AIRE is a critical spindle-associated protein in embryonic stem cells
Figures
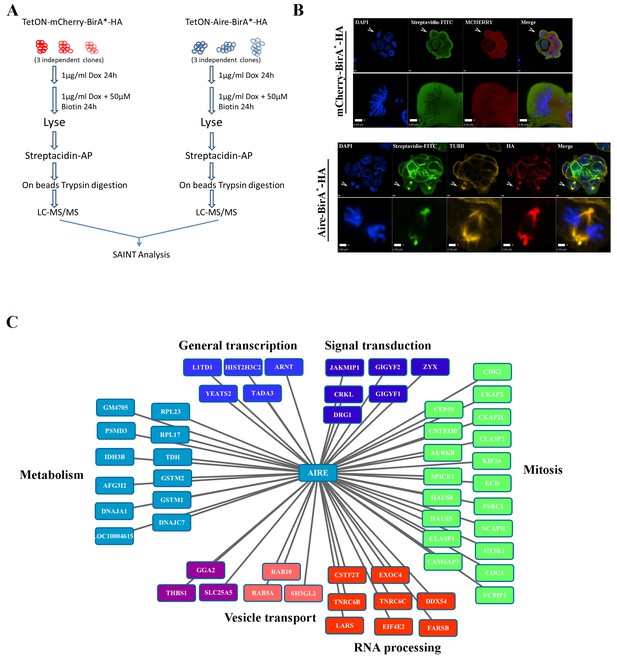
Proximity partners of AIRE in ES cells.
(A) Flow chart of BioID experiments. mCherry-BirA*-HA expressing ES cells were used as control to subtract the effect of promiscuous biotinylation activity of BirA*. (B) Localization of bait proteins (mCherry or HA) and biotin-tagged proteins (Streptavidin-FITC) in mCherry-BirA*-HA control ES cells and Aire-BirA*-HA ES cells shown by immunofluorescence staining. Scale bars: 1 μm. Bottom panel for each group shows magnified views of a mitotic cell, marked by arrows in top panel. Scale bars: 4 μm for mCherry-BirA*-HA, 2 μm for Aire-BirA*-HA. (C) List of proximity partners of AIRE identified by SAINT analysis (n = 3, 1%FDR) of BioID/mass spectrometry data and annotations of functional categories.
-
Figure 1—source data 1
List of proximity partners of AIRE.
- https://doi.org/10.7554/eLife.28131.004
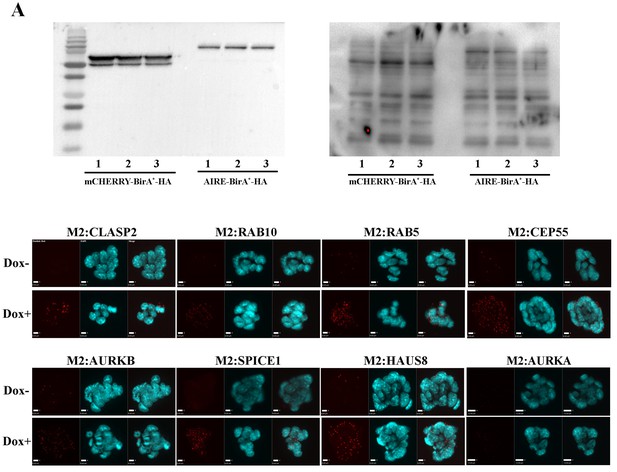
Validations of BioID analysis.
(A) Western Blotting analysis of mCherry-BirA*-HA and Aire-BirA*-HA proteins in mES cells (left: anti-HA antibody) and biotinylated cellular proteins (right: streptavidin-HRP). Each lane corresponds to protein from 105 cells from one biological replicate. (B) Proximity Ligation Amplification validation of the interaction between AIRE and a panel of proximity partners in TetON-3XFlag-Aire mES cells. Red signals: Duolink Orange.
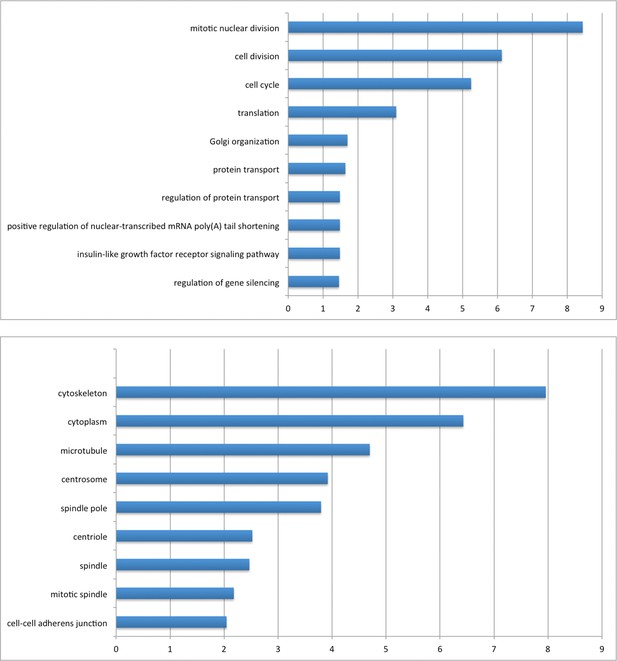
Gene ontology enrichment analysis of AIRE’s proximity partners with DAVID.
