An Aedes aegypti-associated fungus increases susceptibility to dengue virus by modulating gut trypsin activity
Figures
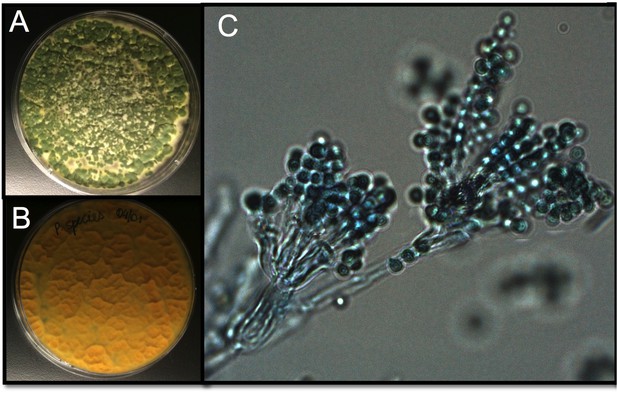
Tsp_PR morphology.
After isolation, the fungus was grown on Sabouraud agar and characterized macroscopically and microscopically. (A) Top and (B) bottom view of the fungus on Sabouraud agar. (C) Microscopic view of the typical brush-like biverticillated conidiophore of Talaromyces sp. fungi.
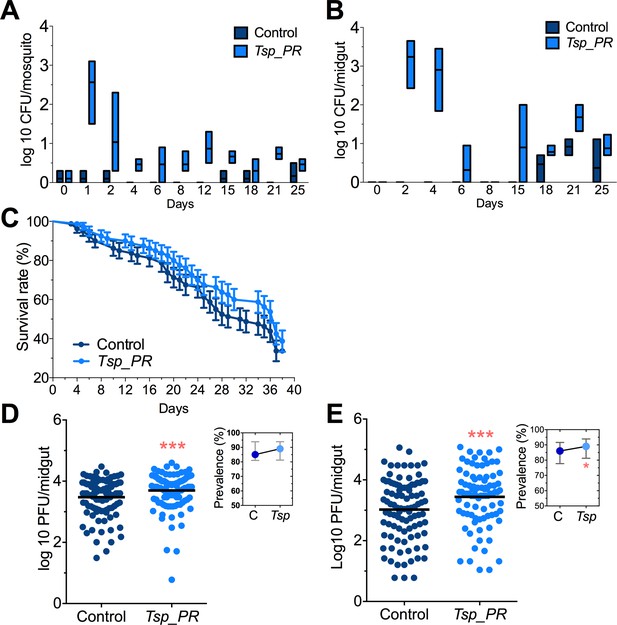
Tsp_PR fungus significantly increases DENV infection in Aedes mosquito midguts.
Aedes mosquitoes were mock-fed or fed for 48 hr with 10% sucrose solution containing 1 × 109 Tsp_PR spores. After spore feeding, (A) Fungus colonization in whole mosquitoes or (B) midguts. The presence of Tsp_PR in the mosquito was monitored for 25 days after introduction by enumerating fungal CFUs on Sabouraud agar with antibiotics cocktail from three independent experiments, the line indicates the mean and bars the maximum and minimum ranges. (C) Survival assays. Female mosquitoes fed with Tsp_PR spores or unfed were monitored in a daily basis for 38 days in three independent experiments (N = 80, p=0.3073). Error bars represent ± SE. (D) Rockefeller strain mosquitoes, (Control, N = 123; Tsp_PR, N = 120) or (E) Orlando strain mosquitoes (Control, N = 113; Tsp_PR, N = 99) were infected with a blood meal containing DENV; at 7 days post-infection (dpi), the midguts were dissected. Each dot represents a plaque-forming unit (PFU) transformed to log10 in individual midguts from three independent experiments. The line indicates the mean. Upper right boxes show the prevalence of infected mosquitoes, error bars represent the 95% confidence interval. *p<0.05, ***p<0.001,.
-
Figure 2—source data 1
Raw data and statistics summary for Figure 2.
