Unique molecular events during reprogramming of human somatic cells to induced pluripotent stem cells (iPSCs) at naïve state
Figures
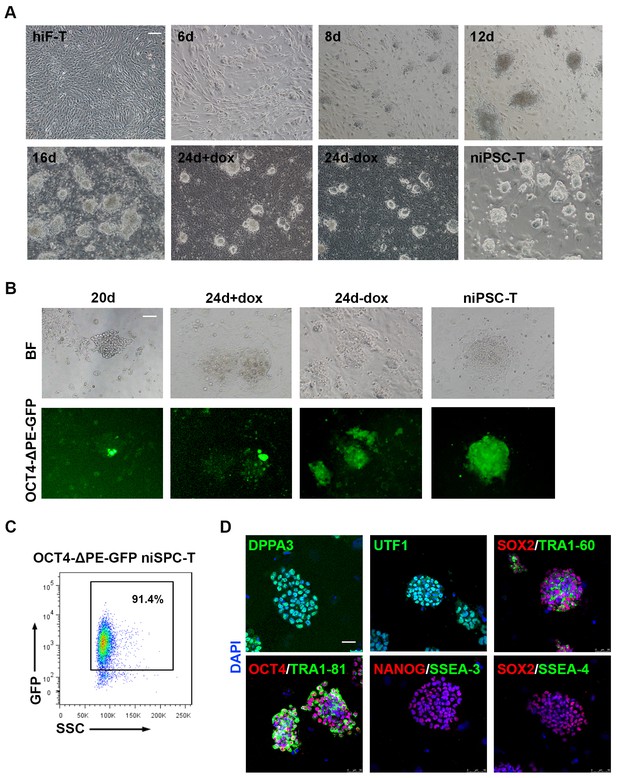
Establishment of the secondary naïve iPSC induction system.
(A) Representative bright field images of hiF-Ts, niPSC-Ts and reprogramming cells at the indicated time points during reprogramming. Scale bar, 100 μm. (B) Phase and OCT4-ΔPE-GFP images of niPSC-Ts and reprogramming cells at the indicated time points during reprogramming. Scale bar, 100 μm. (C) Flow cytometry analysis of the proportion of GFP+ cells in OCT4-ΔPE-GFP niPSC-Ts. (D) Immunostaining images of pluripotency-related marker expression in niPSC-Ts. Scale bar, 50 μm.
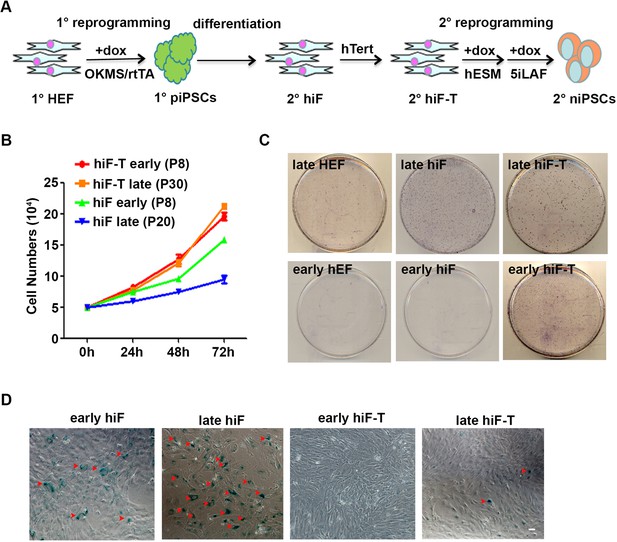
Optimization of secondary human naïve iPSCs reprogramming system.
(A) Schematic overview of secondary naïve reprogramming strategy to generate human naïve iPSCs. 1° HEF, primary human embryonic fibroblasts; 1° piPSCs, primary primed iPSCs; 2° hiF, inducible fibroblasts; 2° hiF-T, immortalized inducible fibroblasts; 2° niPSCs, secondary naïve iPSCs. (B) Growth curves of hiF and hiF-T cells at different passages. (C) Alkaline phosphatase (AP) staining representing the naïve reprogramming efficiencies of HEFs, hiFs and hiF-Ts at different passages. (D) Representative images showing the senescent cells in hiFs and hiF-Ts at different passages by senescence-associated-beta-galactosidase (SA-β-GAL) assay. Scale bar, 50 μm. Senescent cells are stained in blue and indicated with red arrowheads.