Left panel: Biological Processes (BP). Right panel: Cellular Compartment (CC). X axis: -Log(P).
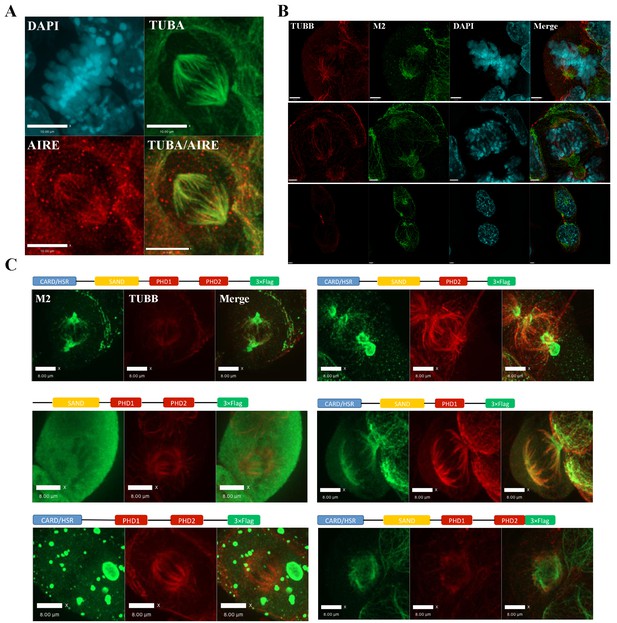
AIRE localizes to mitotic spindles in ES cells.
(A) Immunofluorescence staining of AIRE and α-tubulin (TUBA) in AmES8 cells. Scale bar: 10 μm. (B) Structural Illumination Microscope imaging of spindle localization of AIRE in AIRE-3XFlag expressing ES cells during different phases of mitosis. The Flag-tag was detected using the M2 mouse monoclonal antibody and spindle microtubules were marked by β-tubulin (TUBB). Scale bar: 2.5 μm. (C) Localization of different truncated versions of Aire-3XFlag (shown above each image) in mitotic ES cells by immunofluorescence staining. Scale bar: 5 μm.
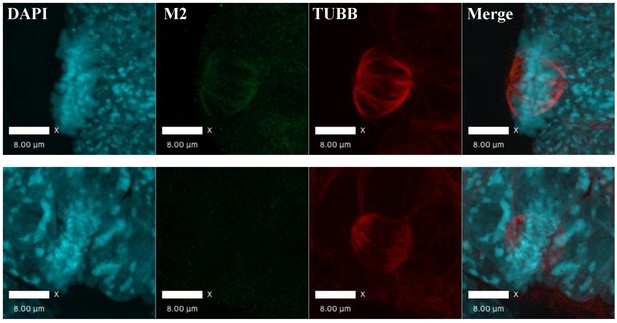
3XFlag-AIRE localized to mitotic spindles in Aire-3XFlag mES cell.
Immunofluorescence image of homozygous Aire-3XFlag mES cell in mitosis (upper panel) and wild type mES control (AmES8) (bottom panel) (Blue: DAPI, Green: M2 (AIRE-3XFlag), Red: TUBA), bar: 8 μm.
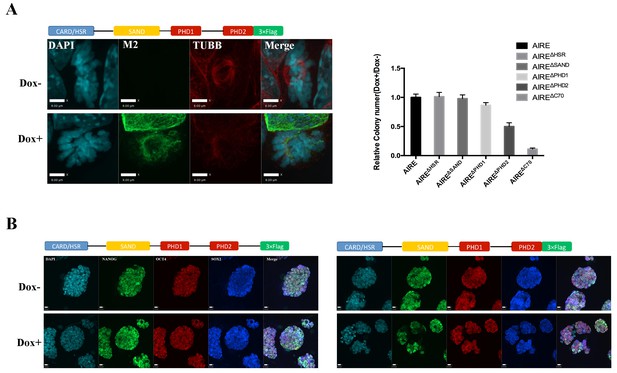
AIREΔc70 functions as a dominant negative, disrupting spindle assembly and inhibiting proliferation in mES cells.
(A) Left: immunofluorescence images of mitotic mES cells after 12 hr of Dox+ or control (Dox-) induction of AIREΔc70 expression. Scale bar: 8 μm. Right: relative ALP positive colony number (Dox+/Dox-) after 72 hr of Dox+ or control (Dox-) induction of AIREΔc70 expression in mES cells. Data presented as mean ± sd of 3 biological replicates. (B) Immunofluorescence images of ES cell markers OCT4, NANOG and SOX2 in mES with (Dox+) or without (Dox-) 12 hr induction of AIREΔc70 expression. Scale bar: 10 μm.
