Genomic regions controlling shape variation in the first upper molar of the house mouse
Figures
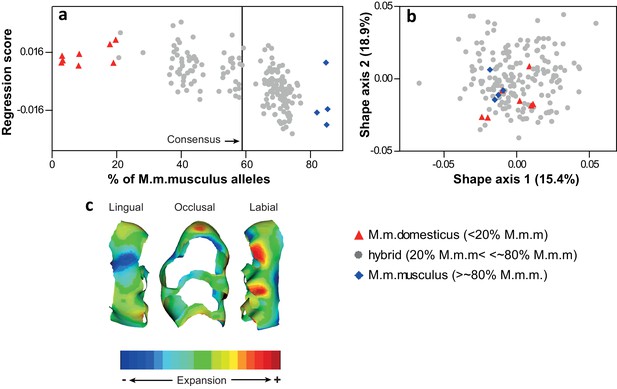
Molar shape variation in the sample.
(a) Multivariate regression of molar shape on the degree of hybridization (M.m.musculus ancestry per individual was obtained from Turner et al. (2012)). (b) Shape variation in the sample depicted on the first two principal axes of a PCA. (c) Transition in molar shape from M.m.domesticus to M.m.musculus. All shape data were obtained from the wear-free template.
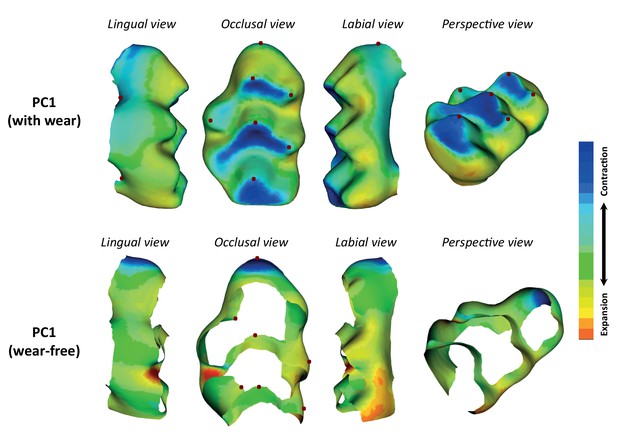
Effect of age and wear in molar shape variation.
The molar shape of all hybrid mice was measured using the complete template, and the wear-free template. The shape reconstruction of the first principal component (PC1) derived from the complete template, and the wear-free template are shown. Abrasion of the cusps is evident in the complete template indicating wear effects. The landmarks used to anchor the template to the tooth surface are shown as red dots.
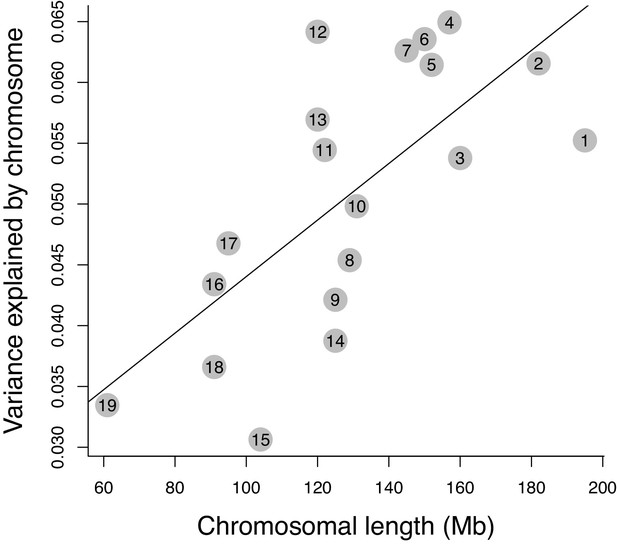
Relative effect of chromosomes on molar shape variance.
The correlation between length and effect size of the 19 autosomes is shown (p=0.001, r = 0.67).
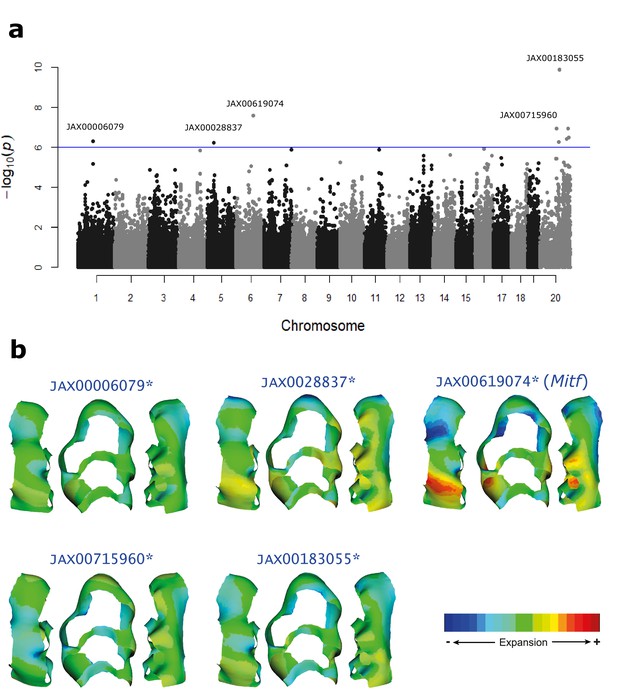
Genomic loci associated with molar shape variation.
