Inferring genetic interactions from comparative fitness data
Figures

All rank orders of the biallelic two-locus system, where the colored rank orders imply epistasis.
Red (top row) indicates positive epistasis and blue (middle row) negative epistasis.
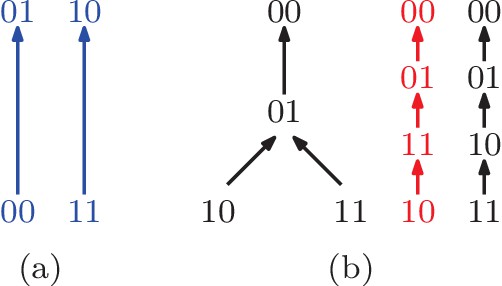
(a) A partial fitness order of genotypes.
The rank orders that extend this partial order are orders (9 , 10 , 12 , 13 , 14), and (16) in Figure 1. All of them imply negative epistasis (). (b) A partial order of genotypes with all its total extensions shown on the right. The first extension shown in red implies positive epistasis (), while the second one in black does not.
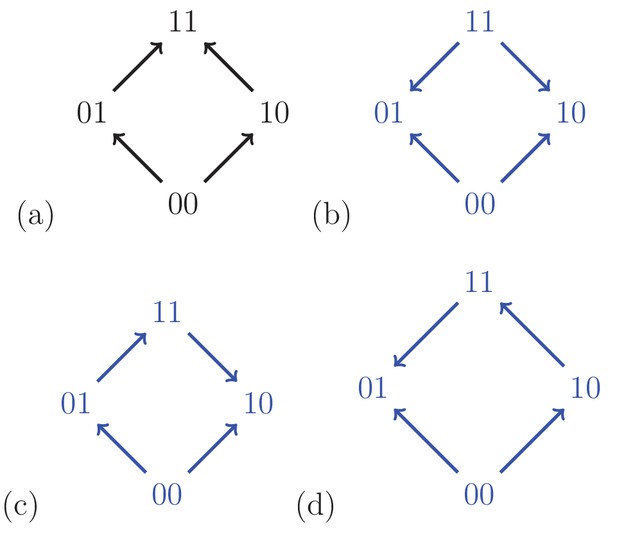
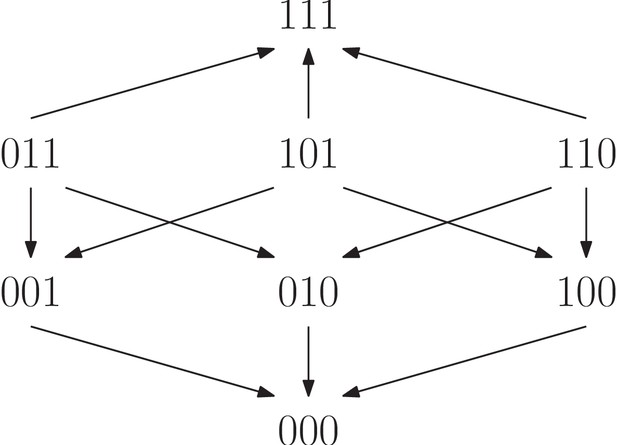
For a biallelic two-locus system where the genotype has the lowest fitness, there are four fitness graphs.
The graph (a) is compatible with additive fitness, whereas the remaining graphs imply negative epistasis.
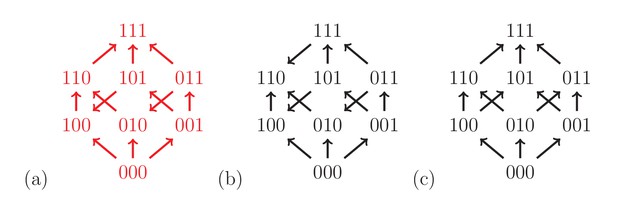
The fitness graph (a) implies three way epistasis, the graph (b) implies epistasis, but not higher order epistasis, and (c) does not imply epistasis, since all arrows point up.
https://doi.org/10.7554/eLife.28629.005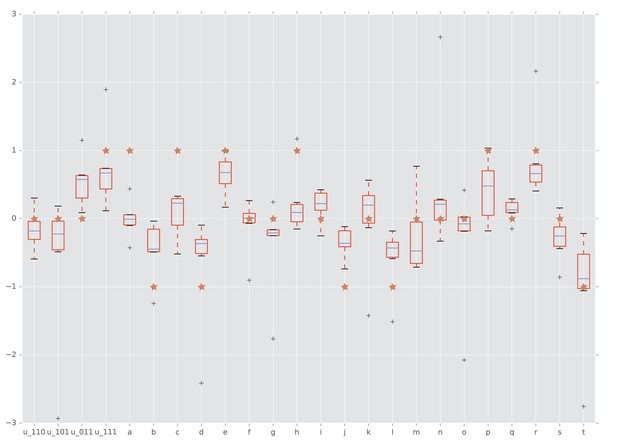
Interactions detected from fitness summary statistics and from rank orders.