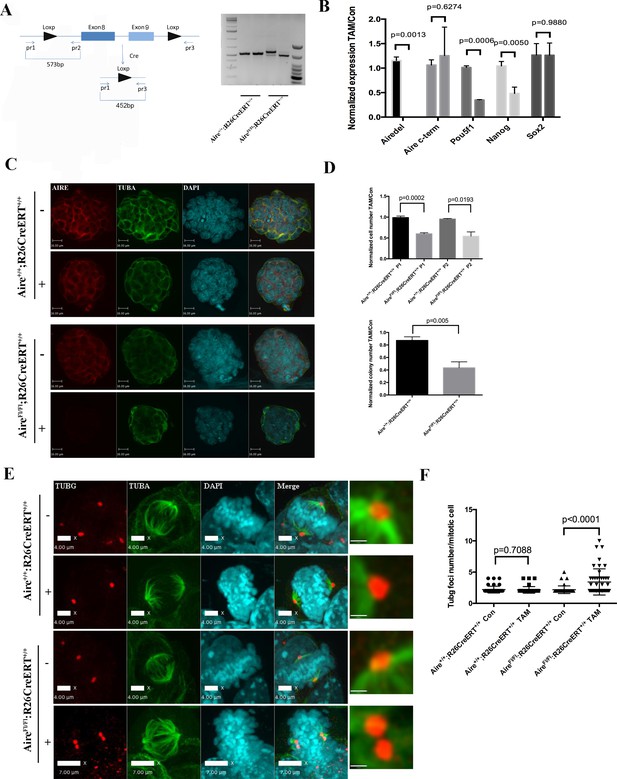
Aire is essential for spindle assembly in ES cells.
(A) Scheme of the AireFl allele and genotyping strategy before and after cre-induced deletion. Genotyping results from Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT+/+ ES cells without tamoxifen (-) and with 5 ng/ml tamoxifen for 24 hr (+). (B) qPCR analysis of gene expression at passage one after tamoxifen treatment in Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT+/+ ES cells. Aire del: primers complementary to deleted region of Aire. Aire c-term: primers complementary to c-terminal region of Aire. Data presented as mean ± sd of 3 biological replicates. p-values were calculated with Wilson’s t-test. (C) Validation of Aire knockout by immunofluorescence imaging in Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT+/+ ES cells without tamoxifen (-) and with 5 ng/ml tamoxifen for 24 hr (+). bar: 16 μm. (D) Proliferation over two passages and colony formation assay showing total cell number (top panel) and ALP positive colony number (bottom panel) in tamoxifen treated over non-treated Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT+/+ ES cells. Data in each panel presented as mean ± sd of 3 biological replicates. p-values were calculated with Wilson’s t-test. (E) Representative immunofluorescence images of γ-tubulin (TUBG) foci on mitotic spindles in passage 1 Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT +/+ ES cells without tamoxifen (-) and with 5 ng/ml tamoxifen for 24 hr (+). Scale bar: 4 μm. Right column shows magnified views of spindle poles. Scale bar: 1 μm. (F) Quantitation of γ-tubulin (TUBG) foci on mitotic spindles in passage 1 Aire+/+R26CreERT+/+ and AireFl/Fl R26CreERT+/+ ES cells without tamoxifen (Con) and with 5 ng/ml tamoxifen for 24 hr (TAM). γ-tubulin (TUBG) foci were counted from 30 mitotic cells from three biological replicates for each group. Each point represents one mitotic cell. Error bars: s.d. of mean. p-values were calculated with Mann–Whitney–Wilcoxon test.
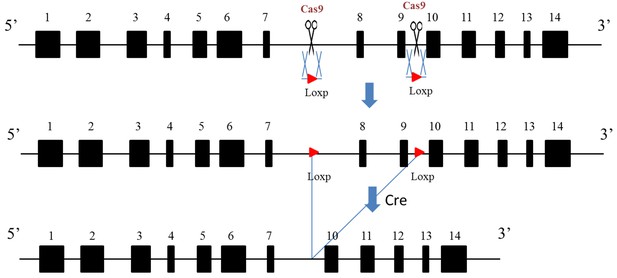
Complete diagram of Aire floxed (Fl) allele and cre induced depletion of critical exons.
