Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase
Figures
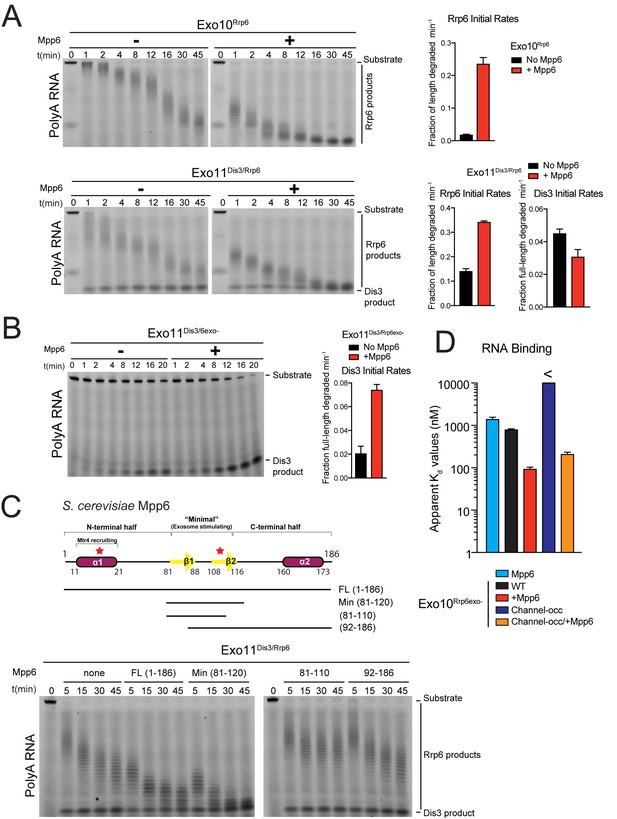
Mpp6 stimulates the nuclear RNA exosome and binds RNA.
(A) Mpp6 stimulates Rrp6 activities when degrading 5’ fluorescein-labeled 49 nt polyA in 10- (top) and 11- subunit exosomes (bottom). Relative positions of RNA substrate, Rrp6 products and the Dis3 product are indicated to the right of gels in panels A, B, and C. Quantitation of initial rates shown to the right of representative gels. For Rrp6 initial rates, the median length of Rrp6 products was determined at the earliest time points and used to calculate initial rates using the equation (1 – (median product length/substrate length)/min) to yield a fraction of length degraded per minute. For Dis3, initial rates were determined by calculating the fraction of full-length substrate degraded based on the accumulation of 4–5 nt product. For the lower gel, Dis3 and Rrp6 initial rates were determined at the earliest time points where distributive products of Rrp6 are easily distinguished/separated from the 4–5 nt processive products of Dis3. (B) Dis3 exoribonuclease activity can be stimulated on 5’ fluorescein-labeled 49 nt polyA in 11-subunit exosomes if Rrp6 is present but catalytically inert. (C) Top: Predicted domain structure of S. cerevisiae Mpp6. Calculated with Jpred (Cole et al., 2008). Previously identified conserved regions (Milligan et al., 2008) are marked with red stars, with Mtr4 recruiting and exosome stimulating domains as described in this work labeled. Below: a minimal fragment of Mpp6 (residues 81 to 120) is necessary and sufficient to stimulate Rrp6 activity in Exo11Dis3/Rrp6. Representative decay assays of Exo11Dis3/Rrp6 on 5’ fluorescein-labeled polyA 49 nt RNA with different Mpp6 constructs added in 2-fold molar excess. (D) Mpp6 enhances RNA binding of Rrp6-containing exosomes on polyA RNA, and alleviates binding defects caused by Exo9 channel occlusions. Fluorescence polarization of Mpp6 and Exo10Rrp6exo- with or without PH-like ring occlusions in the lower half of the Exo9 channel, with binding to 5’ fluorescein-labeled polyA 37 nt RNA. Bar graphs and error bars in panels A, B, and D are the result of triplicate experiments with error bars indicating plus or minus one standard deviation.
