (c) Schematic depicting how constitutive NRF2 activation via KEAP1 mutations or use of small molecule activator, KI696, results in increased glutathione production and high xCT/SLC7A11 expression. Increased GSH production increases cellular demand for glutamate, making KEAP1 mutant cells glutamine dependent and susceptible to glutaminase inhibition. (d) Schematic depicting the balance between cellular production of antioxidants and demand for cysteine versus using glutamate to fuel anaplerosis (left). Activation of Nrf2 results in an imbalance between production of antioxidants and use of glutamate for anaplerosis that can be exploited by CB-839 treatment to further limit glutamate availability (right). All error bars depict s.e.m. ****p<0.0001.
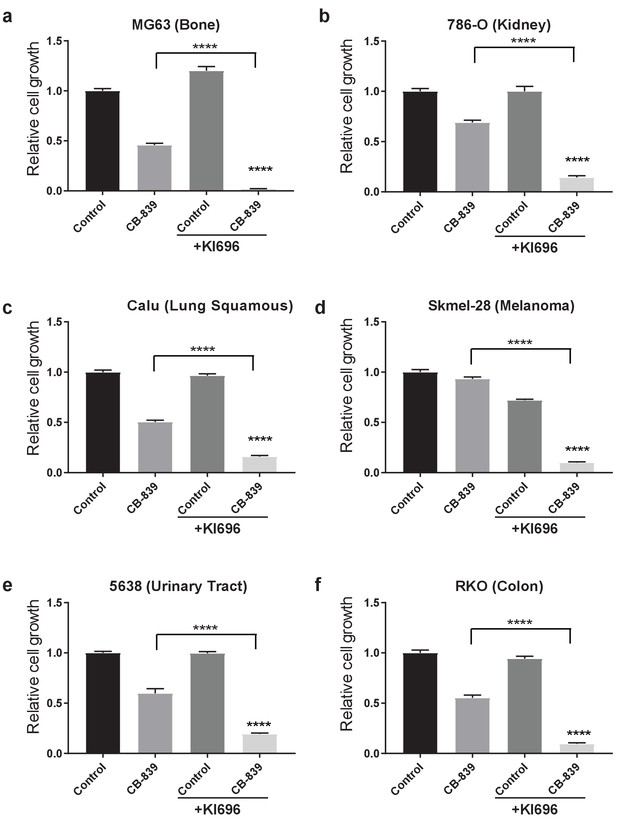
Activation of NRF2 sensitizes KEAP1 wild type cells to glutaminase inhibition in multiple cancer types.
Proliferation of various KEAP1 wild type human cancer cell lines. Cells were pretreated with 1 μM KI696 where indicated followed by 250 nM CB-839 treatment for 5 days. All data is presented as relative to the untreated condition for each cell line (n = 3, triplicate wells). All error bars depict s.e.m. ****p<0.0001.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28083.016