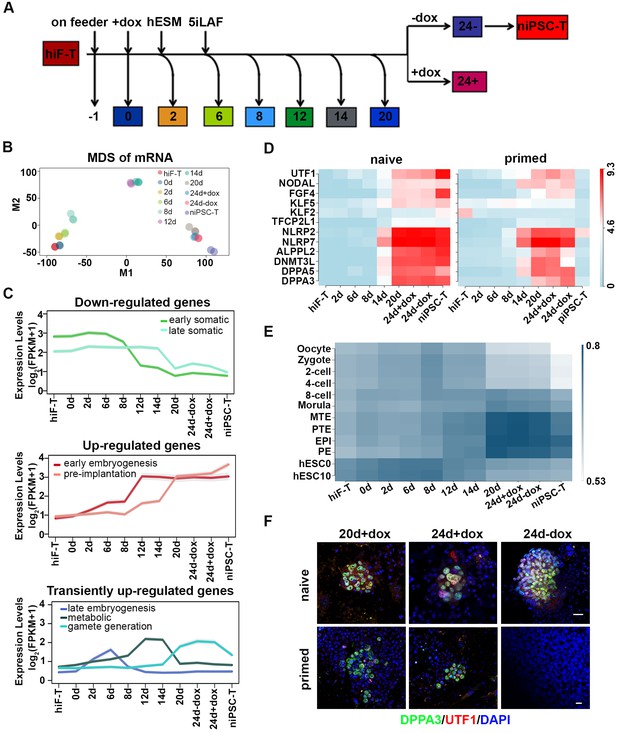
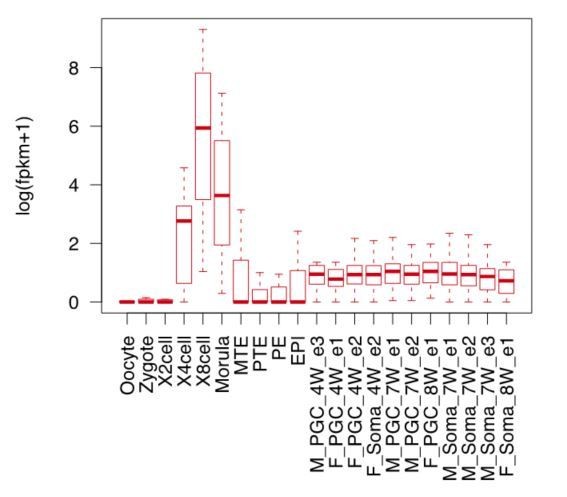
Transcriptional profiling of cells during naïve reprogramming.
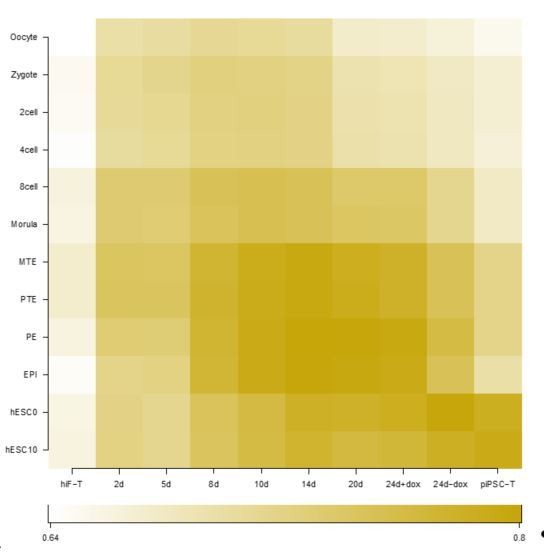
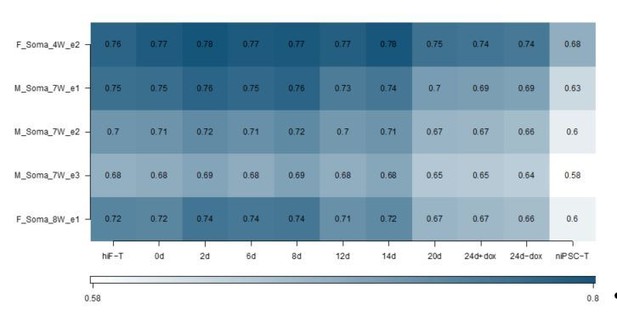
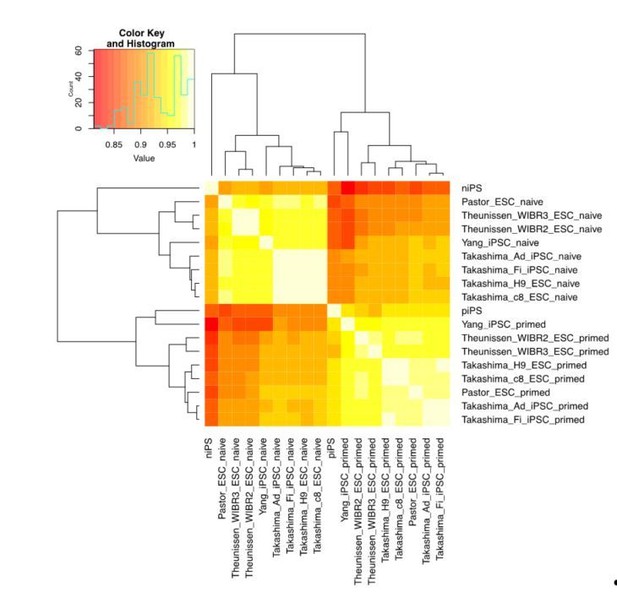
(A) Schematic representation of reprogramming intermediate collection at different time points, as indicated. hiF-T cells were first cultured in conventional hESM with dox for 6 days and then switched to 5iLAF culture medium supplemented with dox until day 20. Cells with or without dox treatment for four additional days were collected. (B) MDS analysis of RNA-seq data during the naïve reprogramming process. (C) Line plots showing transcriptional dynamics of differentially expressed genes during the naïve