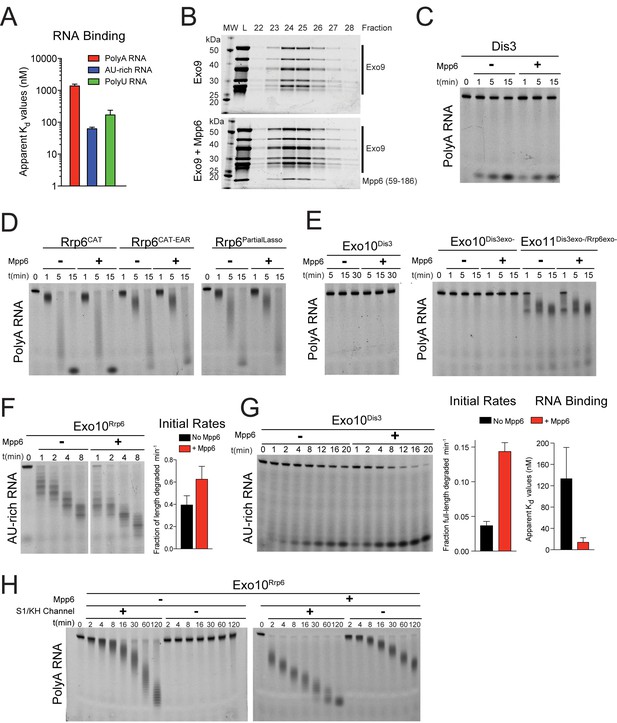
Mpp6 is a RNA-binding protein cofactor of the nuclear RNA exosome that stimulates exosome exoribonuclease activities.
(A) Mpp6 binds various sequences of single-stranded RNA. Fluorescence polarization of Mpp6 bound to 5’ fluorescein-labeled polyA 37 nt, AU-rich 36 nt, and polyU 36 nt RNA. Results of triplicate experiments shown with error bars representing plus and minus one standard deviation. (B) Mpp6 associates with Exo9 by analytical gel-filtration (Superose 6). Fractions were analyzed by SDS-PAGE and stained with Sypro Ruby. A N-terminal truncation corresponding to residues 59–186 was used to better separate Mpp6 from Exo9 subunits with similar molecular weight as full-length Mpp6. (C) Mpp6 does not stimulate free Dis3 on 5’ fluorescein-labeled 49 nt polyA. Reaction products were analyzed by denaturing PAGE. (D) Mpp6 does not stimulate free Rrp6 on 5’ fluorescein-labeled 49 nt polyA. Reaction products were analyzed by denaturing PAGE. (E) Mpp6 does not stimulate either the exoribonuclease or endoribonucease activities of Dis3 in Exo10Dis3 on 5’ fluorescein-labeled 49 nt polyA. Reaction products were analyzed by denaturing PAGE. (F) Mpp6 stimulation of Rrp6 exoribonuclease activity on 5’ fluorescein 36 nt AU-rich RNA in Exo10Rrp6. Initial rates from triplicate experiments, with error bars representing plus and minus one standard deviation. (G) Exo10Dis3 exoribonuclease activity can be stimulated by Mpp6 on 5’ fluorescein-labeled AU-rich 49 nt RNA. Bar graphs represent (left) initial rates and (right) apparent Kd values of Exo10Dis3exo- determined by fluorescence polarization on 5’ fluorescein 36 nt AU-rich RNA. (H) Mpp6 exosomes utilize the S1/KH ring to degrade RNA. Mpp6 can partially, but not completely alleviate the inhibitory effects of point mutations within the S1/KH ring on Rrp6 activity in Exo10Rrp6. Assays performed on 5’ fluorescein-labeled polyA 49 nt RNA, and intermediates analyzed by denaturing PAGE.
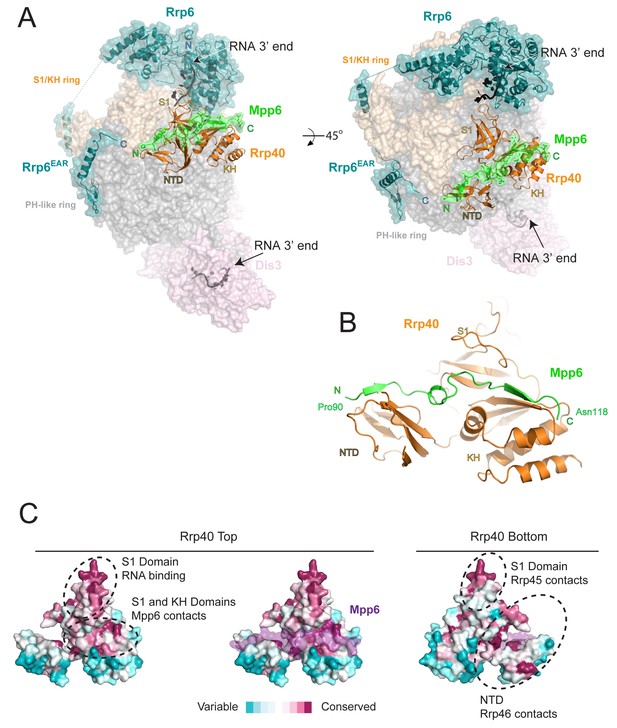
Structure of the 12-subunit Mpp6 nuclear exosome.
