Maternal Gdf3 is an obligatory cofactor in Nodal signaling for embryonic axis formation in zebrafish
Figures
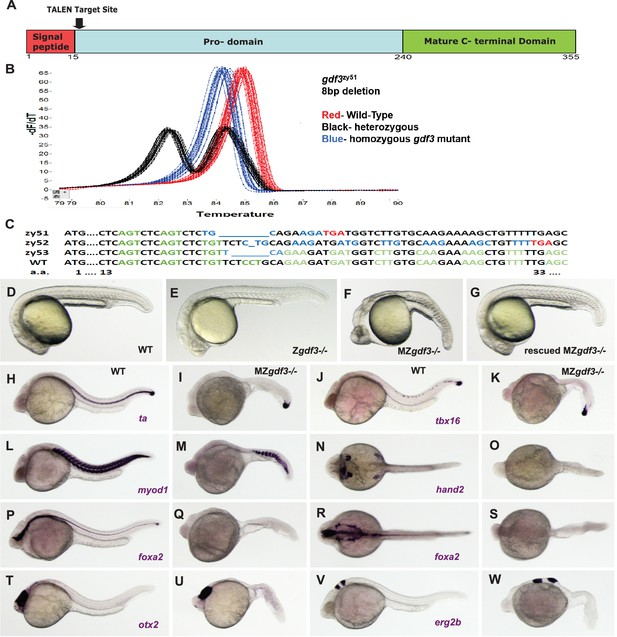
Zygotic gdf3 mutants are viable and maternal zygotic mutants have phenotypes indicative of loss of Nodal signaling.
(A–C) gdf3 mutants were generated using TALEN-mediated mutagenesis. (A) Gdf3, a TGFβ family member, comprises a signal peptide, pro-domain and mature TGFβ domain. TALENs were designed to target genomic sequences located near the amino end of the pro-domain. (B) Mutants were identified by high-resolution melt analysis (HRMA). (C) Three mutant alleles, gdf3zy51, gdf3zy52, gdf3z53, had 8, 1 and 6 bp deletions respectively. (D–G) Morphological phenotypes of gdf3 mutants at 24 hpf. (D) Wild-type (WT) and (E) zygotic (Z) gdf3 mutants were phenotypically indistinguishable. (F) Maternal-zygotic (MZ) gdf3 mutants lacked notochord, pharyngeal endoderm and had reduced anterior neural tissues. (G) MZgdf3 mutants were completely rescued by injection of 100 pg gdf3 RNA at the one-cell stage. (H–W) WISH analysis of gene expression in WT (columns H-T and J-V) and MZgdf3 (columns I-U and K-W) mutants at 24 hpf. (H, I) ta (ntl) expression in the notochord was absent from MZgdf3 although tailbud expression was maintained. (J, K) tbx16 (spt) expression in spinal cord neurons was absent in MZgdf3 while tailbud expression is unaffected. (L, M) Trunk and tail somites expressing myod1 were reduced in number and altered in shape in MZgdf3. (N, O) Expression of hand2 in the heart, pharyngeal arch mesoderm and pectoral fin buds is absent in MZgdf3. (P–S) Ventral neural tissues and pharyngeal endoderm expressing foxa2 (axial) were absent in MZgdf3. Patterns of expression of otx2 in the forebrain and midbrain (T, U), and erg2b (krox20) in hindbrain rhobomeres 3 and 5 (V, W) were altered in MZgdf3 mutants. All embryos (D–W) are shown in lateral view with rostral to the left except N, O, R, S which are dorsal views in the same orientation. Each panel is a representative image from at least 15 embryos.
