Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii
Figures
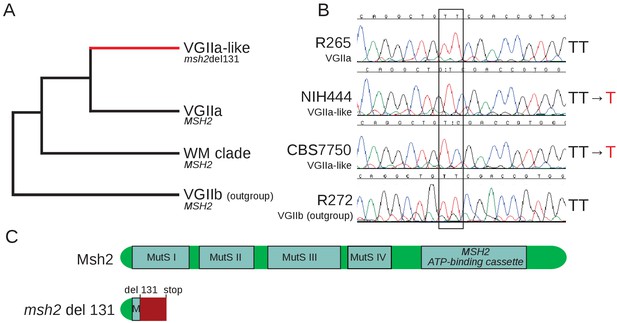
VGIIa-like isolates harbor a frameshift resulting in a predicted nonfunctional protein.
(A) The VGIIa-like clade is part of the clonal radiation that includes the Pacific Northwest Outbreak and shares with the VGIIa clade three ancestral isolates from South America described here as the WM clade. All three isolates are characterized by a frameshift in the MSH2 gene at position 131. (B) Sanger sequencing of the MSH2 gene confirms that NIH444 and CBS7750 have both undergone deletion of a single T within the coding region of MSH2, while the VGIIa R265 strain and the outgroup VGIIb strain have not. (C) This deletion results in a frameshift beginning in the first functional domain of MSH2 and an early premature stop. This truncated protein is predicted to be non-functional.
-
Figure 1—source data 1
Mutations shared by VGIIa-like strains relative to VGIIa.
Mutations identified on the branch separating VGIIa-like from VGIIa strains with predicted effects and functional domain information about genes affected by the changes.
- https://doi.org/10.7554/eLife.28802.004
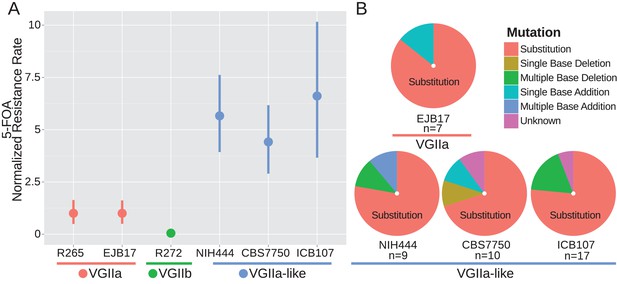
msh2 mutants are hypermutators.
(A) A fluctuation assay for resistance to 5-FOA was carried out for both wildtype VGIIa strains (R265 and EJB17) and hypermutator VGIIa-like strains (NIH444, EJB17, and ICB107), as well as an outgroup VGIIb (R272) strain. Resistance rates were normalized to the rate observed in R265. The VGIIa-like strains demonstrate an increase in mutation rate. Data shown are the mean of ten replicates with 95% confidence interval. (B) The molecular basis of resistance was determined for 5-FOA resistance at the URA5 locus. All isolates tested demonstrated predominantly substitutions as the molecular basis of resistance.
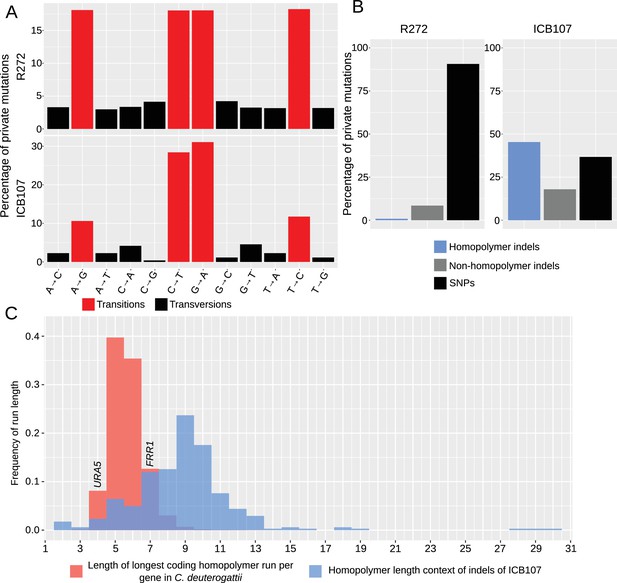
Hypermutators are characterized by high homopolymer instability.