reprograming process. Genes were grouped by k-means clustering. Gray shades represent a 95% bootstrap confidence interval around the mean value. (D) Heatmaps showing the expression patterns of genes with pre-implantation signatures in both the naïve and primed reprogramming process. (E) Correlation analysis of transcriptional profiles between naïve reprogramming and the embryonic development process, with the Pearson correlation coefficient of each pair shown on each cell of the heatmap. (F) Immunostaining images of pluripotency-related marker expression in the reprogramming cells at indicated time points during naïve and primed reprogramming. Scale bar, 50 μm.
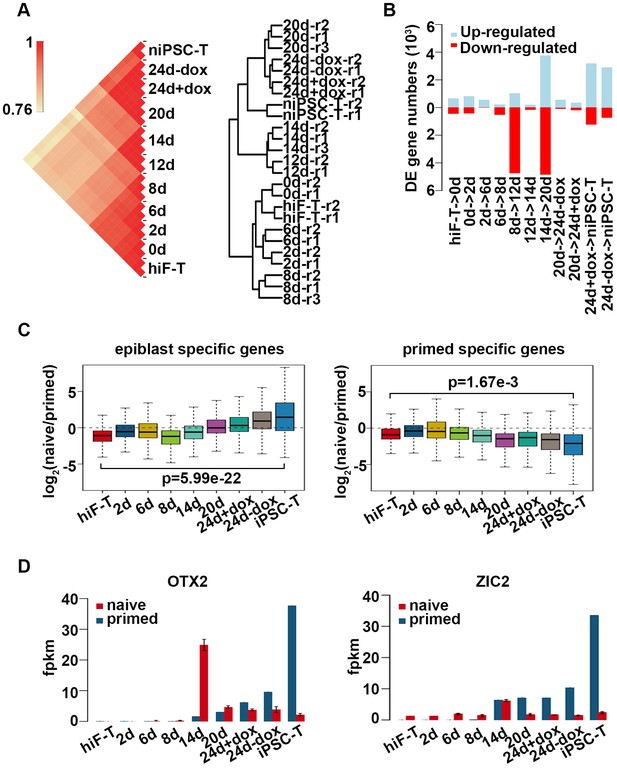
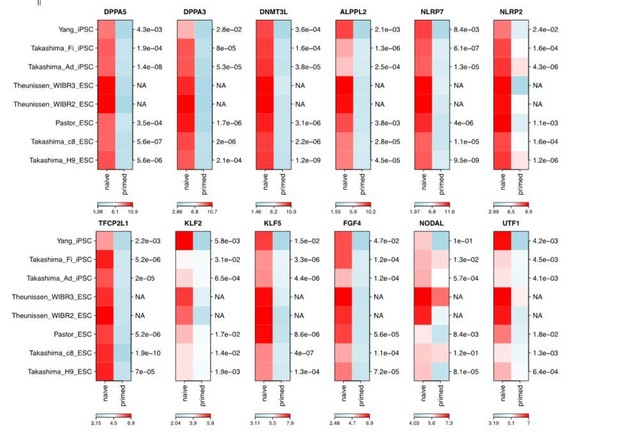
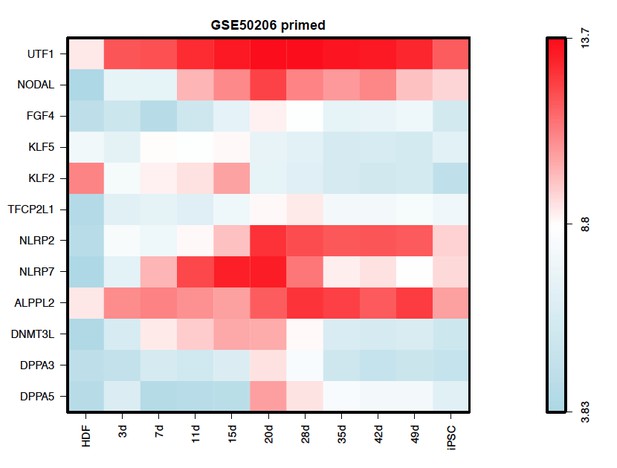