(A) Global view of Exo12Dis3exo-endo-/Rrp6exo-/Mpp6Min bound to a 3’−3’ RNA. Mpp6Minimal interacts with an extended surface across the S1/KH subunit, Rrp40. View from side (left) and top (right). Exosome subunits shown as surface view, Mpp6 in green, Rrp6 in teal, Rrp40 in cartoon (orange), RNA in black sticks. (B) Mpp6Minimal (green) makes extensive contacts to Rrp40 (orange) and spans all three of its domains. (C) Mpp6Minimal (transparent purple surface in middle and right panels) binds to a conserved surface of Rrp40. Other conserved surfaces important for RNA binding and scaffolding interactions to other exosome subunits are indicated. Surface conservation calculated with ConSurf (Ashkenazy et al., 2010).
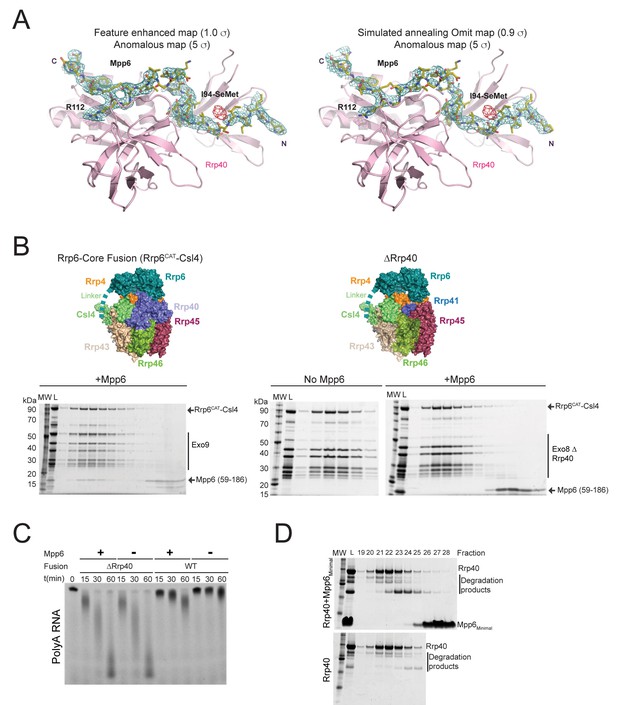
Rrp40 is necessary, but not sufficient for Mpp6 association with the exosome.
(A) Electron densities for Mpp6. On the left, feature enhanced map (blue mesh) contoured at 1 σ covering the Mpp6 peptide. The anomalous peak (red mesh) for selenomethionine substituted Mpp6 (I94M) contoured at 5 σ. Same view on the right but depicting electron densities from a simulated annealing omit map (blue mesh) contoured at 0.9 σ. Proteins and select amino acids labeled, Rrp40 (pink) shown in cartoon depiction, and Mpp6 amino acids shown in stick representation. (B,C) Mpp6 requires Rrp40 for exosome association and stimulation. Exosomes lacking Rrp40 could be formed using Rrp6-CAT fused to Csl4 (Wasmuth and Lima, 2017). While Mpp6 can associate with the fusion complex when Rrp40 was present, Mpp6 failed to associate with the exosome by analytical gel filtration (B) and while Rrp6 activity is higher in the absence of Rrp40, addition of Mpp6 did not stimulate Rrp6 activity (C) when Rrp40 was absent. (D) Mpp6 cannot associate with Rrp40 by analytical gel filtration (Superose 12). Fractions were analyzed by SDS-PAGE and stained by Sypro Ruby.
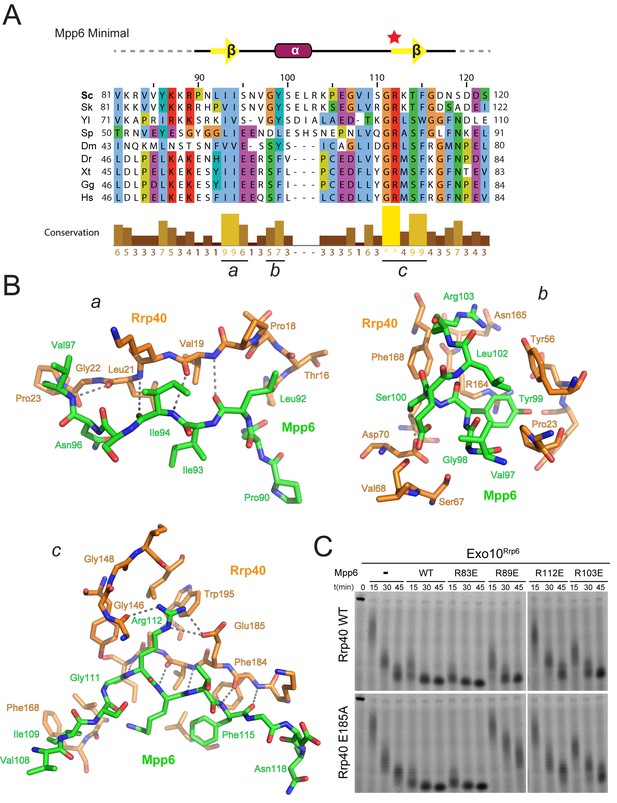
Conserved features of Mpp6 interaction with the S1/KH subunit, Rrp40.