https://doi.org/10.7554/eLife.28131.011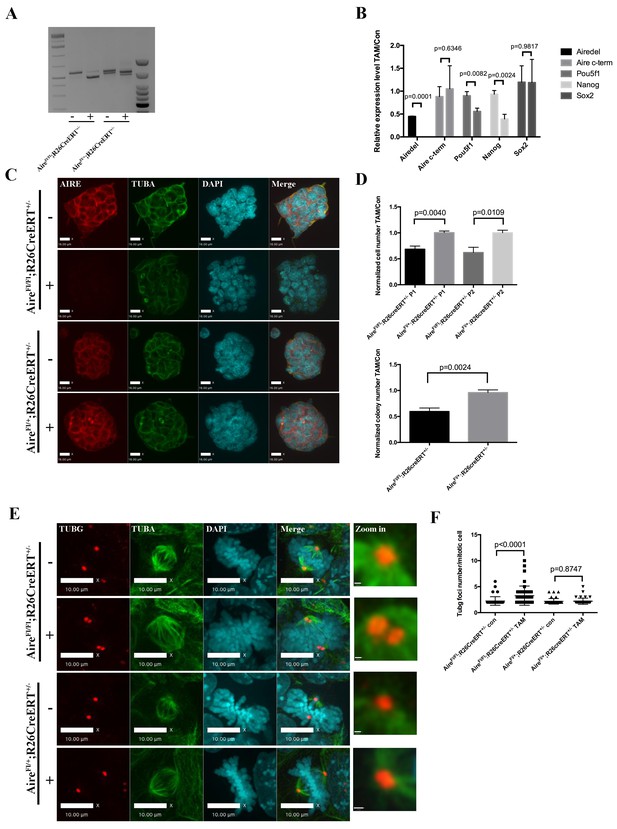
Aire knockout abrogated proliferation and spindle assembly in Pair 2 ES cells.
(A) Genotyping results from Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells without tamoxifen (-) and with 20 ng/ml tamoxifen for 24 hr (+). (B) qPCR analysis of gene expression at passage one after tamoxifen (TAM) treatment in Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells. Aire del: primers complementary to deleted region of Aire. Aire c-term: primers complementary to c-terminal region of Aire. Data presented as mean ± sd of 3 biological replicates. p-values were calculated with Wilson’s t-test. (C) Validation of Aire knockout by immunofluorescence imaging in Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells without tamoxifen (-) and with 20 ng/ml tamoxifen for 24 hr (+). Scale bar: 16 μm. (D) Proliferation over two passages and colony formation assay showing total cell number (top panel) and ALP positive colony number (bottom panel) in tamoxifen treated over non-treated (TAM/Con) Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells. Data in each panel presented as mean ± sd of 3 biological replicates. p-values were calculated with Wilson’s t-test. (E) Representative immunofluorescence images of γ-tubulin (TUBG) foci on mitotic spindles in passage 1 Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells without tamoxifen (-) and with 20 ng/ml tamoxifen for 24 hr (+). Scale bar: 10 μm. Right column shows magnified view of spindle poles. Scale bar: 0.5 μm. (F) Quantitation of γ-tubulin (TUBG) foci on mitotic spindles in passage 1 Aire Fl/Fl R26CreERT ± and AireFl/+R26CreERT ± ES cells without tamoxifen (Con) and with 20 ng/ml tamoxifen for 24 hr (TAM). γ-tubulin (TUBG) foci were counted from 30 mitotic cells from three biological replicates for each group. Each point represents one mitotic cell. Error bars: s.d. of mean. p-values were calculated with Mann–Whitney–Wilcoxon test.
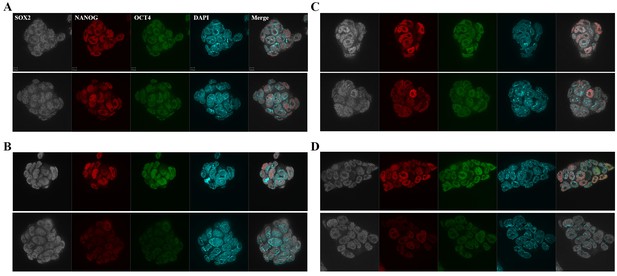
Immunofluorescence images of pluripotency markers in Aire Floxed;R26CreERT cells.
Scale bar: 1 μm. (A) Aire+/+R26CreERT+/+: Upper: Con; Bottom: TAM. (B) AireFl/Fl R26CreERT +/+: Upper: Con; Bottom: TAM. (C) AireFl/+R26CreERT+/-: Upper: Con; Bottom: TAM. (D) Aire Fl/Fl R26CreERT+/-: Upper: Con; Bottom: TAM.