(a) Manhattan plot showing SNPs associated with molar shape variation. The blue line indicates the genome-wide significance threshold (1 × 10−6). However, to determine significance, a threshold was derived by permutations for each PC, and independently for autosomes and X chromosome (see Materials and methods). (b) Molar shape variation associated with the most significant SNP within each locus, estimated as the shape difference between the two homozygous SNP states. Warm colors indicate expansion and cold colors indicate compression of tissue relative to the mean shape. The SNP associated with the gene Mitf shows stronger localized effects. The raw phenotypic data used for the association mapping can be found as Source Data 1.
-
Figure 4—source data 1
Shape data used in the association mapping.
The dimensionality of the molar shape data was reduced using a PCA. The centroid size and PC scores for the first 18 PCs for each mouse are shown. These PC scores were used as phenotypes in the association mapping implemented in GEMMAX.
- https://doi.org/10.7554/eLife.29510.008
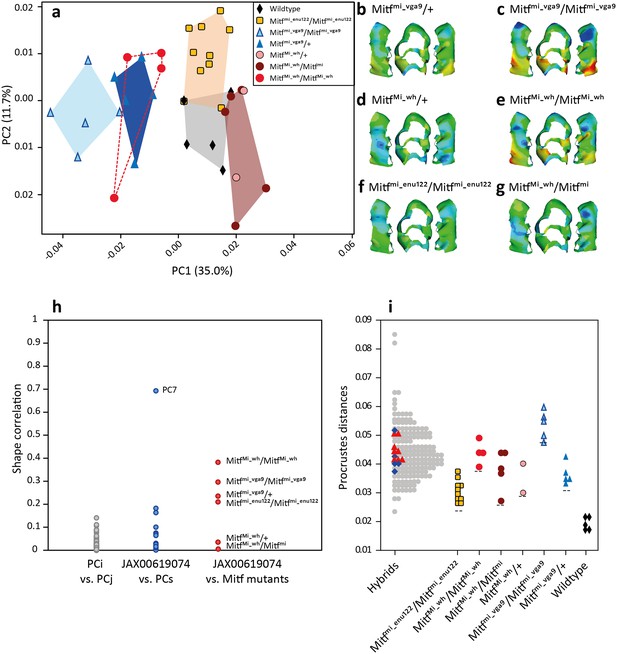
The effects of Mitf mutant alleles on the shape of the upper first molar.
(a) Differentiation of the mutants from the wild-type (B6) in a morphospace (first two axes of a PCA on molar shape descriptors). (b–g) Mean phenotypic effect of each mutation relative to the mean shape of wild-type (C57Bl/J6) mice. The same color scale was used for all shape reconstructions. Warm colors indicate expansion and cold colors compression. (h) Correlation between various effects on tooth shape. Grey dots: pairwise correlations between PCs used for the mapping. They provide a proxy for the expected correlation between orthogonal directions of change. Blue dots: comparison between PCs used for the mapping and the effect of the SNP associated with Mitf -JAX00619074 SNP (most significant SNP in Mo.6 region). Only PC7, associated with Mo.6 in the mapping, resembles the shape effect of the Mitf-associated SNP. Red dots: comparison of the effect of Mitf alleles with the JAX00619074 SNP. Most Mitf mutants display an effect on tooth shape correlated with the effect of Mo.6. (i) Tooth shape variation in the hybrid mice used in the GWAS, and shape variation in the Mitf mutants. Each point corresponds to the Procrustes distance between a mouse tooth and a consensus shape. For hybrid mice the consensus shape is the mean tooth shape of all hybrids; for mutant mice it corresponds to the mean shape of wild-type mice. Within the hybrid group, blue diamonds and red triangles represent individuals with more than 80% alleles from M.m.musculus or M.m.domesticus, respectively. The Mitf mutations studied here generate shape changes in the range of magnitude (Procrustes distance) of natural variation observed within wild hybrids.
Tables
Mitf alleles used in this study.
The effect on gene expression as well as the organismal phenotype associated with each allele is shown. All mutants are on C57Bl/6J background.
| Phenotype | |||||||
|---|---|---|---|---|---|---|---|
| Allele | Symbol | Mode of induction | Lesion | Effect | Heterozygote | Homozygote | |
| micropthalmia | Mitfmi | X-irradiation | 3 bp deletion in basic domain | Affects Mitf DNA binding affinity | Iris pigment less than in wild type; spots on belly, head and tail | White coat, eyes small and red; deficiency of mast cells, basophils, and natural killer cells; spinal ganglia, adrenal medulla, and dermis smaller than normal; incisors fail to erupt, osteopetrosis; inner ear defects | |
| White | MitfMi-wh | Spontaneous or X-irradiation | I212N | Affects Mitf DNA binding affinity | Coat color lighter than dilute (d/d); eyes dark ruby; spots on feet, tail and belly; inner ear defects | White coat; eyes small and slightly pigmented; spinal ganglia, adrenal medulla, and dermis smaller than normal; inner ear defects; reduced fertility | |
| VGA-9 | Mitfmi-vga9 | Transgene insertion | Transgene insertion and 882 bp deletion | Loss-of-function | Normal | White coat, eyes red and small; inner ear defects | |
| enu-22(398) | Mitfmi-enu22(398) | Ethylnitroso-urea | C205T, Q26STOP in exon 2A, | Affects splicing | Normal | Normal eyes, white belly and large unpigmented spots in coat | |
-
This table was modified from Steingrimsson et al. (2004). Information for the allele enu-22(398) comes from Bauer et al. (2009).