(A) Top: revised domain structure of Mpp6Minimal based on the crystal structure of Exo12Dis3exo-endo-/Rrp6exo-/Mpp6Min. Bottom: sequence alignment among S. cerevisiae Mpp6Minimal and other eukaryotes reveals conserved residues within Mpp6 Minimal. Sequences for S. kudriavzevii (Sk), Y. lipolytica (Yl), S. pombe (Sp), D. melanogaster (Dm), D. rerio (Dr), X. tropicalis (Xt), G. gallus (Gg), and H. sapiens (Hs). Sequence alignment and conservation calculated with Clustal Omega (Larkin et al., 2007) and Jalview (Waterhouse et al., 2009). Three regions (a, b, c) of high conservation are noted. (B) Stick representation of conserved contacts between Mpp6 (green) and Rrp40 (orange) are noted. Region a includes a hydrophobic patch that forms between the Rrp40 NTD and N-terminal residues of Mpp6Minimal. Region b highlights an aromatic residue in Mpp6 that nestles between the NTD and KH domains of Rrp40. Region c focuses on the network of contacts between the ‘arginine anchor’ of Mpp6 and the S1 and KH domains of Rrp40. (C) Mpp6 arginine residues were individually substituted to glutamate residues to interrogate the importance of the arginine anchor. Representative RNA degradation assays of 5’ fluorescein-labeled 49 nt polyA RNA and Exo10Rrp6 with Mpp6Minimal WT and arginine mutants. Mutations disrupting the arginine anchor, Arg112, are most detrimental to Mpp6-mediated stimulation of Rrp6 activity. An additive effect is observed when a distal Mpp6 mutation, Arg89, is combined with a mutation in Rrp40 (E185A) that disrupts coordination of the Mpp6 arginine anchor.
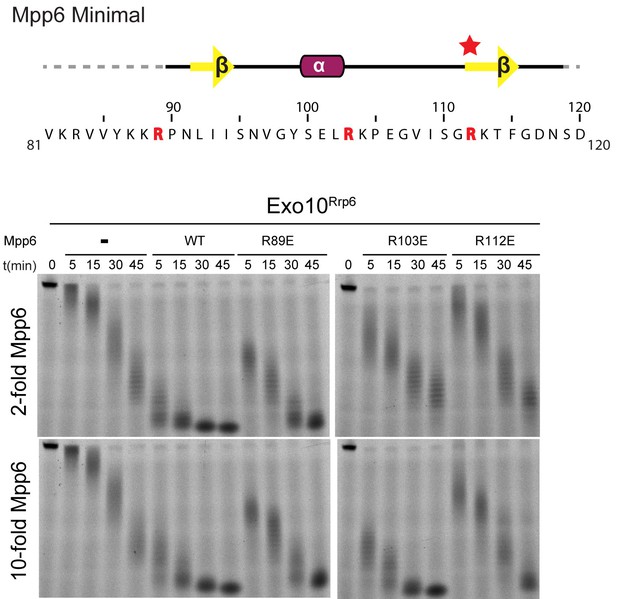
Conserved residues in Mpp6 interact with Rrp40.
Top: Sequence of Mpp6Minimal as shown in Main Figure 3A. Bottom: Representative RNA degradation assays of 5’ fluorescein-labeled 49 nt polyA RNA and Exo10Rrp6 with Mpp6Minimal WT and arginine mutants added in 2-fold and 10-fold molar excess. Defects in Rrp6-dependent RNA decay can be overcome by addition of excess Mpp6 R103E, but not the other Mpp6 mutations.