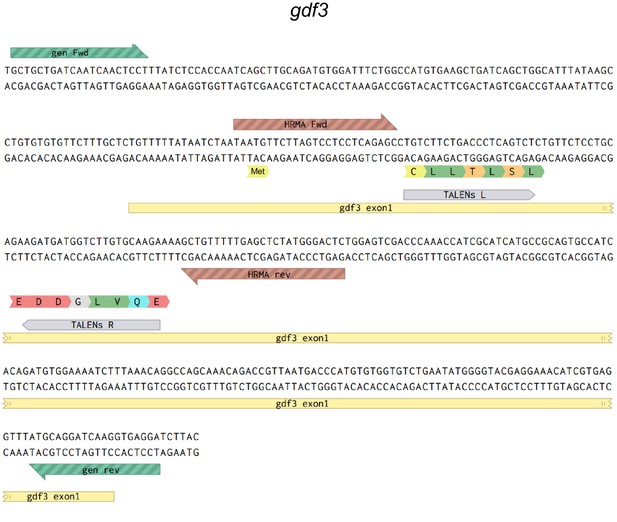
Map of the genomic sequences around the first coding exon of gdf3.
Primers used to sequence genomic DNA flanking the TALEN target site are in green, HRMA primers are in brown and left (L) and right (R) TALENS are in grey.
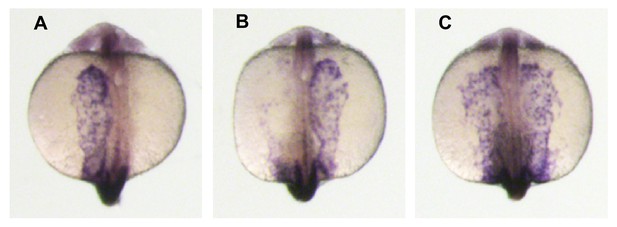
The Left-Right patterning marker spaw is expressed in zygotic gdf3 mutants.
(A–C) WISH analysis of spaw expression in 18–20 somite-stage embryos. The nodal paralogue spaw is normally expressed in the left lateral plate mesoderm. (A), but can also be observed in the right LPM (B) or expressed bilaterally (C). In embryos derived from crosses of gdf3zy51 heterozygous parents 90% had left-sided, 2% right-sided, and 8% bilateral expression (five clutches; n = 163 embryos total). The distribution of normal left-sided expression in embryos derived from gdf3 heterozygotes was not significantly different to that in embryos derived from wild-type AB strain parents (93% left, 2.5% right, 4.5% bilateral; n = 197, from four clutches; Pearson X2 test – p=0.463). Midline staining is myod1, which identifies somites and is used as a staging marker.
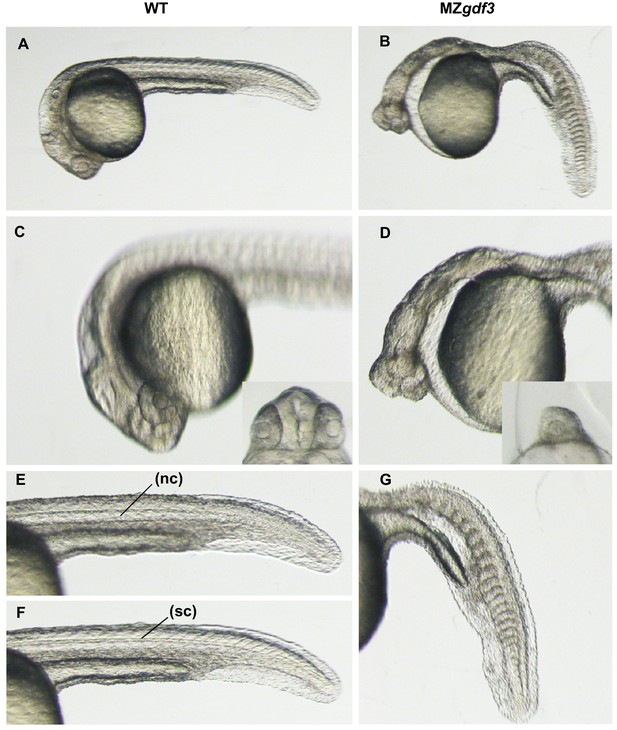
High magnification views of wild-type and MZgdf3 mutant embryos.