The horizontal axis is labeled by the four interaction coordinates , , and twenty circuits , , . The boxplots show the distributions of the various interactions induced by the empirical fitness distribution. The red star indicates whether the interaction has been detected by our rank order method. Specifically, a star with vertical coordinate , 0, and one means negative, no, and positive interaction, respectively.
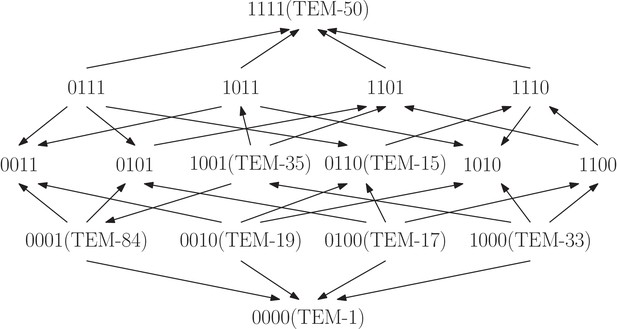
The TEM-family of -lactamase contributes to antibiotic resistance problems in hospitals.
The fitness graph shows a four-locus system consisting of the wild type, TEM-1, the quadruple mutant, TEM-50, and all intermediate mutants, including six clinically found mutants in the TEM family. The mutation M69L corresponds to , E104K to , G238S to , and N276D to . Growth rates were measured for the 16 genotypes under exposure to the antibiotic FEP Cefepime, and the fitness graph was determined accordingly (Mira et al., 2015). The graph reveals higher order epistasis.
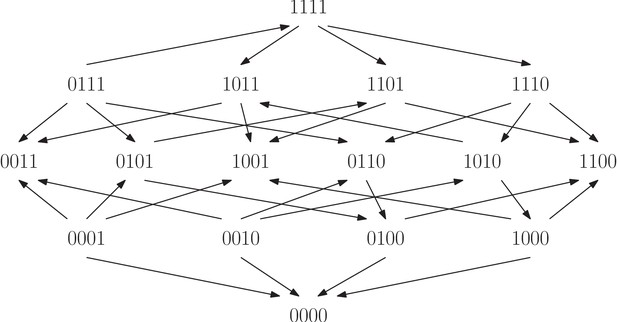
The fitness graph shows a four-locus system for the filamentous fungus Aspergillus Niger.
The system consists of all combinations of the four individually deleteriouis mutations fwnA1, leuA1, oliC2 and crnB12. The landscape has in total four peaks, labeled and .
The fitness graph is compatible with the two rank orders (5) and (6).
https://doi.org/10.7554/eLife.28629.009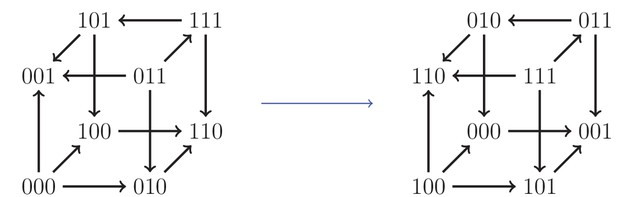
An example of an isomorphism.
Here, the allele labels ‘0’ and ‘1’ in the first locus have been interchanged, as well as the second and third loci.
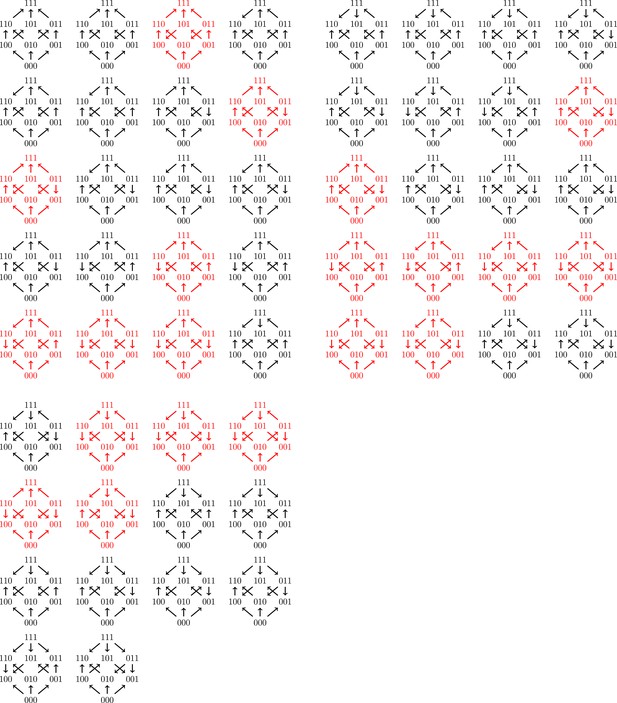
All 54 fitness graph types.
Those depicted in red imply three-way epistasis.
Tables
Numbers and fractions of rank orders that imply -way epistasis.
https://doi.org/10.7554/eLife.28629.011| Loci | Rank orders | Imply epistasis | Fraction |
|---|---|---|---|
| 2 | 24 | 16 | |
| 3 | 40,320 | 16,128 | |
| 4 | 20,922,789,890,000 | 4,649,508,864,000 |
Comparison of the rank order method with t-test.