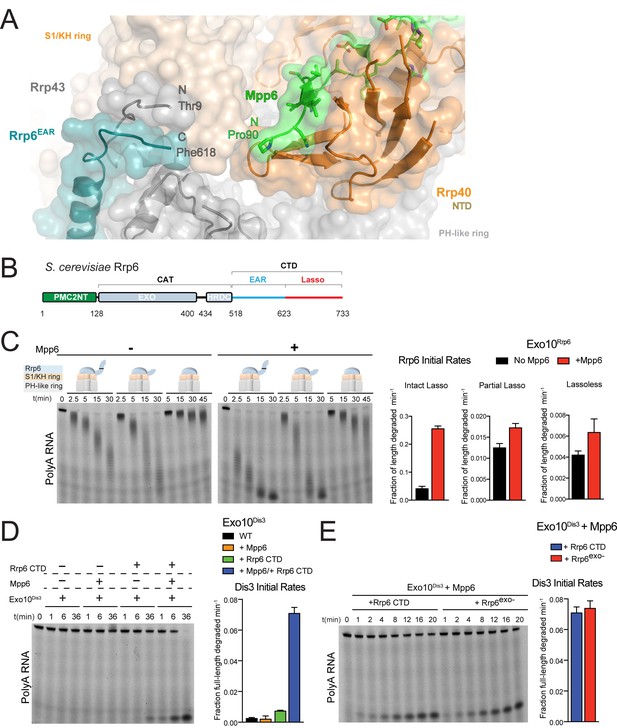
Mpp6 cooperates with the Rrp6 lasso to stimulate exosome activities.
(A) Close-up view to show the C-terminal residue (Phe618) of the Rrp6 EAR (teal), the Rrp43 loop (residues 321–326 below Rrp6) and N-terminal residue (Thr9) (gray), and the N-terminal residue (Pro90) of Mpp6Minimal (green). Rrp6, Rrp43, Mpp6 and Rrp40 shown in cartoon and stick representation under a transparent surface. (B) Schematic domain structure of Rrp6. (C) Exo10Rrp6 lacking part (128-685) or all of the Rrp6 lasso (128-634) are less stimulated by Mpp6 than Exo10Rrp6 containing an intact lasso (128-733). Decay of 5’-fluorescein-labeled polyA 49 nt RNA. (D) Mpp6-dependent stimulation of Dis3 exoribonuclease activity in Exo10Dis3 requires the Rrp6 CTD. Decay of 5’-fluorescein-labeled polyA 49 nt RNA. (E) The Rrp6 CTD stimulates Dis3 exoribonuclease activity in Exo10Dis3 in a Mpp6-dependent manner similar to levels observed in the presence of Rrp6exo-. Decay of 5’-fluorescein-labeled polyA 49 nt RNA. Bar graphs and error bars in panels C, D, and E are the result of triplicate experiments with error bars indicating plus or minus one standard deviation.
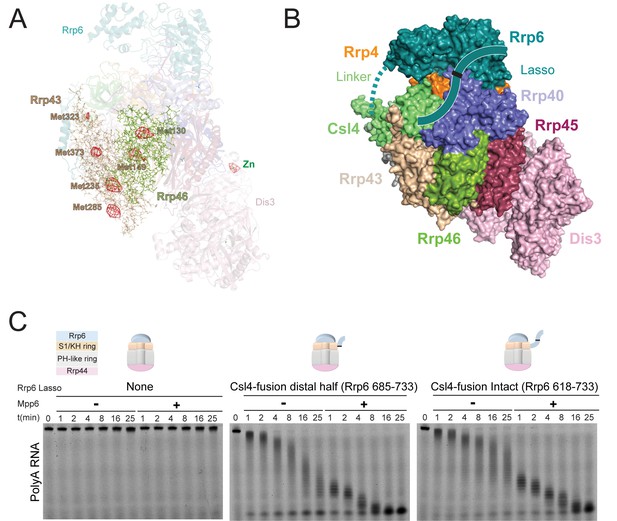
Stimulation by Mpp6 depends on the Rrp6 lasso.
(A) Anomalous Fourier difference map derived from crystals of Exo12Dis3exo-endo-/Rrp6exo-/Mpp6Min reconstituted with selenomethionine containing Rrp46/Rrp43. Anomalous peaks corresponding to SeMet positions in Rrp46 and Rrp43 are shown, including Rrp43 Met323. Electron density (red mesh) contoured at 4 σ. (B) Model of Exo10Dis3/Rrp6CAT-Csl4 similar to Figure 2—figure supplement 1, but containing the Rrp6 lasso on the C-terminus of Csl4. (C) Mpp6 stimulates Exo10Dis3/Rrp6CAT-Csl4-Rrp6Lasso complexes that include the intact and distal half of the Rrp6 lasso fused to Csl4, indicating that Rrp6 residues 519–684 are dispensible for Mpp6-mediated stimulation of Rrp6 activity. Representative decay of 5’ fluorescein-labeled polyA 49 nt RNA.