Overview of WT (A) and MZgdf3 (B) embryos at 26 hpf. (C) A focal plane at the anterior neural tube of WT embryos shows well developed eyes and complex brain morphology including ventricles and a fold at the midbrain-hindbrain junction. The eyes (inset) are widely separated by the tissue of the forebrain. (D) In contrast to WT embryos MZgdf3 embryos have a narrow simple neural tube, and the eyes are cyclopic (inset). The large space between the anterior neural tube and yolk is created by the loss of pharyngeal endoderm and cranial mesoderm including the anterior lateral plate that gives rise to the heart. (E, F) The trunk and tail of a WT embryo shows a well-defined notochord (nc) in (E) and spinal cord (sc) in (F) and chevron-shaped somites. (G) The notochord and spinal cord are absent from the trunk and tail of MZgdf3 and the somites are U-shaped and malformed.
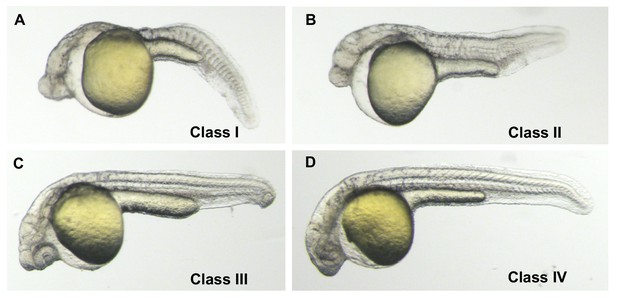
Phenotypic classes of MZgdf3 rescued with gdf3 RNA.
(A) Class I − 27 hpf un-injected MZgdf3 mutant. Embryos had greatly reduced anterior neural tube, a cyclopic eye, no notochord and poorly formed somites. (B) Class II – ‘minimal’ rescue of MZgdf3 with gdf3 RNA. Embryos showed rescue of posterior notochord and somites but no change in anterior neural tube or eye compared to un-injected mutants (C) Class III – ‘partial’ rescue of MZgdf3 with gdf3 RNA. In addition to rescue of posterior morphology embryos showed rescue of anterior neural tube including complex brain folds such as the midbrain-hindbrain junction but the eyes remained fused or incompletely separated. (D) Class IV - ‘complete’ rescue of MZgdf3. Embryos had morphologically normal rostral structures including fully separated eyes. The above classification scale was used to determine the optimal dose of gdf3 used in the rescue experiments presented in this report. Injecting MZgdf3 embryos with 25 pg of gdf3 RNA resulted in 0% Class 1, 39% Class II, 36% Class II and 25% Class IV embryos (n = 56). Injecting MZgdf3 embryos with 50 pg of gdf3 RNA resulted in 0% Class 1, 11% Class II, 39% Class II and 50% Class IV embryos (n = 54). Injecting MZgdf3 embryos with 100 pg of gdf3 RNA resulted in 0% Class 1, 0% Class II, 9% Class II and 91% Class IV embryos (n = 68). Doses of 200 pg gave rescue percentages similar to 100 pg (0%, 0%, 3%, 88%, Class I-IV respectively), but 9% of injected embryos (n = 65) showed tissue necrosis associated with anterior neural structures suggesting toxicity due to too much injected RNA. A dose of 100 pg was chosen for the reported rescue experiments.