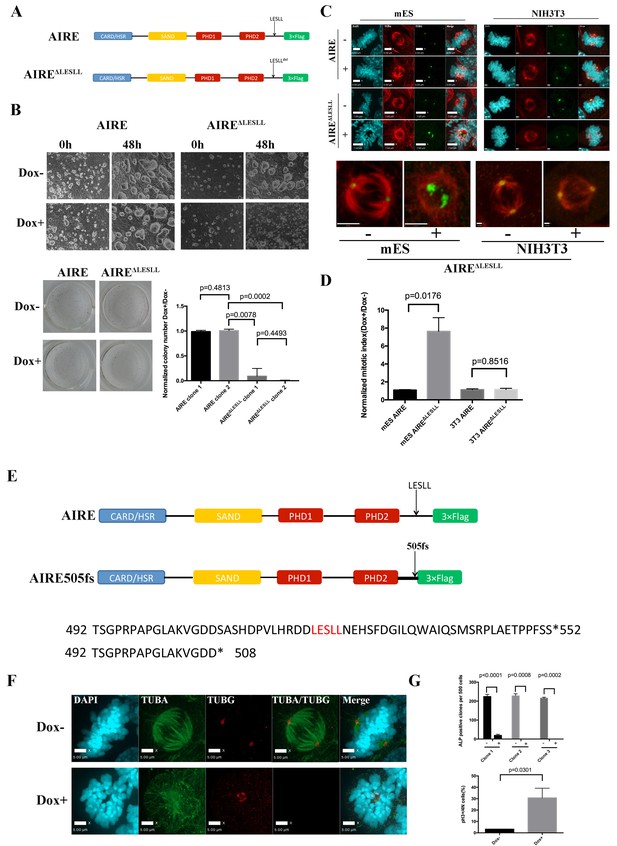
The last LxxLL(LESLL) motif is essential for the mitotic functions of Aire in ES cells.
(A) Diagram showing Aire and Aire ΔLESLL transgene structures. (B) Characterization of proliferation and colony formation of ES cells upon doxycycline-induced overexpression of AIRE or AIREΔLESLL. Representative bright field images of ES cell cultures 48 hr after doxycycline induction from three biological replicates (top panel). Representative images (bottom left panel) and quantification (bottom right panel) of ALP-stained colonies of AIRE or AIRE ΔLESLL-overexpressing ES cells 3 days after seeding. Quantification of ALP positive colonies was done from two independent clones for each transgene and normalized to Dox- controls. Data presented as mean ± sd of 3 biological replicates. p-values were calculated with Wilson’s t-test. (C) Immunofluorescence staining of spindles in control (Dox-) and AIRE or AIRE ΔLESLL-overexpressing (Dox+) ES cells (left panel) and NIHT3T cells (right panel). Scale bar: 7 μm. Magnified images of spindles from AIRE ΔLESLL-overexpressing groups. Scale bar: 4 μm. (D) Mitotic index (number of cells in mitosis in Dox+/Dox- conditions) of AIRE and AIRE ΔLESLL-overexpressing ES cells and NIH3T3 cells. Number of cells in mitosis was determined by flow analysis for PI-4N/pH3 markers. Data presented as mean ± sd of 3 replicates. p-values were calculated with Wilson’s t-test. (E) Diagram showing domain structure and partial amino acid sequence of human AIRE and AIRE505fs. (F) Immunofluorescence staining of spindles in control (Dox-) and AIRE505fs-overexperssing (Dox+) ES cells. Scale bar: 5 μm. (G) Characterization of colony formation and mitotic index of ES cells upon doxycycline-induced overexpression of Aire505fs. Top panel: number of ALP-positive colonies per well analyzed for 3 AIRE505fs transgenic ES cell clones without (Dox-) and with (Dox+) addition of doxycycline. Data presented as mean ± sd of 3 biological replicates for each clone, p-values were calculated with Wilson’s t-test. Bottom Panel: percent of cells in mitosis in control (Dox-) and AIRE505fs overexpressing (Dox+) ES cells. Number of cells in mitosis was determined by flow analysis for PI-4N/pH3 markers (bottom panel). Data presented as mean ± sd of 3 replicates. p-values were calculated with Wilson’s t-test.
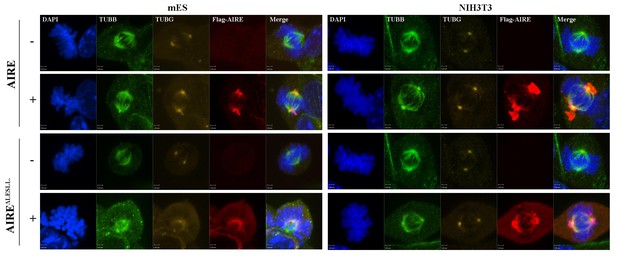
Both AIRE and AIREΔLESLL localized to mitotic spindles in mES and NIH3T3 cells.
Immunofluorescence images of spindle microtubules (β-tubulin (TUBB)), spindle pole (γ-tubulin (TUBG)) in AIRE and AIREΔLESLL-overexperssing (Flag-AIRE) mES cells and NIH3T3 cells.
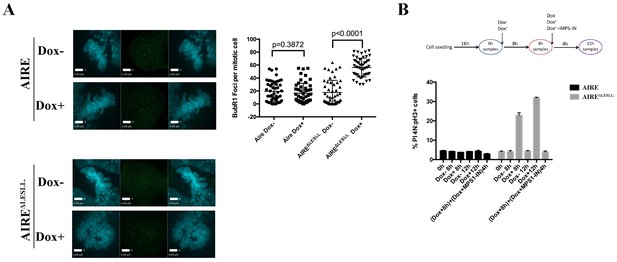
AIREΔLESLL activates the Spindle Assembly Checkpoint in ES cells.