The first column lists the four interaction coordinates and twenty circuits. The second column shows p-values returned by Student’s t-test based on fitness measurements. The third column shows which interactions are significant based on 0.03 threshold and their signs. For comparison, the last column displays the results obtained from rank order methods.
| Interaction | p-value | Result | From rank order |
|---|---|---|---|
| 1.13e-31 | |||
| 2.67e-12 | |||
| 1.20e-24 | |||
| 1.50e-29 | |||
| 7.10e-16 | |||
| 5.23e-32 | |||
| 7.62e-04 | |||
| 8.36e-68 | |||
| 1.39e-38 | |||
| 2.59e-01 | |||
| 3.10e-59 | |||
| 2.22e-02 | |||
| 7.97e-05 | |||
| 2.20e-32 | |||
| 1.96e-05 | |||
| 7.50e-51 | |||
| 4.88e-07 | |||
| 9.87e-37 | |||
| 8.83e-03 | |||
| 7.18e-19 | |||
| 1.94e-01 | |||
| 5.02e-50 | |||
| 7.10e-27 | |||
| 8.49e-61 |
Data from (Franke et al., 2011) on a 5-locus system determined by the mutations fwnA1, argH12, pyrA5, leuA1, and pheA1.
We consider 5-way epistasis for the system. The first column lists the ranking of the genotypes, where ”?” means missing measurement. The eighth column indicates whether the genotype is odd or even. The ninth and tenth columns show the cumulative number of ’s and ’s, respectively. The last column indicates whether the number of ’s exceeds the number of ’s () or vice versa (). We see that if genotype has higher fitness than genotype , genotypes are ranked arbitrarily, the missing genotype has rank , and rank , then the last column would change to all +’s, so the rank order would imply .
| Rank | fwn | arg | pyr | leu | phe | #mutations | / | # cumul. | # cumul. | neg. vs pos. |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | + | |
| 2 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 2 | + | |
| 3 | 0 | 1 | 0 | 1 | 1 | 3 | 1 | 2 | + | |
| 4 | 0 | 1 | 0 | 0 | 1 | 2 | 1 | 3 | + | |
| 5 | 1 | 1 | 0 | 1 | 1 | 4 | 1 | 4 | + | |
| 6 (or 7) | 1 | 1 | 0 | 0 | 0 | 2 | 1 | 5 | + | |
| 7 (or 6) | 1 | 0 | 0 | 1 | 0 | 2 | 1 | 6 | + | |
| 8 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 6 | + | |
| 9 | 1 | 1 | 1 | 0 | 0 | 3 | 3 | 6 | + | |
| 10 | 0 | 1 | 0 | 0 | 0 | 1 | 4 | 6 | + | |
| 11 | 0 | 1 | 1 | 0 | 1 | 3 | 5 | 6 | + | |
| 12 | 1 | 0 | 1 | 0 | 0 | 2 | 5 | 7 | + | |
| 13 | 0 | 0 | 0 | 1 | 0 | 1 | 6 | 7 | + | |
| 14 | 1 | 0 | 0 | 0 | 0 | 1 | 7 | 7 | + | |
| 15 | 1 | 1 | 0 | 0 | 1 | 3 | 8 | 7 | ||
| 16 | 0 | 1 | 1 | 0 | 0 | 2 | 8 | 8 | + | |
| 17 | 0 | 0 | 1 | 1 | 1 | 3 | 9 | 8 | ||
| 18 (or 19) | 1 | 1 | 1 | 0 | 1 | 4 | 9 | 9 | ||
| 19 (or 18) | 1 | 0 | 0 | 1 | 1 | 3 | 10 | 9 | ||
| 20 | 0 | 0 | 0 | 1 | 1 | 2 | 10 | 10 | + | |
| 21 | 0 | 1 | 0 | 1 | 0 | 2 | 10 | 11 | + | |
| 22 | 0 | 0 | 1 | 0 | 1 | 2 | 10 | 12 | + | |
| 23 | 0 | 0 | 1 | 0 | 0 | 1 | 11 | 12 | + | |
| 24 | 0 | 1 | 1 | 1 | 1 | 4 | 11 | 13 | + | |
| 25 | 0 | 1 | 1 | 1 | 0 | 3 | 12 | 13 | + | |
| 26 | 0 | 0 | 1 | 1 | 0 | 2 | 12 | 14 | + | |
| 27 | 1 | 0 | 1 | 0 | 1 | 3 | 13 | 14 | + | |
| 28 | 1 | 1 | 1 | 1 | 0 | 4 | 13 | 15 | + | |
| 29 | 1 | 1 | 1 | 1 | 1 | 5 | 14 | 15 | + | |
| 30 | 1 | 0 | 1 | 1 | 0 | 3 | 15 | 15 | + | |
| ? | 1 | 1 | 0 | 1 | 0 | 3 | ? | ? | ? | |
| ? | 1 | 0 | 1 | 1 | 1 | 4 | ? | ? | ? |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28629.015