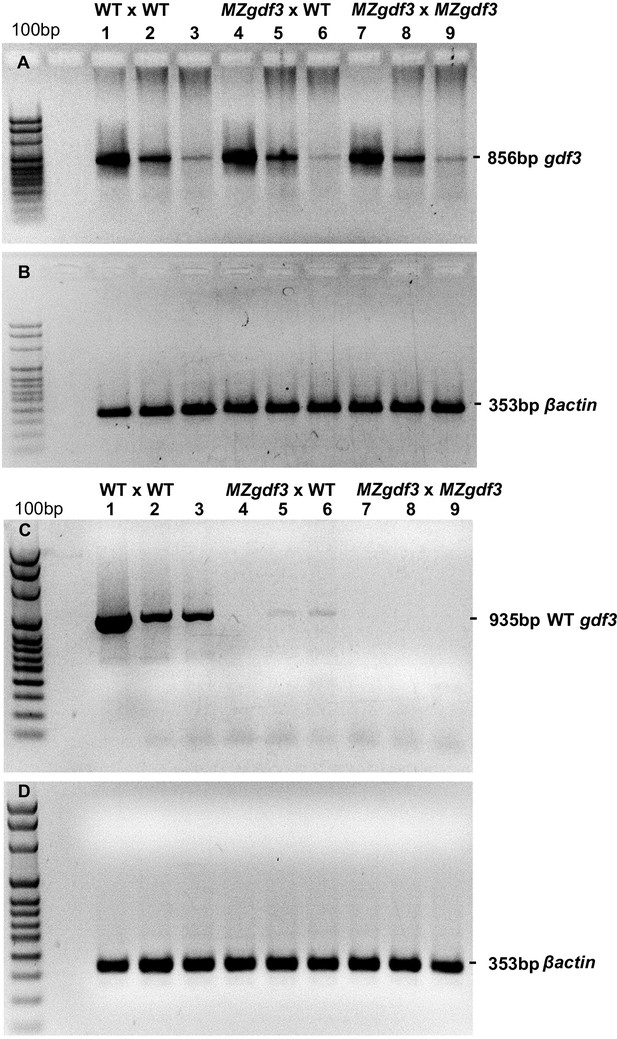
(A, B) RT-PCR analysis of cDNA prepared from 1000 cell-, 90% epiboly-, and 18 somite-stage embryo RNA, respectively, from: (Lanes 1–3) WT embryos, (Lanes 4–6) embryos derived from a female MZgdf3zy51 X WT male, and (Lanes 7–9) MZgdf3zy51 embryos.
PCR primers: gdf3 cDNA fwd ACCCAAACCATCGCATCATG; gdf3 cDNA rev TTTGATGGGAACGCAACAGG. (B) Control RT-PCR of replicate samples of those in panel A using βactin (act2b) primers: βactin cDNA fwd CCCAAGGCCAACAGGGAAA; βactin cDNA rev GGTGCCCATCTCCTGCTCAA. (C, D) RT-PCR analysis of cDNA prepared from the same stages and genotypes as in panels A, B. PCR Primers: (C) WT-specific gdf3 fwd CCTCAGTCTCTGTTCTCCTGC; gdf3 cDNA rev TTTGATGGGAACGCAACAGG. (D) βactin cDNA fwd CCCAAGGCCAACAGGGAAA; βactin cDNA rev GGTGCCCATCTCCTGCTCAA.