- https://doi.org/10.7554/eLife.28844.004
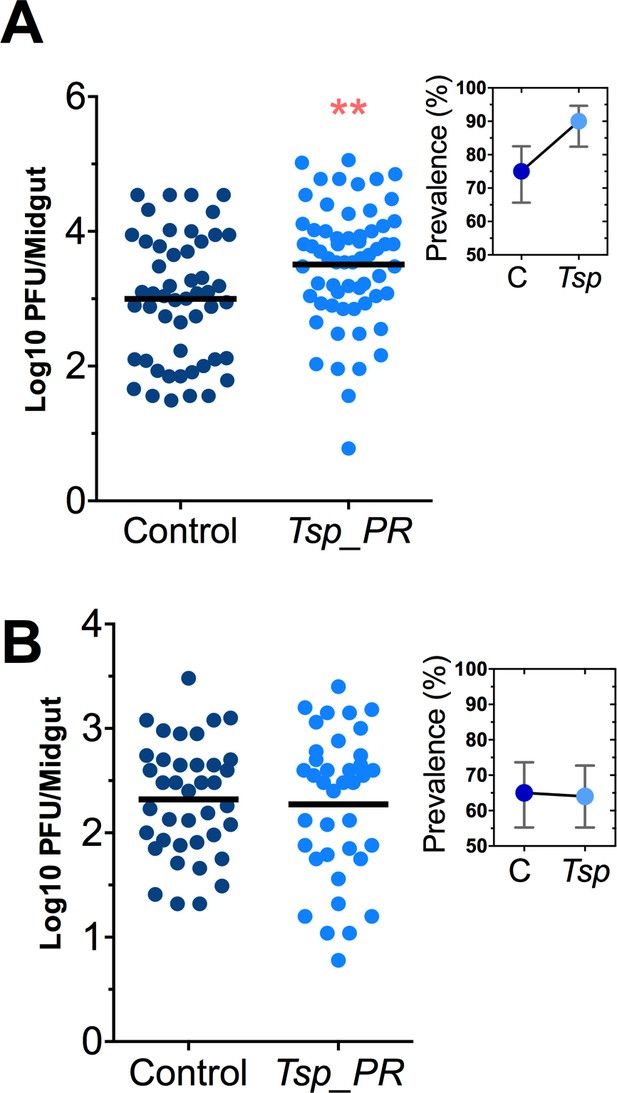
Heat-sensitive Tsp_PR secreted molecule(s) render mosquitoes more susceptible to DENV infection.
DENV titers by plaque assay. Orlando strain mosquitoes were mock-fed or fed for 48 hr with a 10% sucrose solution a Tsp_PR filtered solution, which contained only (A) the fungus-secreted molecules (Control, N = 68; Tsp_PR, N = 68), or (B) a heat-treated Tsp_PR fungus-secreted molecules (Control, N = 60; Tsp_PR, N = 61). Mosquitoes were infected with a blood meal containing DENV, and midguts were dissected at 7 dpi. Each dot represents a log 10 PFU in individual midguts from three independent experiments. The line indicates the mean. Upper right boxes show the prevalence of infected mosquitoes, error bars represent the 95% confidence interval. **p<0.01.
-
Figure 3—source data 1
Raw data and statistics summary for Figure 3.
- https://doi.org/10.7554/eLife.28844.006
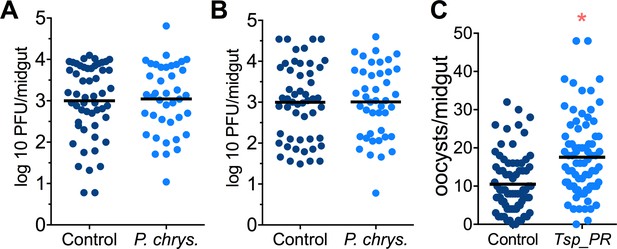
Penicillium chrysogenum does not modulate DENV infection in Aedes mosquito midguts, while Tsp_PR render An. gambiae more susceptible to Plasmodium infection.