(A) Left: representative immunofluorescence images of BubR1(Green) in mitotic ES cells without (Dox-) or with (Dox+) induction of AIRE or AIREΔLESLL. Scale bar: 5 μm. Right: quantification of BUBR1 foci number/mitotic cell. 50 cells from two biological replicates were quantified for each group. Each point represents one mitotic cell. Error bars: s.d. of mean. p-values were calculated with Mann–Whitney–Wilcoxon test. (B) Top: scheme of MPS1 treatment experiment. Bottom: quantification of mitotic index (the percentage of PI-4N/pH3 +cells) by flow cytometry after respective treatment shown below the x-axis. Data presented as mean ± sd of 3 biological replicates.
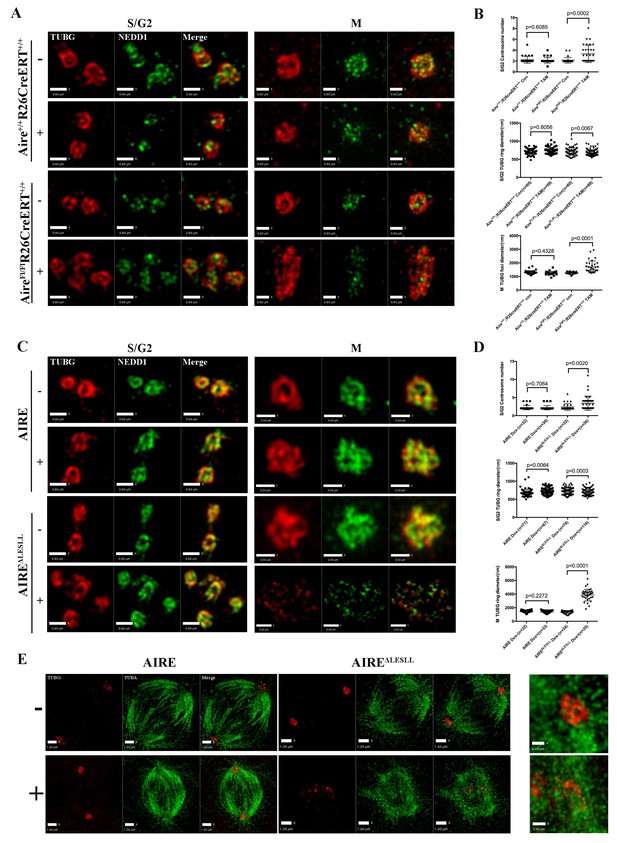
Aire is critical for centrosome number sustenance and the integrity of mitotic spindle poles.
(A) Representative immunofluorescence SIM images of γ-tubulin (TUBG) and NEDD1 in control (Aire+/+;R26CreERT+/+ tamoxifen-/+and AireFL/FL;R26CreERT+/+ tamoxifen-) and Aire-/- (AireFL/FL;R26CreERT+/+ tamoxifen +) mES cells during S/G2 and M phases of the cell cycle. Scale bar: 0.7 μm. (B) Quantification of centrosome parameters in control and Aire-/- ES cells (groups same as in panel (A)). Centrosome number in 30 S/G2 cells (top panel) from three biological replicates were counted for each group, mean ± sd is shown. Each point represents one mitotic cell. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin(TUBG) ring diameter in S/G2 cells (middle panel). Each point represents one centrosome. Mean ± sd was presented. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin(TUBG) foci diameter in M phase cells (bottom panel). 30 spindle poles from three biological replicates were quantified for each group. Each point represents one spindle pole. Mean ± sd is presented. p-values were calculated with Mann–Whitney–Wilcoxon test. (C) Representative immunofluorescence SIM images of γ-tubulin (TUBG) and NEDD1 in control (Dox-) and AIRE or AIRE ΔLESLL-overexpressing mES cells (Dox+) during S/G2 and M phases of the cell cycle. Scale bar: S/G2 Figure 0.6 μm; M images in top three panels 0.5 μm, and bottom panel 0.9 μm. (D) Quantification of centrosome parameters in AIRE or AIRE ΔLESLL-overexpressing mES cells (groups same as in panel (C)) Centrosome number in S/G2 cells (top panel). Each point represents one mitotic cell. Mean ± sd is presented. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin (TUBG) ring diameter in S/G2 cells (middle panel). Each point represents one centrosome. Mean ± sd was presented. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin(TUBG) diameter in M phase cells(bottom panel). Each point represents one spindle pole. Mean ± sd is presented. p-values were calculated with Mann–Whitney–Wilcoxon test. (E) Representative immunofluorescence SIM images of spindles and MOTCs (γ -tubulin and α-tubulin) in control (Dox-) and AIRE or AIRE ΔLESLL-overexpressing mES cells (Dox+) during M phase. Scale bar: 1.3 μm. Magnified view of MTOC in AIRE ΔLESLL (Dox-) (top scale bar: 0.6 μm) and AIREΔLESLL (Dox+) (bottom scale bar: 0.4 μm).