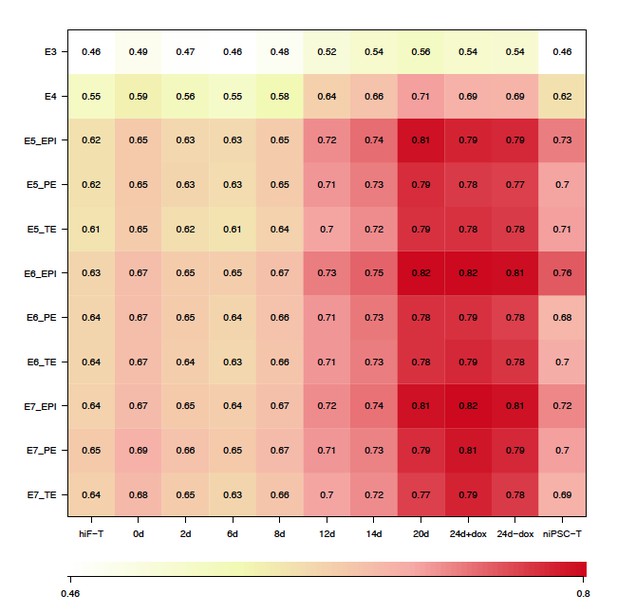
Transcriptional profiling of naïve pluripotency reprogramming cells.
(A) Correlation analysis (left panel) ad cell clustering analysis (right panel) of transcriptional profiles at different time points during naïve reprograming. (B) Bar plot showing dynamic changes in differentially expressed (DE) gene numbers of two adjacent time points during naïve reprogramming. (C) Expression dynamics of epiblast-specific genes and primed-specific genes during naïve reprogramming relative to those in the primed reprogramming process. (D) Bar plot showing the absolute expression values of OTX2 and ZIC2 genes in the naïve and primed reprogramming processes. Error bars represent a 95% confidence interval around the mean value.