Penicillium chrysogenum was isolated from field-caught Anopheles sp. mosquitoes and re-introduced into Aedes mosquitoes to test the modulation of DENV infection. Aedes Orlando strain was mock-fed or fed for 48 hr with a 10% sucrose solution containing (A) 1 × 109 P. chrysogenum spores (Control, N = 61; P. chrysogenum, N = 47) or (B) fungus-secreted molecules (Control, N = 68; P. chrysogenum, N = 53). After fungus feeding, the mosquitoes were infected with a blood meal containing DENV; at 7 days post-infection (dpi), the midguts were dissected. Each dot represents a PFU value in individual midguts from three independent experiments. The line indicates the mean. (C) Influence of Tsp_PR on P. falciparum infection of An. gambiae, as a measured by oocyst numbers 7 days after feeding on a P. falciparum gametocyte culture (infection intensity). The mosquito cohort (N = 79) that had been exposed to a Tsp_PR -laced sucrose solution for 48 hr prior to parasite infection had a significantly higher P. falciparum infection than did the non-fungus-exposed control cohort (N = 76). Graphs show three independent experiments. Each dot represents a single midgut, and bars represent the mean. *p<0.05.
-
Figure 4—source data 1
Raw data and statistics summary for Figure 4.
- https://doi.org/10.7554/eLife.28844.008
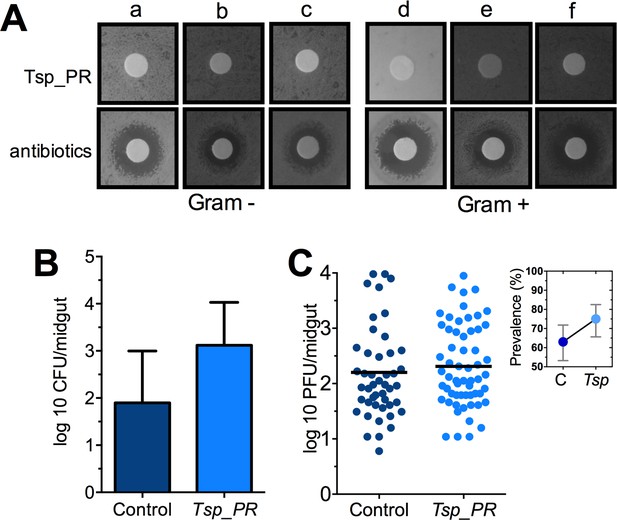
The Tsp_PR secreted molecule(s) do not affect bacterial load or DENV infection in aseptic mosquitoes.
(A) Bacterial growth inhibition assay. Six bacterial isolates of field-caught mosquitoes (Ramirez et al., 2012) were independently plated on LB agar and covered with a disk soaked in a Tsp_PR secretome solution or antibiotic cocktail. Three isolates were Gram-negative bacteria: Serratia marcescens (a), Chromobacterium haemolyticum (b), and Enterobacter hormaeche (c). Three were Gram-positive bacteria: Bacillus subtilis (d), Staphilococcus capprae (e), and Lactococcus lactis (f). Bacterial inhibition was indicated by the presence of a bacterial inhibition zone around the disk. (B) Total bacterial loads. Midguts of secretome solution-exposed and unexposed mosquitoes were collected, homogenized, and plated on LB agar. Bacteria were counted as CFU. Error bars represent ± SD of three independent experiments p=0.202. (C) DENV titers in aseptic mosquitoes. Mosquitoes were treated with an antibiotic cocktail via a sugar meal 4 days before the fungal treatment and were mock-fed or fed for 48 hr on a Tsp_PR secretome solution prior to DENV infection. Each dot represents the PFU after 7 dpi in individual midguts from three independent experiments (Control, N = 75; Tsp_PR, N = 78). The line indicates the mean, p=0.867. Upper right box shows the prevalence of infected mosquitoes, error bars represent the 95% confidence interval.
-
Figure 5—source data 1
Raw data and statistics summary for Figure 5B,C.
- https://doi.org/10.7554/eLife.28844.010
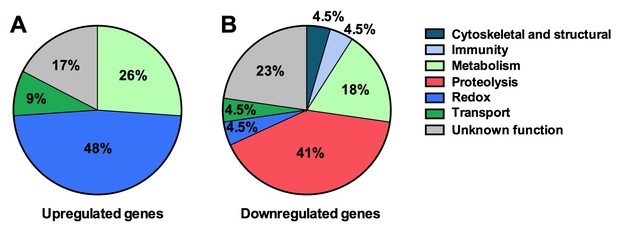
Tsp_PR secreted-molecule(s) –induced gene regulation.