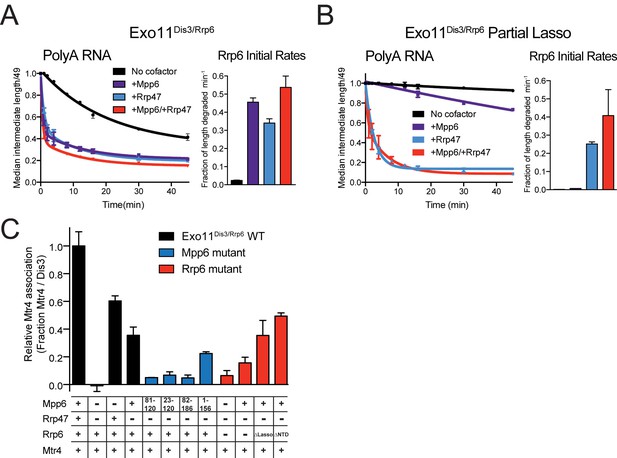
Mpp6 and Rrp47 stimulate Rrp6 activity and contribute to Mtr4 recruitment.
(A) Both Mpp6 and Rrp47 stimulate Rrp6 activity in degradation of a single-stranded 5’ fluorescein-labeled 49 nt polyA RNA in Exo11Dis3/Rrp6. Initial rates from triplicate experiments shown, with error bars representing plus or minus one standard deviation. (B) Mpp6, but not Rrp47, requires the Rrp6 lasso to stimulate Rrp6 activity on 5’ fluorescein-labeled 49 nt polyA RNA. Initial rates from triplicate experiments are shown, with error bars representing plus or minus one standard deviation. (C) Mpp6 contributes to Mtr4 recruitment in Exo11Dis3/Rrp6. Analytical gel filtration experiments were performed with Exo10Dis3 and Exo11Dis3/Rrp6, Rrp47, Mtr4, with various truncations of Mpp6 and Rrp6 as indicated. Bar graphs represent the ratio of Mtr4 (122 kDa) to Dis3 (114 kDa) in peak fractions of the complex (Figure 5—figure supplement 1C), as calculated by densitometric analysis of the fractions on SDS-PAGE, with error bars representing plus or minus one standard deviation.
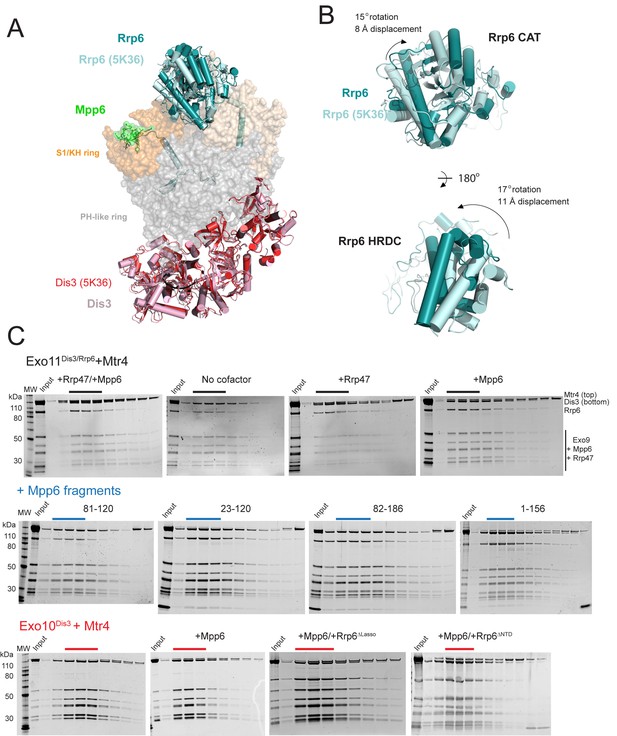
Superposition of Exo12Dis3exo-endo-/Rrp6exo-/Mpp6Minonto Exo11Dis3exo-endo-/Rrp6exo- (PDB 5K36) and Mtr4 interactions with exosomes as analyzed by gel filtration and SDS-PAGE.
(A) Overall structures with the S1/KH ring in orange (Rrp40) and wheat (Csl4 and Rrp4), the PH-like ring in grey, Dis3 in pink or red, and Rrp6 in teal or cyan. Select subunits are labeled. (B) Close up of the Rrp6 CAT and HRDC domains to illustrate rotations and displacements relative to the two structures. (C) Representative SDS-PAGE of analytical gel filtration runs (Superdex 200 Increase) of the indicated Mtr4-exosome combinations presented in Figure 5C, with each lane representing an independent fraction. Gels were Sypro Ruby stained and the ratio of Mtr4 to Dis3 was calculated via densitometry within lanes indicated by solid lines above each gel.