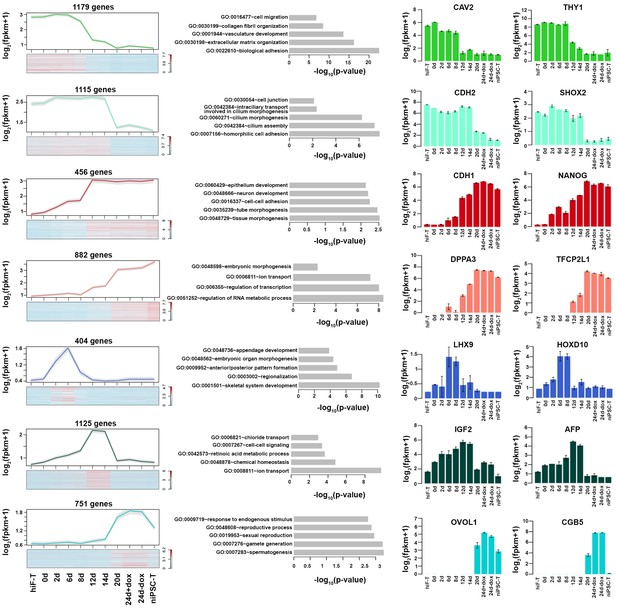
Expression dynamics of gene clusters in Figure 2C.
Detailed description of Figure 2D gene clusters with specific expression patterns. Line plot and heatmap (left panels) showing expression patterns of each cluster during naïve reprograming. Grey shades represent a 95% confidence interval around the mean value; Bar plot (middle panels) showing enrichment of p-value of representative GO term of each cluster; Bar plot (right panels) showing expression dynamics of selected genes across naïve reprogramming time course.
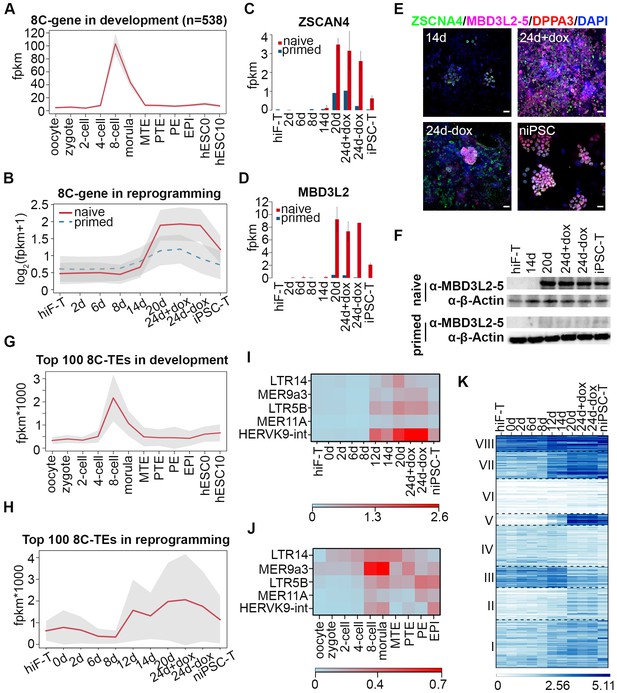
Transient activation of transcripts with 8C-stage-like signatures during naïve reprogramming.
(A) Line plot showing expression dynamics of 8C-stage-specific genes during human embryonic development. Gray shades represent a 95% confidence interval around the mean value. (B) Line plot showing transcriptional dynamics of 8C-specific genes across naïve and primed reprogramming. (C–D) Bar plot showing the absolute expression values of ZSCAN4 (C) and MBD3L2 genes (D) in the naïve and primed reprogramming processes. Error bars represent a 95% confidence interval around the mean value. (E) Immunostaining images of ZSCAN4, MBD3L2-5 and DPPA3 expression in cells during naïve reprogramming. Scale bar, 50 μm. (F) Western blot results of MBD3L2-5 expression in naïve and primed reprogramming cells, niPSC-Ts and piPSC-Ts. β-ACTIN was used as endogenous control. (G) Line plot showing the expression patterns of 8C-specific TEs during human embryonic development. Gray shades represent a 95% confidence interval around the mean value. (H) Line plot showing expression dynamics of 8C-specific TEs during naïve reprogramming. (I–J) Heatmap of expression patterns of 8C-specific HERVK integrants across naïve reprogramming (I) and pre-implantation development (J). (K) Heatmaps showing different expression patterns of KRAB-ZNF genes in the naïve reprogramming process. K-means clustering was performed on KRAB-ZNF genes with k = 8 using R library ‘amap’. Distance between genes was measured based on their correlation.