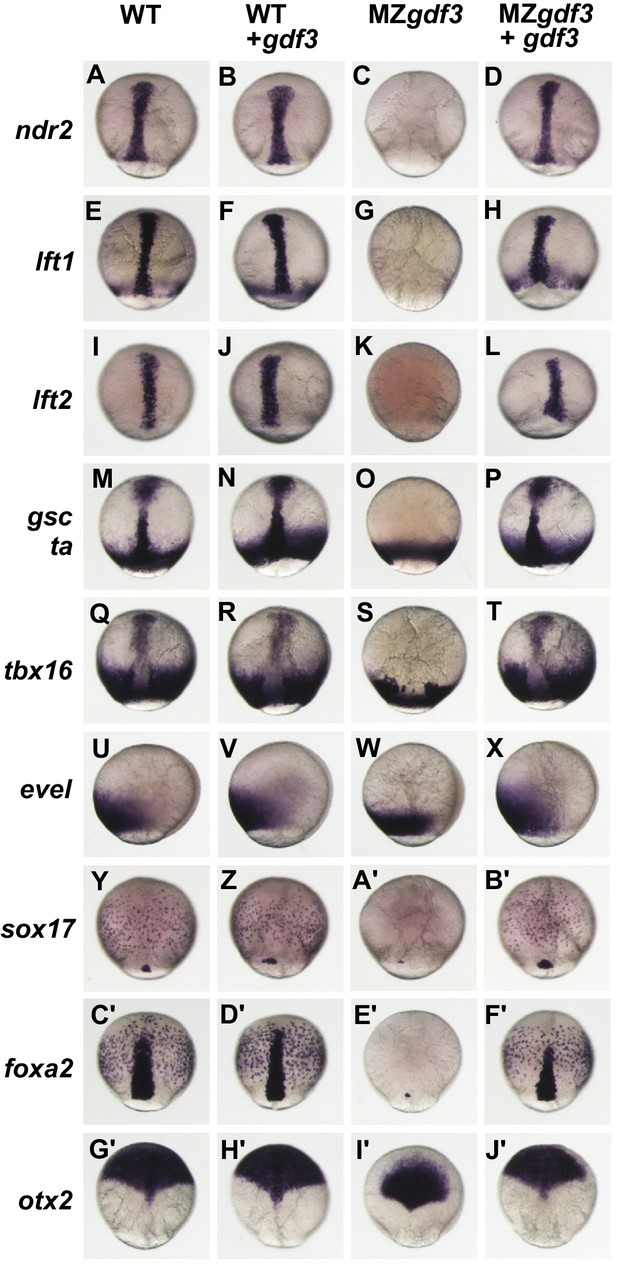
Gdf3 is required for mesoderm, endoderm and neural patterning.
(A–J’) WISH analysis of gene expression in embryos at 90% epiboly. Columns from left to right show WT embryos, WT embryos injected with 100 pg of gdf3 RNA, MZgdf3 mutants and MZgdf3 injected with gdf3 RNA. Each panel is a representative image of at least 15 embryos examined. (A–L) Midline and margin expression of Nodal signaling pathway genes ndr2 (A–D) and Lefty family members lft1 (E–H) and lft2 (I–L) were absent in MZgdf3 mutants and restored by gdf3 mRNA injection. (M–X) Analysis of early mesoderm transcription factor genes. (M–T). Expression domains of gsc, ta (ntl) and tbx16 (spt) were absent from the midline of MZgdf3 mutants, but restored by gdf3 RNA injection. (Q–X) Lateral and ventral mesendoderm expression domains of tbx16 (Q–T), and eve1 (U–X), which were reduced in width in MZgdf3, were restored to wild-type levels by gdf3 RNA. (Y–F’) Endoderm expression domains of transcription factors sox17 (Y–B’) and foxa2 (C’–F’) were absent in MZgdf3, and restored by gdf3 RNA, as was expression of foxa2 in midline neural tissues. (G’–J’). otx2 expression in the anterior neural plate is reduced in MZgdf3 but rescued to its normal extent by gdf3 RNA. (Second column from left) Strikingly, although injection of gdf3 RNA was capable of rescuing mesoderm, endoderm and neural tissues in MZgdf3 mutants it had no effect on gene expression in WT embryos.