Functional classification in real numbers of the differentially expressed genes in mosquito midguts treated with Tsp_PR secretome for 48 hr, as compared to those of untreated mosquitoes. The fungus treatment-responsive genes are presented in Table 1 and Supplementary file 1.
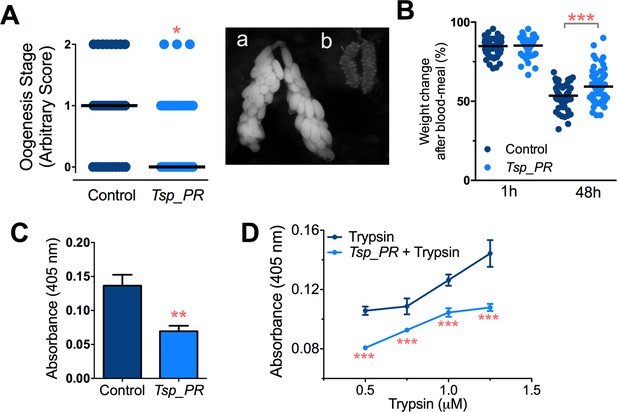
Inhibition of Ae.aegypti midgut trypsin activity by Tsp_PR-secreted molecule(s).
(A) (left) Ovary development based on an arbitrary score of the ovary size at 6 days after a blood meal, 0 for small round follicles, 1 for intermediate size follicles, and 2 for fully developed follicles, with the elongated shape of normal mature eggs. Control, N = 34; Tsp_PR, N = 34, line represents the median, of three independent experiments. (right) Light microscopy picture of (a) a completely developed ovary follicle, which represents a score 2 (b) small round follicles, represent score 0. (B) Change in mosquito body weight after 1 hr (Control, N = 66; Tsp_PR, N = 74), (p=0.784) and 48 hr (Control, N = 58; Tsp_PR, N = 74) (p=0.001) of a non-infected blood meal (C) Trypsin in vivo enzymatic activity in midguts of mosquitoes treated or mock-treated with Tsp_PR secretome. Error bars represent ± SEM of three independent experiments. (D) Trypsin in vitro enzymatic assays of Tsp_PR’s ability to inhibit commercial trypsin activity. The activity was measured at various concentrations of trypsin. Tsp_PR represents the control group in which the fungus filtrate was added but no trypsin, and the absence of trypsin activity was experimentally confirmed (not shown). Error bars represent ± SEM of three independent experiments. *p<0.05, **p<0.01, ***p<0.01.
-
Figure 7—source data 1
Raw data and statistics summary for Figure 7.
- https://doi.org/10.7554/eLife.28844.014
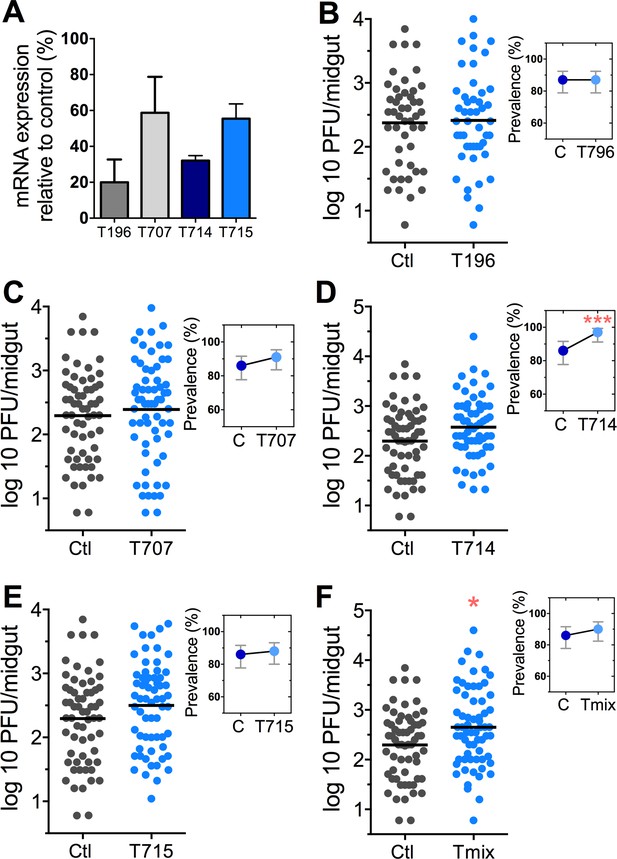
DENV infection after dsRNA-mediated silencing of trypsin genes.