(A) Percentage of transitions and transversions among SNPs private to R272, an outgroup, and ICB107, a hypermutator, are shown. Transitions are more common in both strains, while one type of transition is modestly reduced in the hypermutator. (B) All private mutations within R272 and ICB107 were characterized either as SNPs, indels, or homopolymer indels. The cutoff used to distinguish a homopolymer indel was longer than four bases. ICB107 had a substantial increase in homopolymer run shifts. (C) Frequency of indels within all homopolymers longer than one base from the private ICB107 mutations are shown in blue. All genes from the C. deuterogattii transcriptome were grouped by frequency of the longest homopolymer run within the coding region of that gene and shown in red. URA5 only has a run of 4 bases, while FRR1, which encodes FKBP12, the target of FK506 and rapamycin, has a run of 7 bases.
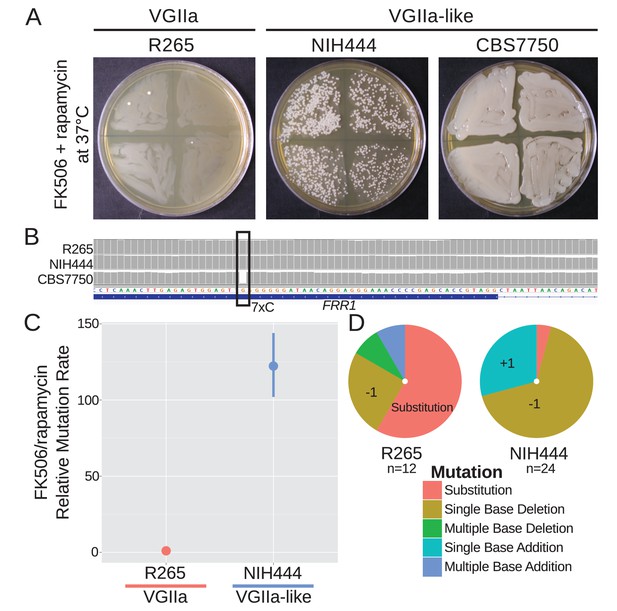
A homopolymer run within the FRR1 gene allows rapid inactivation and resistance to FK506 and rapamycin.
(A) Swab assays of the VGIIa and VGIIa-like strains were conducted to determine whether VGIIa-like stains develop resistance to FK506 and rapamycin at 37°C at an elevated rate compared to VGIIa. NIH444 demonstrated a large increase in resistance rate, while CBS7750 was completely resistant. (B) The resistance in CBS7750 is attributable to a single base deletion within the coding 7C run in the FRR1 gene that has been fixed in this strain. (C) Fluctuation assays for NIH444 and R265 show that the hypermutator conferred a greater than 100-fold increase in mutation rate compared to the wildtype VGIIa strain. Data shown are the mean of ten replicates with 95% confidence intervals and are normalized to the rate in R265. (D) Analysis of the molecular basis of resistance shows that substitutions are still the predominant mechanism for resistance in R265, but in the hypermutator strain single base additions and single base deletions within the homopolymer run are responsible for the vast majority of resistance.
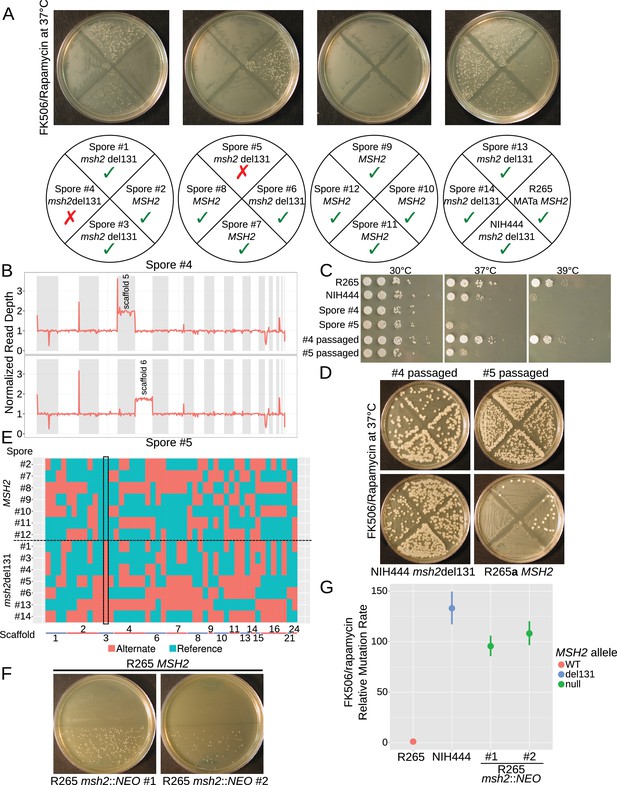
The hypermutator phenotype is linked to the frameshift in MSH2.