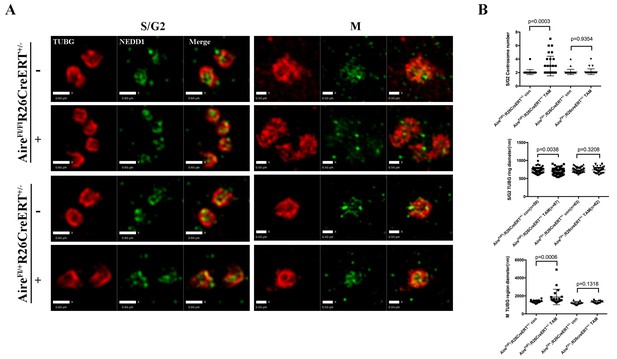
Aire is critical for centrosome number sustenance and the integrity of mitotic spindle poles.
(A) Representative immunofluorescence SIM images of γ-tubulin (TUBG) and NEDD1 in control (AireFl/+;R26CreERT ± tamoxifen-/+and AireFL/FL;R26CreERT ± tamoxifen-) and Aire-/- (AireFl/Fl;R26CreERT+/- tamoxifen +) mES cells during S/G2 and M phases of the cell cycle. Scale bar: 0.6 μm for S/G2 images, 0.5 μm for M images. (B) Quantification of centrosome parameters in control and Aire-/- ES cells (groups same as in panel (A)). Centrosome number in 30 S/G2 cells (top panel) from three biological replicates were counted for each group, mean ± sd is shown. Each point represents one mitotic cell. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin(TUBG) ring diameter in S/G2 cells (middle panel). Each point represents one centrosome. Mean ± sd was presented. p-values were calculated with Mann–Whitney–Wilcoxon test. γ-tubulin(TUBG) foci diameter in M phase cells (bottom panel). 20 spindle poles from three biological replicates were quantified for each group. Each point represents one spindle pole. Mean ± sd is presented. p-values were calculated with Mann–Whitney–Wilcoxon test.
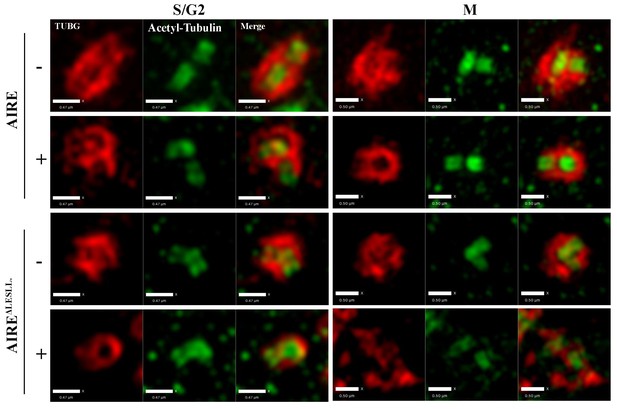
AIREΔLESLL overexpression caused defects in the pericentiole matter (PCM) around centrioles.
Representative SIM images of centrioles (Acetyl-Tubulin) and PCM (γ-tubulin (TUBG)) without (-) or with (+) 12 hr induction of the expression of AIRE or AIREΔLESLL. Left panels: Images of cells in S/G2 phase, scale bar: 0.47 μm. Right panels: Images of cells in M phase, scale bar: 0.5 μm.
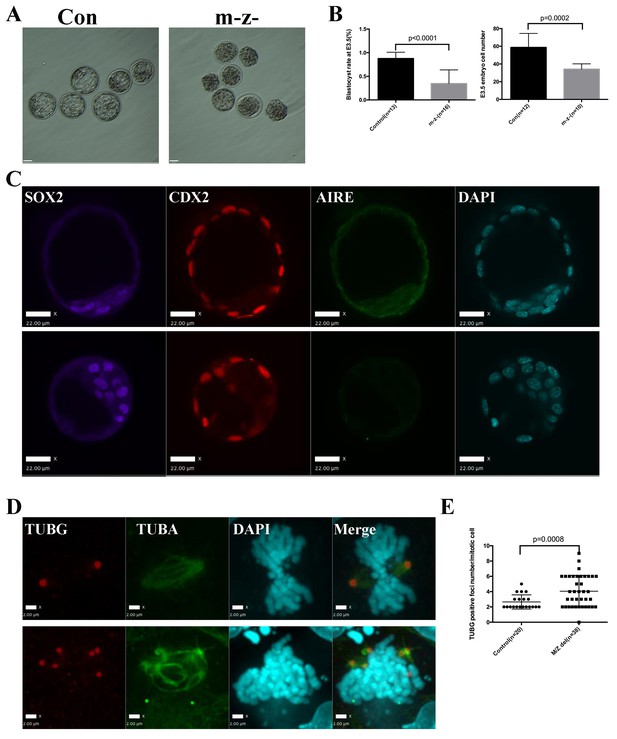
Aire m-/z- preimplantation embryos showed proliferation delay and spindle defects.