(A) Trypsin genes abundance after dsRNA-mediated gene silencing, (AAEL010196 (T196), AAEL013707 (T707), AAEL013714 (T714), AAEL013715 (T715). (B–F) DENV infection intensity of trypsin genes-silenced mosquitoes are compared to GFP dsRNA-treated control mosquitoes (B) T196 (Control, N = 54; T196, N = 54), (C) T707 (Control, N = 71; T707, N = 69), (D) T714 (Control, N = 71; T714, N = 62), (E) T715 (Control, N = 71; T715, N = 72), (F) Simultaneous silencing of all trypsins (Tmix) (Control, N = 71; Tmix, N = 71). The line indicates the mean, each dot represents the log10 PFU after 7 dpi in individual midguts from four independent biological experiments, except T196 which represents three independent experiments. Upper right boxes show the prevalence of infected mosquitoes, error bars represent the 95% confidence interval. *p<0.05, ***p<0.001.
-
Figure 8—source data 1
Raw data and statistics summary for Figure 8.
- https://doi.org/10.7554/eLife.28844.016

Trypsin phylogeny.
Phylogenetic tree of the nucleotide alignment of trypsins regulated by Tsp_PR and others associated with the Aedes midgut. Branch support values represent approximate likelihood ratios, constructed using the program PhyML 3.0 approximate likelihood-ratio test (Dereeper et al., 2008).
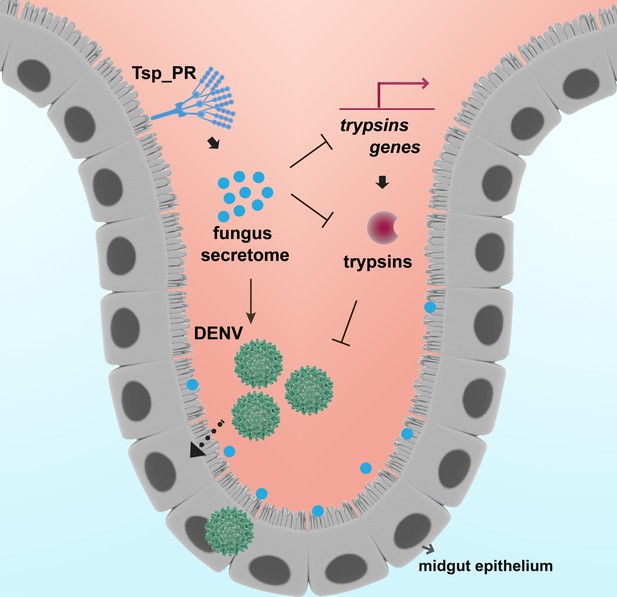
Model of Tsp_PR-mediated increased Ae. aegypti permissiveness to DENV.
Tsp_PR secreted factors render Ae. aegypti more permissive to DENV through a mechanism that involves the down-regulation of gut trypsin transcripts and inhibition of enzymatic activity in the midgut epithelium. Trypsins have an antagonistic role in DENV infection. Decrease of trypsins abundance results in increased susceptible to DENV infection. Additional files.
Tables
Significantly regulated genes in Tsp_PR secretome-exposed mosquitoes.
Log2 values of differential mRNA abundances (Tsp_PR exposed/non-exposed) of genes.