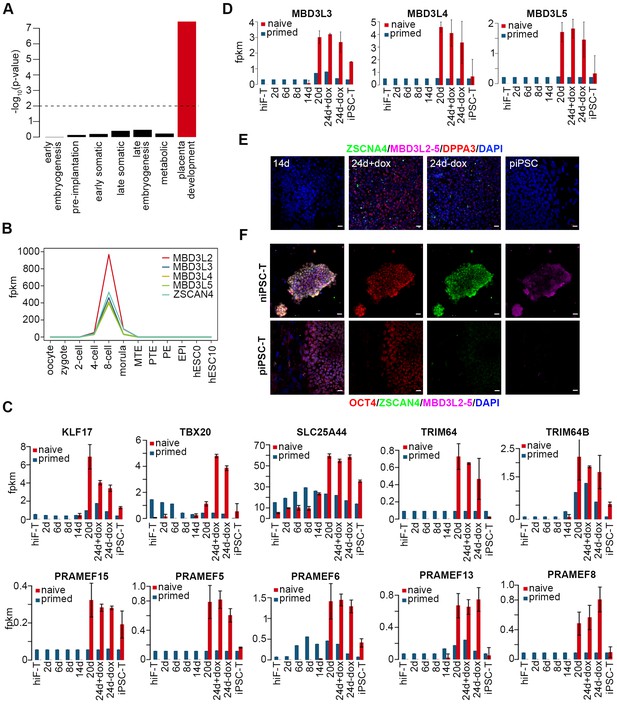
Dynamics of 8C-genes during naïve reprogramming.
(A) Bar plot showing the p-value of 8C-gene enrichments in each cluster measuring with Fisher’ exact test. (B) Line plot showing expression patterns of selected genes during human pre-implantation development. (C) Bar plot showing the absolute expression values of several 8C-genes in the naïve and primed reprogramming processes. Error bars represent a 95% confidence interval around the mean value. (D) Bar plot showing the absolute expression values of MBD3L3/4/5 genes in the naïve and primed reprogramming processes. Error bars represent a 95% confidence interval around the mean value. (E) Immunostaining images of ZSCAN4, MBD3L2-5 and DPPA3 expression in cells during primed reprogramming. Scale bar, 50 μm. (F) Immunostaining for OCT4, ZSCAN4 and MBD3L2-5 in niPSC-Ts and piPSCTs respectively. Scale bar, 20 μm.
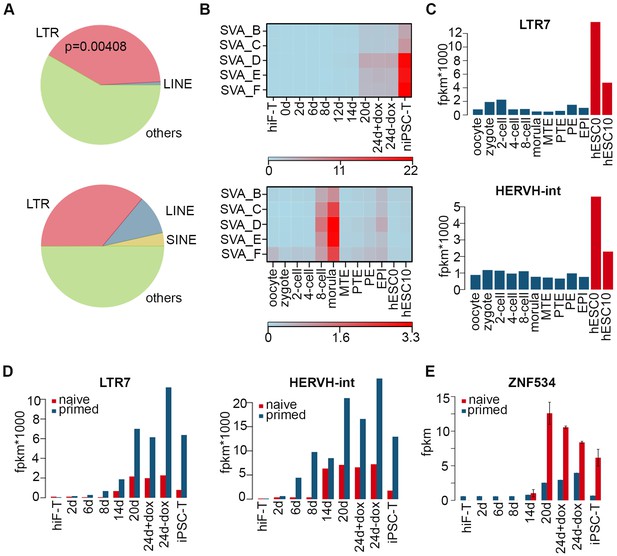
Dynamics of TEs during naïve reprogramming.
(A) Pie charts of genomic distribution of top 100 8C-TEs distribution (upper panel) and all TEs (lower panel), showing significant enrichment (Fisher’s exact test p-value=0.00408) in LTRs. (B) Heatmaps of expression patterns of SVA integrants during naïve reprogramming (upper panel) and embryonic development (lower panel). (C) Expression dynamics of LTR7-HERVH during embryonic development. (D) Bar plot showing expression dynamics of LTR7-HERVH during naïve and primed reprogramming. (E) Transcriptional dynamics of ZNF534 in naïve and primed reprogramming. Error bars represent a 95% confidence interval around the mean value.