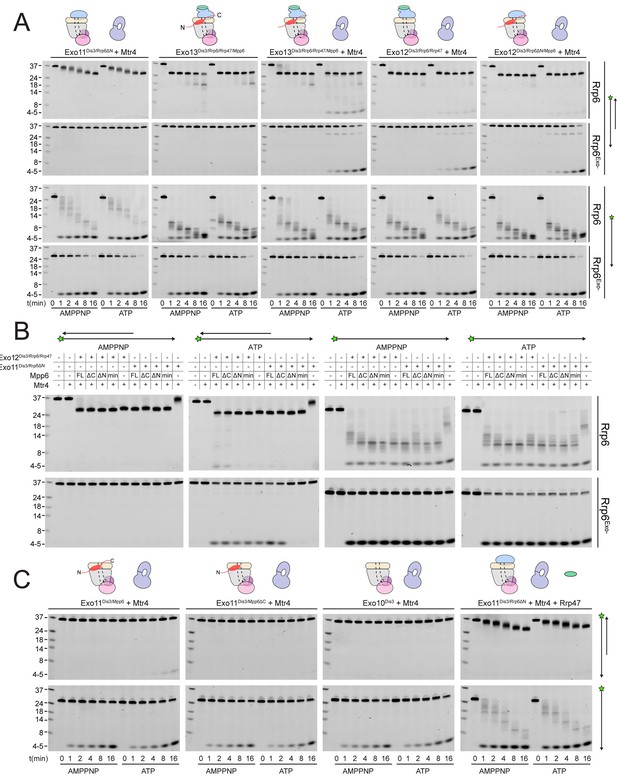
Mtr4-dependent RNA degradation requires either Mpp6 or Rrp47.
Urea-PAGE analysis of RNA decay products by indicated exosome complexes and cofactors. In reactions labeled ‘Rrp6Exo-’, Rrp6 contains a D238N mutation to render its exonuclease site catalytically inactive. 5’ fluorescein-labeled polyA RNA of indicated lengths are present in the leftmost lane of each gel with the Dis3 product labeled ‘4–5’. Samples were not heated prior to gel electrophoresis, so the dsRNA substrate runs as a duplex at approximately 37 nt (A) Activity of reconstituted RNA exosome complexes in the presence or absence of equimolar exogenous Mtr4. (B) Mix-in of 1.5-fold molar excess of full-length Mpp6 or indicated truncations in RNA decay reactions containing reconstituted Exo12Dis3/Rrp6/Rrp47 or Exo11Dis3/Rrp6ΔN and equimolar Mtr4 (when present). Results shown for an end point assay after 1 min incubation with exosomes containing Rrp6 or 8 min for exosomes containing Rrp6exo-. (C) RNA degradation activities of reconstituted RNA exosome complexes and indicated mutations in the presence of equimolar exogenous Mtr4 and/or Rrp47. Cartoons shown above gels in panels A and C depict the exosome core in grey and wheat, Rrp6 in blue, Dis3 in pink, Mpp6 in red and Rrp47 in green. The central body of Mpp6 is shown as an ellipse with N- and C-terminal tails labeled or removed to reflect the protein used in the assay. The N-terminal PMC2NT domain of Rrp6 is shown as an appendage to the Rrp6 protein.
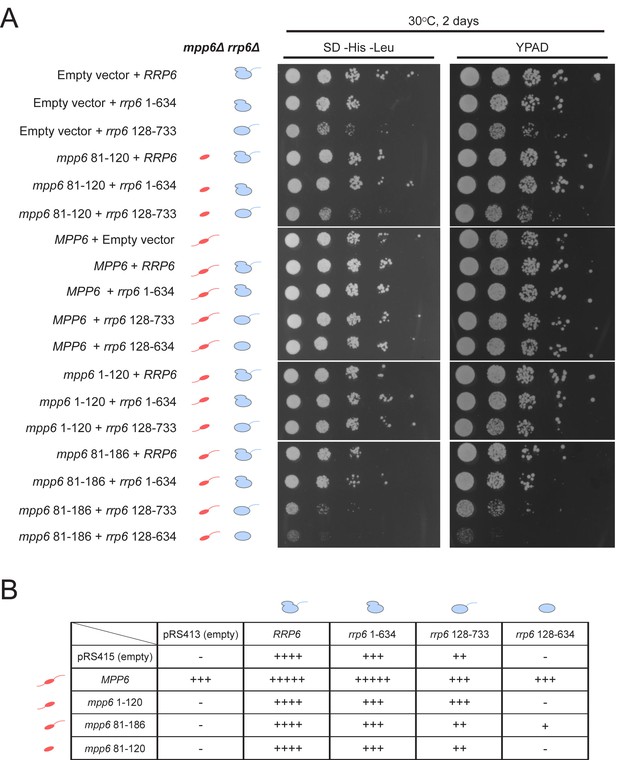
Optimal cell growth depends on unique domains in Rrp6 and Mpp6.