(A) Progeny from a cross between NIH444 and R265 show co-segregation of the msh2 del131 allele with the hypermutator phenotype in all except two cases. (B) Whole genome depth of coverage plots show that scaffolds five and six are aneuploid in spores #4 and #5, respectively. (C) Temperature sensitivity assay for the incongruent spores before and after serial passaging at 37°C. (D) After passage at 37°C, the non-congruent spores now properly demonstrate linkage with the msh2 del131 allele. (E) Whole genome sequencing of NIH444 x R265a) progeny, with variant regions indicated as reference in blue or alternate in red. The boxed region indicates a SNP in close linkage to the msh2 del131 allele. (F) Two independent de novo deletions of MSH2 via biolistic transformation demonstrate elevated mutation rates in an FK506/rapamycin swabbing assay at 37°C. (G) Fluctuation assays for R265, NIH444, and two independent deletions of msh2 in R265 show that the hypermutator phenotype is recapitulated in the null mutants. Data shown are the mean of ten replicates with 95% confidence intervals and are normalized to the rate observed in R265.
-
Figure 5—source data 1
Aneuploidy in NIH444 x R265a cross.
Predicted aneuploid scaffolds for 14 spore progeny from an NIH444 by R265a cross based on read depths from whole genome sequencing. 8 of 14 demonstrate aneuploidy for at least one scaffold.
- https://doi.org/10.7554/eLife.28802.009
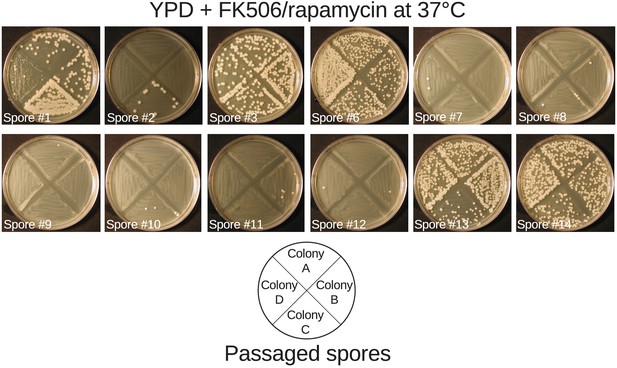
Passaged spore progeny demonstrate linkage between msh2del131 allele and mutator phenotype.
Four independent overnight cultures of spore progeny passaged at 37°C to select against aneuploidies that confer temperature sensitivity were struck on YPD media containing FK506 and rapamycin at 37°C. All fourteen passaged spore progeny demonstrated behavior predicted by the state of the MSH2 locus for the majority of the four samples tested. Spores #4 and #5 and controls are depicted in Figure 5.
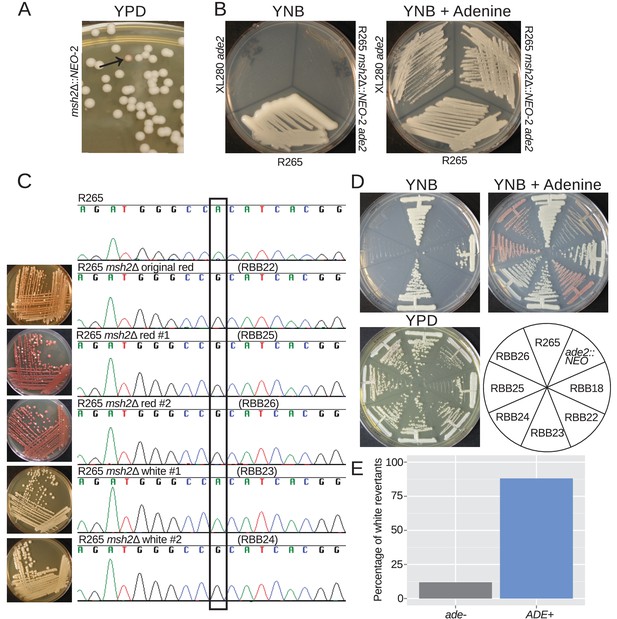
Hypermutation allows inactivation and reactivation of adenine biosynthetic pathway.
(A) A spontaneous red colony was isolated from the de novo msh2 deletion (RBB18). (B) This colony (RBB22) demonstrated adenine auxotrophy, suggesting that it was an ade2 mutant. (C) Sequencing of the ADE2 locus confirmed that the original colony was an ade2 mutant. In addition, two red (RBB25 and RBB26) and two white derivatives (RBB23 and RBB24) were tested. One white derivative had reverted the original mutation (RBB23), while the second had eliminated production of the red intermediate but had not reverted the original ade2 mutation (RBB24). (D) One revertant colony (RBB23) demonstrated adenine prototrophy, while the other (RBB24) remained an auxotroph despite losing the red pigmentation. (E) An assay to test direct reversion frequency versus secondary mutation to eliminate the red toxic intermediate demonstrated that the most common mutations were direct reversions and restoration of adenine prototrophy.