SNP heritability estimates per principal component axis.
The standard error of the estimate derived from LMM in GEMMA is shown. The heritability of molar shape is a weighted sum of the heritability per PC, the weights being the percentage of total variation represented by each PC.
| PC | %var | Heritability per PC | Error | Molar herit |
|---|---|---|---|---|
| 1 | 18.9 | 0.83 | 0.09 | 15.8 |
| 2 | 15.4 | 0.95 | 0.08 | 14.7 |
| 3 | 9.9 | 0.78 | 0.10 | 7.7 |
| 4 | 7.1 | 0.62 | 0.14 | 4.4 |
| 5 | 5.9 | 0.49 | 0.12 | 2.9 |
| 6 | 5.1 | 0.83 | 0.10 | 4.2 |
| 7 | 3.9 | 0.53 | 0.12 | 2.1 |
| 8 | 3.2 | 0.89 | 0.11 | 2.8 |
| 9 | 2.8 | 0.89 | 0.09 | 2.5 |
| 10 | 2.5 | 0.86 | 0.13 | 2.2 |
| 11 | 2.1 | 0.72 | 0.14 | 1.5 |
| 12 | 1.8 | 0.60 | 0.15 | 1.1 |
| 13 | 1.7 | 0.68 | 0.13 | 1.2 |
| 14 | 1.5 | 0.56 | 0.16 | 0.8 |
| 15 | 1.4 | 0.53 | 0.15 | 0.7 |
| 16 | 1.3 | 0.26 | 0.16 | 0.3 |
| 17 | 1.2 | 0.14 | 0.20 | 0.2 |
| 18 | 1 | 0.51 | 0.14 | 0.5 |
| Total Var | 86.7% | |||
| pve for molar shape | 65.50 | |||
Association mapping of molar shape variation.
The name of each significant region is defined by Mo (molar) and chromosomal location. The SNP with lowest p-value, its position in the genome, and p-value are shown. Effect size is calculated as the percentage of molar shape variation explained by the SNP. *All protein coding genes in the significant regions are shown, except for region Mo.X.2 where only genes relevant to the discussion are included; in total it contains 306 protein-coding genes. **Only this region was associated with more than one SNP. The five significantly associated SNPs spam a 55 Mb region (ChrX:104533418–15959832).
| QTL | Chr | Position | Best SNP | p-value | Effect size | PC axis | Genes* |
|---|---|---|---|---|---|---|---|
| Mo.1 | chr1 | 84306638 | JAX00006079 | 5.11E-07 | 1.1% | PC11 | Pid1, Dner |
| Mo.5 | chr5 | 36723779 | JAX00128837 | 5.79E-07 | 3.2% | PC18 | Psapl1, Tada2b, Ccdc96, Grpel1, Tbc1d14, D5Ertd579e, Sorcs2 |
| Mo.6 | chr6 | 97980057 | JAX00619074 | 2.71E-08 | 2.8% | PC7 | Gm765, Mitf |
| Mo.X.1 | chrX | 92638616 | JAX00715960 | 1.18E-07 | 1.6% | PC16 | Fam123b, Zc4h2, Asb12, Arhgef9 |
| Mo.X.2** | chrX | 104533418 | JAX00183055 | 1.28E-10 | 2.2% | PC7 | Rps6ka3, Dach2, Ap1s2, Itm2a and 301 other genes |
Additional files
-
Supplementary file 1
(A) Shape comparison between Mitf mutants and wild-type B6 mice.
A Hotelling T2 test was performed to evaluate the difference in mean shape between mutant and wildtype groups; p-value, test statistic, and sample size (N) are shown. The Procrustes distances between mutant and wild type mean shapes are also indicated. *The comparison between heterozygous and homozygous mice for the Mitfmi-vga9 mutation is also shown. (B) Missense variants found in the gene Mitf of wild mice. Nine populations of wild mice were screened for SNPs causing coding changes in Mitf: Mus musculus musculus from Kazakhstan, Check Republic, and Afganistan; M. m. domesticus from Iran, Heligoland, France, and Germany; Mus castaneus; and Mus spretus. Data available in the UCSC browser → MyData - > Public Sessions - > wildmouse (Harr et al., 2016). In addition, eight hybrids between M.m. musculus and M.m. domesticus from the German hybrid zone (Turner, Tautz and Harr unpublished data), the same population used in this study, were also screened for coding changes. Reference and variant alleles are shown. nVar = number of chromosomes with the variant allele, n = number of chromosomes per population.
- https://doi.org/10.7554/eLife.29510.011
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29510.012