(A) Growth analysis of Saccharomyces cerevisiae strains carrying viable combinations of Rrp6 and Mpp6 alleles. Ten-fold dilutions series of the rrp6Δ mpp6Δ strains transformed with the indicated pRS415 mpp6 and pRS413 rrp6 plasmids. Spotting was performed on SD-His-Leu or YPAD solid media and cells were incubated at 30°C for 2 days. Cartoons depict mpp6 and rrp6 alleles with respect to N- and C-terminal deletions. (B) Scoring table of the yeast growth phenotypes established in panel A and Figure 7—figure supplement 1B after 1 day growth. Scoring is based on a subjective five-point system where five ‘+' symbols (‘+++++') correspond to the fastest observed growth rate and one ‘+' symbol to the slowest growth rate. Plasmid combinations indicated with the '- 'symbol resulted in synthetic lethality (see Figure 7—figure supplement 1A).
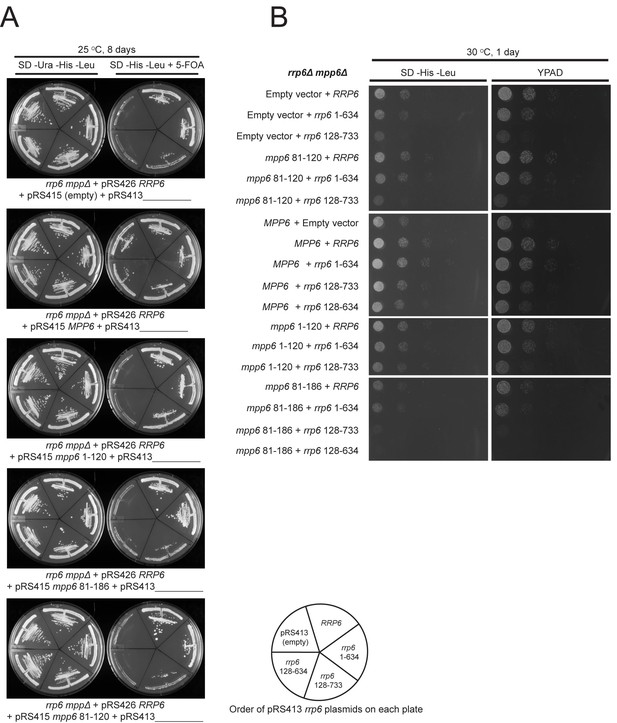
Selection of viable yeast strains carrying different combinations of pRS415 mpp6 and pRS413 rrp6 plasmids.
(A) After transforming the rrp6 mpp6Δ + pRS426 RRP6 strain with the indicated pRS415 mpp6 and pRS413 rrp6 plasmids, single clonal yeast cells were isolated and streaked either on SD-Ura-His-Leu solid medium (streaking control) or SD-His-Leu+5 FOA solid medium, where viable cells lacking the 2-micron cover plasmid (pRS426 RRP6) are selected. Cells on each plate contain a single pRS415 mpp6 plasmid (shown below each panel) and each divided section within a plate contains a different pRS413 rrp6 plasmid (order of pRS413 rrp6 plasmids is indicated at the bottom of the figure). Yeast plates shown were incubated at 25°C for 8 days. (B) Ten-fold dilutions series of the rrp6 mpp6Δ strains transformed with the indicated pRS415 mpp6 and pRS413 rrp6 plasmids. Spotting was performed on SD-His-Leu or YPAD solid media and cells were incubated at 30°C for 1 day.
Tables
Crystallographic data and refinement statistics.
One crystal was used. Highest resolution shell is shown in parenthesis.
| Exo12Dis3exo-endo-/Rrp6exo-/RNA/Mpp6Min | |
|---|---|
| Data collection | |
| X-ray Source | APS GM/CA 23IDD |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 141.1, 213.6, 225.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Wavelength (Å) | 1.0332 |
| Resolution (Å) | 44.3–3.3 (3.42–3.3) * |
| Rmerge | 0.086 (0.613) |
| I/σI | 9.1 (1.7) |
| CC1/2 | 0.997 (0.161) |
| Completeness (%) | 97.0 (95.0) |
| Redundancy | 3.4 (2.6) |
| Wilson B factor (Å2) | 99.7 |
| Refinement | |
| Resolution (Å) | 44.3–3.3 |
| No. reflections observed | 341339 |
| No. unique reflections | 100440 |
| Rwork/Rfree | 0.217/0.266 |
| No. atoms | 29498 |
| Protein | 29147 |
| RNA | 249 |
| Ligands | 38 |
| Water | 64 |
| Average B-factors | |
| Protein | 138 |
| RNA | 139 |
| Ligands | 141 |
| Water | 61 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.001 |
| Bond angles (°) | 0.41 |
| Ramachandran plot | |
| % favored | 93.5 |
| % allowed | 6.5 |
| % outliers | 0 |
| Molprobity | |
| Clashscore/Percentile | 5.44/100th |
| MolProbity Score/Percentile | 1.72/100th |