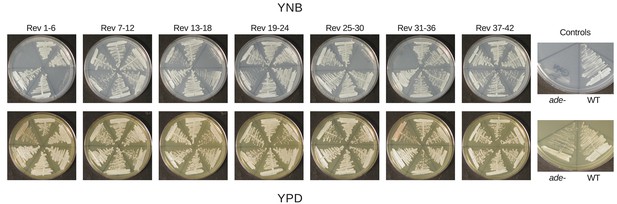
Reversion of ade2 mutants primarily occurs through repair of ADE2.
42 white colonies were selected as independent revertants from a red ade2 mutant (RBB22). Colonies were struck to both YPD and YNB to test for auxotrophy indicative of either direct reversion or a second site mutation upstream of ade2 in the adenine biosynthetic pathway.
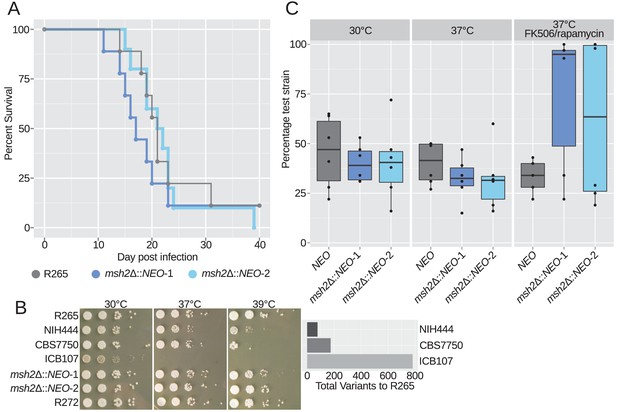
Hypermutation does not have immediate virulence defects but may potentiate long term deficits.
(A) Virulence tested in the murine inhalation model was not strongly affected by deletion of the MSH2 gene. (B) All three VGIIa-like strains demonstrated defects in high temperature growth via a spot dilution assay in comparison with the VGIIa R265 strain and the VGIIb R272 strain as an outgroup. Strains with longer branches exhibited larger high temperature growth defects. (C) Competition experiments between a tester strain with the neomycin resistance marker and the wildtype R265 strain. (Strain used: SEC501, RBB17, RBB18). Original cultures were mixed in a 1:1 ratio and then grown overnight in liquid YPD. Both hypermutators showed a modest growth defect at 30°C and 37°C but a dramatic growth advantage in the high stress FK506/rapamycin 37°C condition. Boxplots show minimum, first quartile, median, third quartile, and maximum values. Points represent the results from six individual replicates summarized by the box plot. The NEO vs WT competition is shown in gray, while the two msh2Δ::NEO competitions are shown in dark and light blue.
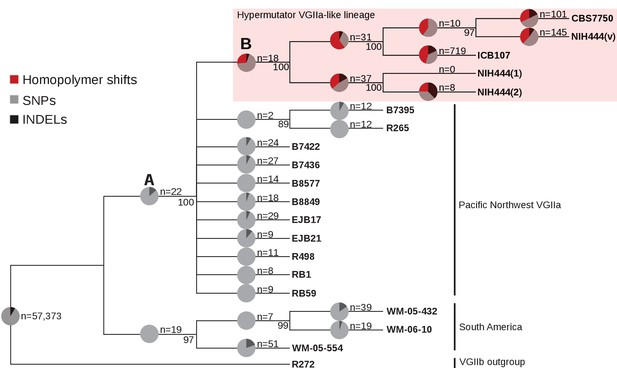
The VGIIa-like hypermutator is derived and not ancestral to the Pacific Northwest outbreak.
A maximum parsimony phylogeny of the VGIIa group with the VGIIb R272 genome as an outgroup demonstrates that the VGIIa-like group is a branch parallel to the VGIIa group. To test for the presence of a defect in MMR throughout the tree, the mutation spectrum was examined on each branch. High rates of homopolymer run shifts were observed throughout the VGIIa-like group, but no evidence was apparent at branch A). Instead it appears that the hypermutator first arose on branch B).
Additional files
-
Source code file 1
Custom Perl script
- https://doi.org/10.7554/eLife.28802.015
-
Supplementary file 1
Strains used in this study.
- https://doi.org/10.7554/eLife.28802.016
-
Supplementary file 2
Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.28802.017
-
Supplementary file 3
SNP data used to build phylogeny
- https://doi.org/10.7554/eLife.28802.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28802.019