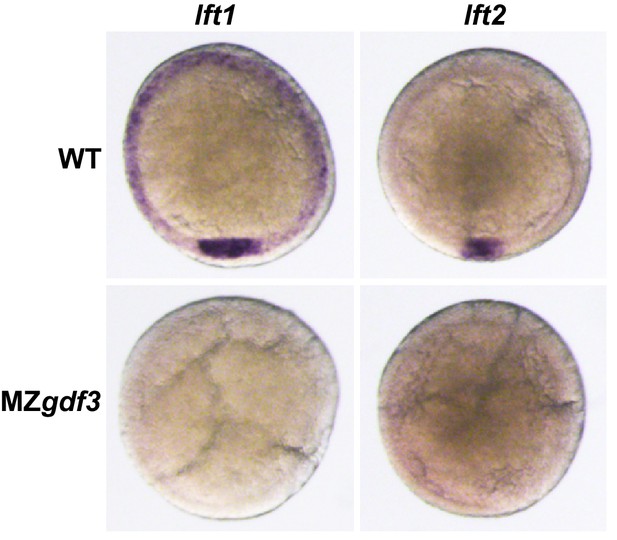
Nodal antagonists lft1 and lft2 are not expressed in MZgdf3.
(A, B) Wild-type embryos at shield stage expressed lft1 (A) in dorsal mesoderm at the dorsal shield and in mesendoderm precursors at the margin while lft2 expression (B) was limited to the dorsal shield. Shield-stage embryos of MZgdf3 did not express either lft1 (C) or lft2 (B).
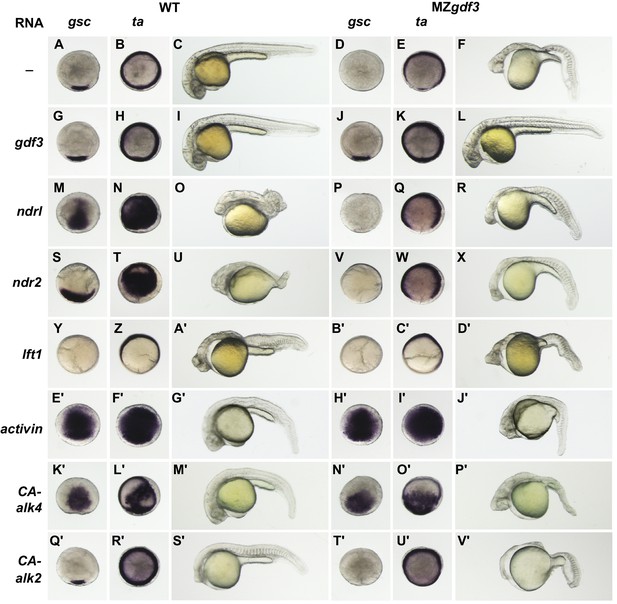
MZgdf3 mutants are refractory to Nodal ligands and inhibitors, yet respond to other TGFβs and have an intact downstream response pathway.
(A–V’) WT and MZgdf3 embryos injected at 1–2 cell stage with RNAs encoding gdf3, other TGFβ pathway members, inhibitors of Nodal, and downstream constitutively active (CA) Alk4 and Alk2 receptors. Panels show WISH expression patterns of gsc and ta in animal pole views of shield stage embryos (dorsal toward bottom) and light micrographs of morphology at 24 hpf. Each panel is a representative image of at least 15 embryos examined. (A–C) Un-injected WT and (D–F) MZgdf3 embryos. (G–I) Wild-type embryos injected with 100 pg gdf3 RNA were unaffected (J–L). Injected gdf3 RNA completely rescued gsc and ta expression patterns and morphology at 24 hpf in MZgdf3 embryos. Injected Nodal RNAs ndr1 (10 pg) (M–O) and ndr2 (10 pg) (S–U) caused ectopic expression of gsc and ta at shield, and dorsalized embryos at 24 hpf. In contrast, (P–R) ndr1 RNA or (V–X) ndr2 RNA injection into MZgdf3 embryos failed to rescue dorsal gsc or ta expression at shield stage and did not alter mutant morphology at 24 hpf. (Y–A’) Injection of 25 pg RNA encoding the Nodal inhibitor lft1 into WT embryos resulted in a loss of dorsal gsc and ta expression at shield stage similar to that seen in MZgdf3 embryos, and with phenotypes resembling MZgdf3 at 24 hpf. (B’–D’) In MZgdf3 injected with lft1 the gsc and ta expression patterns and 24 hpf mutant phenotype were similar to un-injected MZgdf3. Phenotypes of lft1-injected embryos appear more severe than MZgdf3 because these embryos are slightly older. Injection of 2.5 pg RNA encoding Xenopus Activin, a general TGFβ pathway activator, induced ectopic expression of gsc and ta in both (E’–G’) WT and (H’–J’) MZgdf3 embryos, and dorsalization of embryos that survived to 24 hpf. (K’–P’) Injection of 20 pg RNA encoding human constitutively active (CA)-Alk4 receptor, which mediates Nodal/GDF signaling during mesoderm induction, resulted in ectopic expression of gsc and ta and dorsalization of (K’–M’) WT embryos and (N’–P’) MZgdf3 mutants. (Q’–V’) Injection of 25 pg RNA encoding Xenopus CA-Alk2, which mediates BMP signaling for ventral patterning, had little or no effect on the dorsal expression of gsc or ta at shield stage but lead to ventralization of both WT and MZgdf3 embryos at 24 hpf.