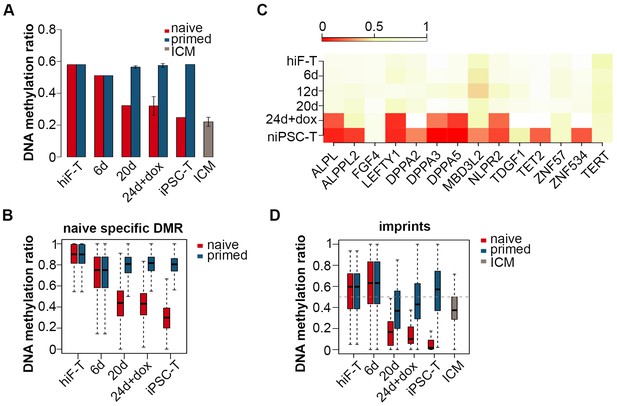
Changes in DNA methylation during naïve reprogramming.
(A) Bar plot showing changes in average DNA methylation ratios of all covered C sites during naïve and primed reprogramming. Error bars represent a 95% confidence interval around the mean value. (B) Box plot showing DNA methylation ratio dynamics of naïve specific DMRs during naïve and primed reprogramming. The middle lines of the boxes indicate the median, the outer edges represent the first and the third quartiles, and the whiskers indicate the 1.5 × interquartile range below the lower quartile and above the upper quartile. (C) Dynamics in the DNA methylation levels of naïve-specific DMR-related genes during naïve reprogramming. (D) DNA methylation over stable primary imprints during naïve and primed reprogramming. The middle lines of the boxes indicate the median, the outer edges represent the first and the third quartiles, and the whiskers indicate the 1.5 × interquartile range below the lower quartile and above the upper quartile.
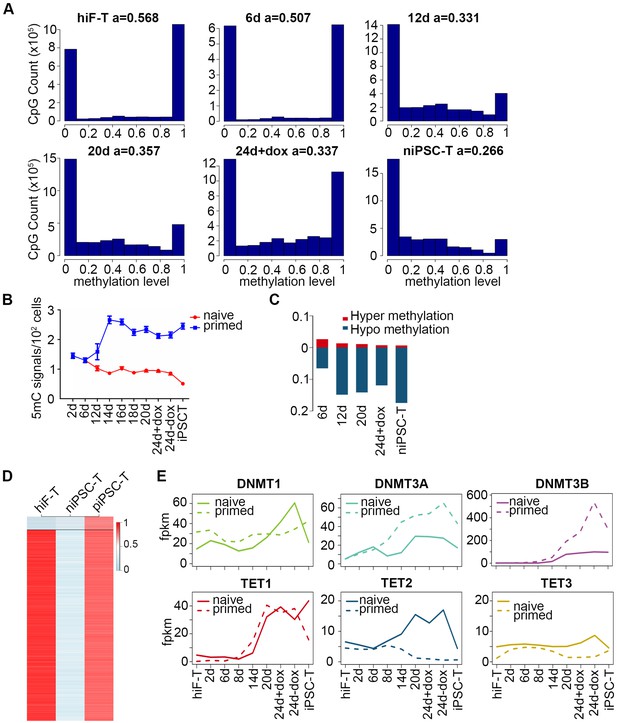
Dynamics of DNA methylation in naïve reprogramming.
(A) Histograms showing the distribution of methylation levels (%) across all CpGs at indicated time points during naïve reprogramming. a, average methylation level. (B) 5mC dynamics across naïve and primed reprogramming by HPLC-MS analysis. (C) Bar plot showing DMC dynamics during naïve reprograming. The ratio of hyper-DMCs divided by the number of all C sites (red bars) and the ratio of hypo-DMCs divided by the number of all C sites (blue bars) were plotted separately. (D) Heatmap showing dynamics of DNA methylation on hiF-T with niPSC-T and piPSC-T. Each line is a DMR called using mcomp from MOABS. Naïve specific DMRs were defined as those only detected in niPSC-Ts compared with hiF-Ts and piPSC-Ts. (E) Line plot showing the expression dynamics of selected genes related to DNA methylation (upper panel) and TET family genes (lower panel) in naïve reprogramming (solid line) and primed reprogramming (dash line) process.