(A) Brightfield images of control (Con) and Aire m-/z- embryos at embryonic day (E) 3.5. (B) Quantification of blastocyst-formation rate (left panel) and cell number at E3.5 (right panel) in control (Con) and Aire m-/z- embryos. Data is presented as mean ± sd. p-values were calculated with Wilson’s t-test. (C) Immunofluorescence staining for Aire in control (Con) (upper) and Aire m-/z- (down) E3.5 embryos. Scale bar: 22 μm. (D) Representative immunofluorescence image of spindles (γ –tubulin (TUBG), α-tubulin (TUBA)) in E3.5 control (Con) (top) and Aire m-/z- (bottom) embryo cells. Scale bar: 2 μm. (E) Quantification of γ –tubulin (TUBG) positive foci per mitotic cell in control and Aire m-/z- embryos. Each point represents one mitotic cell. Mean ± sd is presented. p-values were calculated with Wilson’s t-test.
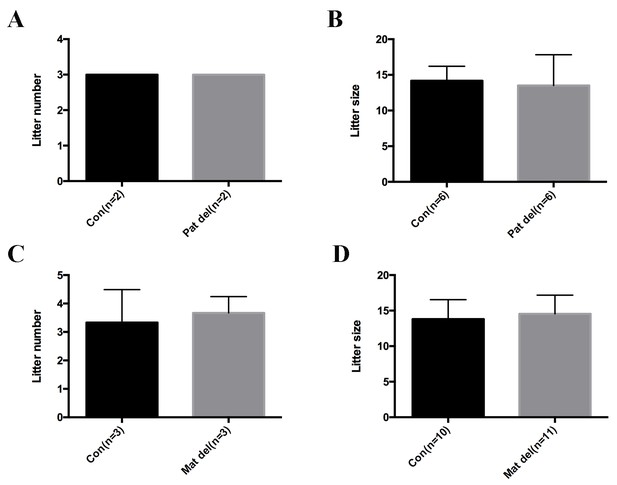
No obvious infertility phenotype was observed in paternal or maternal Aire knockout mice.
Data presented as mean ± sd. (A) Litter numbers produced in 3 months in control and paternal Aire knockout mice. (B) Litter sizes produced in 3 months in control and paternal Aire knockout mice. (C) Litter numbers produced in 3 months in control and maternal Aire knockout mice. (D) Litter sizes produced in 3 months in control and maternal Aire knockout mice.
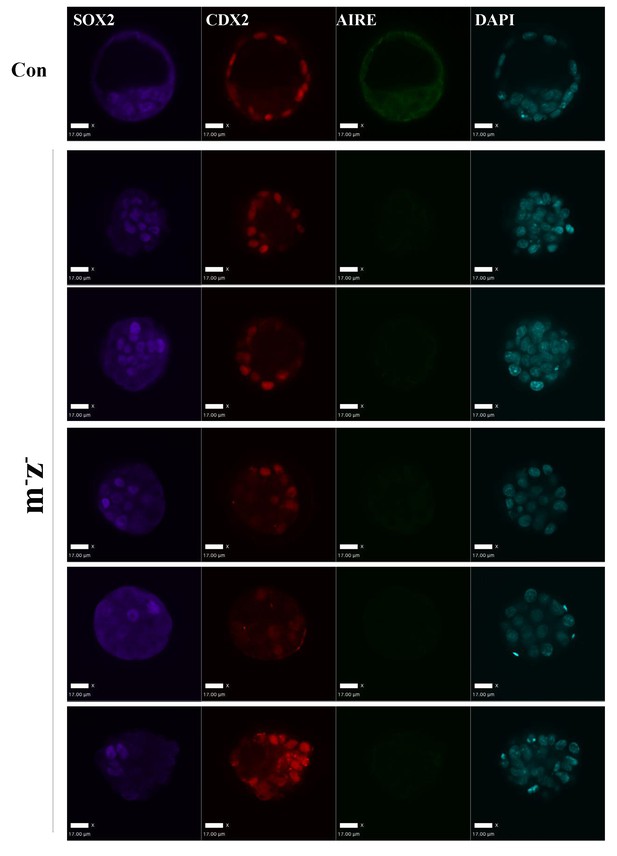
Aire m- z- embryos didn’t show obvious early lineage defects.
Representative immunofluorescence images of control (Con) and Aire m- z- E3.5 embryos for the epiblast marker SOX2, a trophectoderm marker CDX2 and AIRE. Legends for supplementary files.
Additional files
-
Supplementary file 1
Reagents and resources used in this study
- https://doi.org/10.7554/eLife.28131.023
-
Supplementary file 2
Oligos and primers sequences
- https://doi.org/10.7554/eLife.28131.024