| Gene description | Gene ID | Log2 |
|---|---|---|
| trypsin | AAEL010196 | −2.33 |
| trypsin, putative | AAEL013714 | −2.25 |
| trypsin | AAEL010203 | −2.07 |
| Catalytic activity, serine-type endopeptidase activity, proteolysis | AAEL017520 | −1.99 |
| trypsin | AAEL013715 | −1.95 |
| serine-type enodopeptidase, putative | AAEL001690 | −1.64 |
| saccharopine dehydrogenase | AAEL014734 | −1.35 |
| Sialin, Sodium/sialic acid cotransporter, putative | AAEL004247 | −1.25 |
| hypothetical protein | AAEL013835 | −1.22 |
| alkaline phosphatase | AAEL000931 | −1.19 |
| trypsin | AAEL013707 | −1.19 |
| hypothetical protein | AAEL007591 | −1.08 |
| carboxypeptidase | AAEL010776 | −1.07 |
| triosephosphate isomerase | AAEL002542 | −1.03 |
| leucinech transmembrane proteins | AAEL005762 | −0.90 |
| serine-type enodopeptidase, putative | AAEL001701 | −0.90 |
| Conserved hypothetical protein (chitin-binding domain type 2) | AAEL017334 | −0.89 |
| sterol carrier protein-2, putative | AAEL012697 | −0.82 |
| hypothetical protein | AAEL002875 | −0.82 |
| lysosomal acid lipase, putative | AAEL004933 | −0.81 |
| hypothetical protein | AAEL002963 | −0.78 |
| lysosomal alpha-mannosidase (mannosidase alpha class 2b member) | AAEL005763 | −0.76 |
| ornithine decarboxylase | AAEL000044 | 1.70 |
| glucosyl/glucuronosyl transferases | AAEL003099 | 1.23 |
| conserved hypothetical protein(salivary protein [Culex]) | AAEL009985 | 1.05 |
| cytochrome P450 | AAEL014607 | 1.01 |
| cytochrome P450 | AAEL014609 | 0.99 |
| cytochrome P450 | AAEL006811 | 0.97 |
| cytochrome P450 | AAEL014616 | 0.96 |
| cytochrome P450 | AAEL014608 | 0.95 |
| hypothetical protein | AAEL004317 | 0.94 |
| hypothetical protein | AAEL005669 | 0.92 |
| hypothetical protein | AAEL002263 | 0.90 |
| glucosyl/glucuronosyl transferases | AAEL010386 | 0.88 |
| CRAL/TRIO domain-containing protein | AAEL003347 | 0.87 |
| alpha-amylase | AAEL010537 | 0.85 |
| hypothetical protein | AAEL011203 | 0.83 |
| glucose dehydrogenase | AAEL004027 | 0.82 |
| hypothetical protein | AAEL009198 | 0.81 |
| glutamate decarboxylase | AAEL010951 | 0.80 |
| cytochrome P450 | AAEL008846 | 0.79 |
| Vanin-like protein 1 precursor, putative | AAEL006023 | 0.78 |
| cytochrome b5, putative | AAEL012636 | 0.77 |
| cytochrome P450 | AAEL009131 | 0.76 |
| cytochrome P450 | AAEL014893 | 0.75 |
| Reagent type (species) or resource | Designation | Source or reference | Additional information |
|---|---|---|---|
| strain, strain background (Aedes aegypti Rockefeller strain) | Rock | other | From Johns Hopkins University |
| strain, strain background (Aedes aegypti Orlando strain) | Orl | other | From Johns Hopkins University |
| cell line (Aedes albopictus C6/36) | C6/36 | ATCC CRL-1660 | |
| cell line (Baby hamster kidney cells (BHK-21)) | BHK-21 | ATCC CCL-10 | |
| biological sample (Talaromyces sp.) | Tsp_PR | this paper | Collected from a wild-caught mosquito from Naguabo, Puerto Rico |
| biological sample (Penicillium chrysogenum) | P. chrysogenum | PMID 27678168 | |
| biological sample (Dengue virus 2 strain New Guinea C (NGC) | DENV | PMID 18604274 | |
| biological sample (Plasmodium falciparum) | P. falciparum | PMID 27678168 | From Johns Hopkins Malaria Research Institute |
| Low Input Quick Amp Labeling kit | Agilent Technologies | ||
| RNeasy Mini Kit | QIAGEN | ||
| MMLV Reverse Transcriptase kit | Promega |
Additional files
-
Supplementary file 1
Table shows all the genes that had differential mRNA abundances (Tsp_PR exposed/non-exposed) over or under the significance cutoff value of ±0.75 Log2.
- https://doi.org/10.7554/eLife.28844.019
-
Supplementary file 2
Primer sequences used for dsRNA synthesis and qPCR.
Sequences underlined corresponds to T7 promoter
- https://doi.org/10.7554/eLife.28844.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28844.021