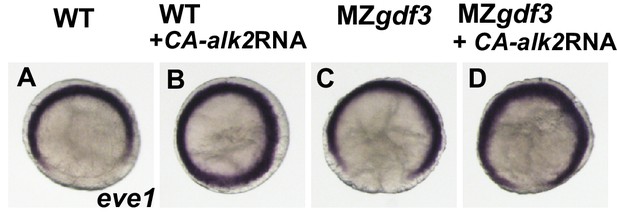
(A-D) The transcriptional repressor eve1 is expressed at the ventral and lateral margin at shield stage in WT and MZgdf3 embryos.
Injection of 25 pg of CA-alk2 RNA into 1-2 cell WT or MZgdf3 embryos expands this expression domain onto the dorsal side.
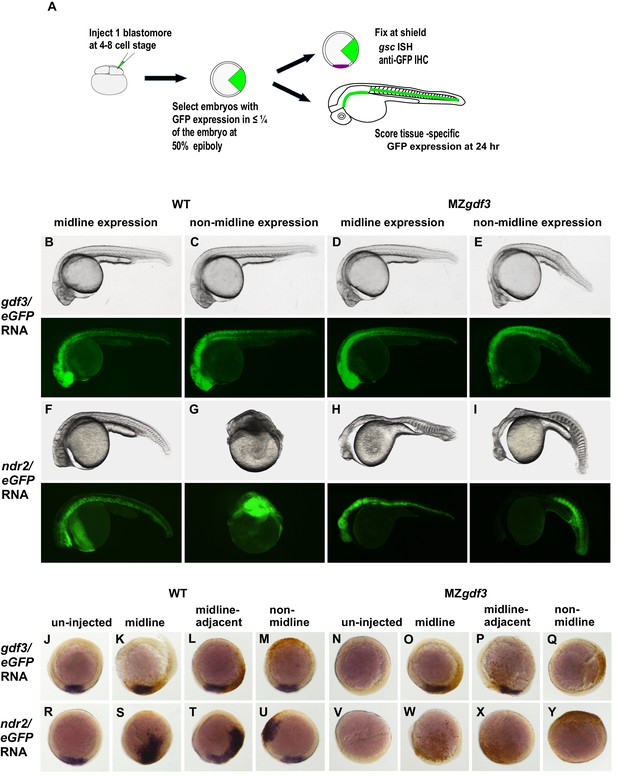
Gdf3 and Nodal must be co-expressed in lineages fated to become dorsal midline tissues.
The site of ectopic Nodal/Gdf3 signaling influences the efficacy of MZgdf3 mutant rescue and the severity of overexpression phenotypes in WT embryos. (A) Experimental Approach: 4–8 cell WT and MZgdf3 embryos were injected with 5 pg ndr2 RNA +25 pg eGFP RNA or with 50 pg gdf3 RNA +25 pg eGFP RNA. At 50% epiboly embryos expressing eGFP in 25% or less of the blastoderm were selected. These embryos were grown until 24 hpf and photographed with transmitted light and fluorescent illumination or were grown until shield stage and processed by WISH with a gsc probe then by IHC with anti-GFP. (B, C) 24 hpf WT embryos injected with gdf3/eGFP RNA had normal phenotypes, regardless of whether (B) midline or (C) non-midline lineages were targeted. (D) MZgdf3 embryos expressing Gdf3/eGFP in dorsally-derived midline lineages including the notochord and polster showed complete morphological rescue of notochord, somites, brain and eyes. (E) MZgdf3 embryos with expression of Gdf3/eGFP in non-midline lineages including the skin and somites showed rescue of somites and notochord but lacked normal development of anterior neural tissues and eyes. (F) Expression of Ndr2/eGFP in dorsal midline lineages in WT embryos resulted in embryos that were predominantly normal but some exhibited slightly narrower head and trunk and kinked notochords. This is strikingly distinct from (G) expression of Ndr2/eGFP outside midline lineages, which strongly dorsalized the embryos, or from ubiquitous expression in WT embryos injected with ndr2 RNA at the 1–2 cell stage (Figure 3S–U). (H, I) MZgdf3 embryos injected with ndr2/eGFP RNA showed no rescue of the MZgdf3 mutant phenotype regardless of what lineages were targeted. (J–Y) Shield stage embryos that were injected at the 4–8 cell stage with the indicated RNAs were processed by WISH for gsc and by IHC for GFP. Purple cells express gsc RNA; brown cells express GFP from RNA injection. Panels show embryos representative of each phenotypic class. (J–M) WT embryos at shield stage that were injected with gdf3/eGFP RNA showed no alteration in, or ectopic expression of gsc regardless of the location of expressing cells. (N–Q) MZgdf3 embryos injected with gdf3/eGFP RNA showed rescue of gsc expression when the expressing cells were located at (O), or adjacent to the presumptive dorsal shield (P). (R–U) WT embryos injected with ndr2/eGFP RNA showed ectopic gsc expression associated with the clone of expressing cells, regardless of the location of these cells within the embryo. (V–Y) MZgdf3 embryos injected with ndr2/eGFP RNA were unresponsive to this nodal ligand and showed no gsc expression. Note: Due to the lack of gsc expression, the location of the GFP-expressing clone of cells relative to the dorsal axis could not be reliably assigned in these embryos.
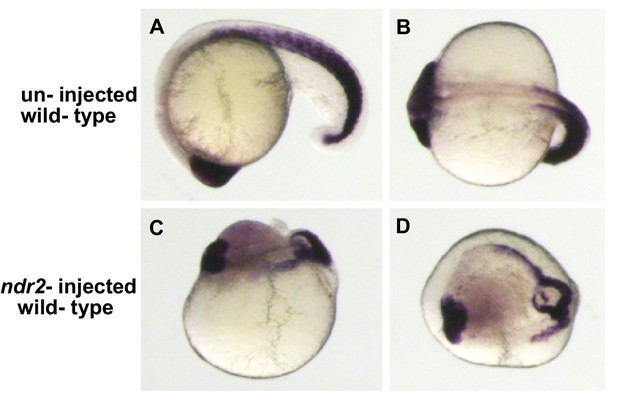
Morphological alterations caused by ndr2 overexpression.
WISH of 18 somite-stage embryos using probes to six3b which marks forebrain and retina and myod1 which marks somites in the trunk and tail. (A) Lateral and (B) ventral views of an un-injected WT embryo. Lateral (C) and dorsal (D) views of a wild-type embryo injected at the 4–8 cell stage with ndr2 showing a substantial disruption in the organization of embryonic axes and tissues.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28534.015