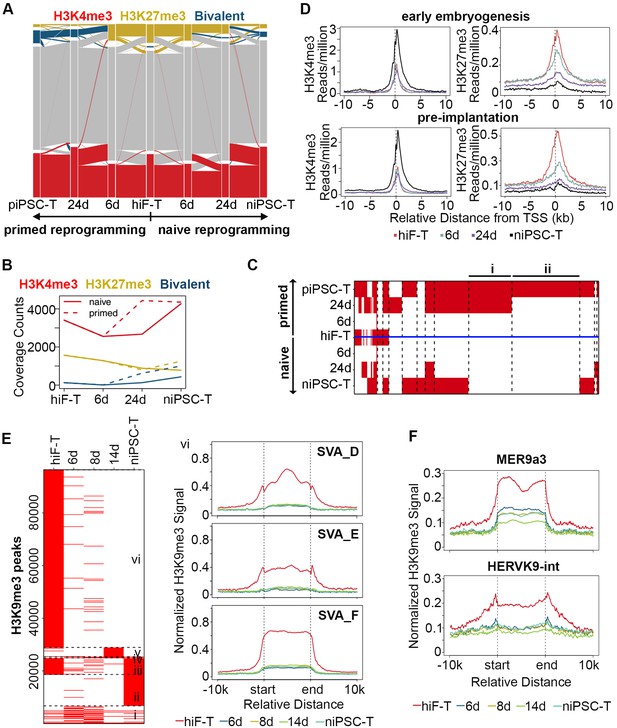
Histone modification profiles during naïve reprogramming.
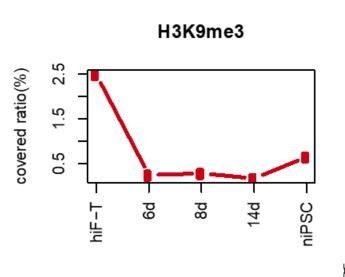
(A) Alluvial plots showing the global dynamics of genes covered by different chromatin states during naïve reprogramming. Each line represents a gene. Red bar represents genes with promoter that covered only by H3K4me3. Yellow bar represents genes with promoter that covered only by H3K27me3. Blue bar represents genes with promoter that covered by both H3K4me3 and H3K27me3. Grey bar represents genes with promoter that covered by neither H3K4me3 nor H3K27me3. (B) Line plot showing dynamics of different histone modification signals across naïve and primed reprogramming. (C) Heatmap showing clusters of genes with different bivalency patterns across naïve and primed reprogramming. (D) Average profiles of H3K4me3 and H3K27me3 signals surrounding the TSS of genes characteristic for early embryogenesis and pre-implantation during naïve reprogramming. (E) Heatmap showing six clusters of H3K9me3 peaks with different patterns (left panel) and average H3K9me3 profiles around integrants of SVA family in cluster vi of the heatmap (right panel) during naïve reprogramming. (F) Average H3K9me3 profiling around MER9a3-HERVK-9 TE during naïve reprogramming.
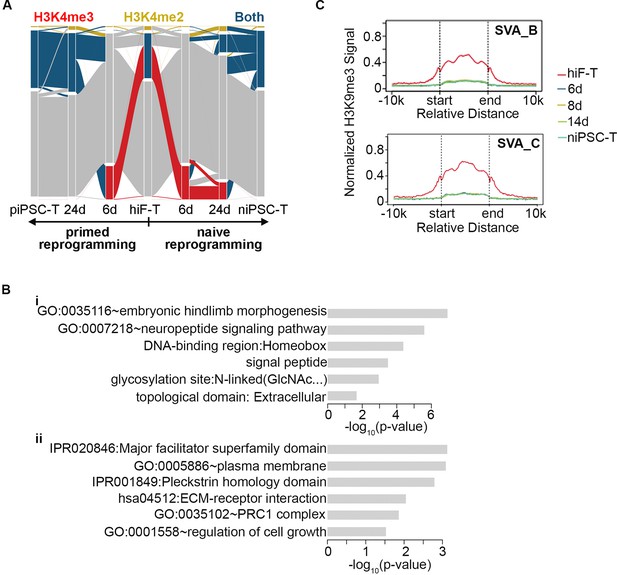
Dynamics of histone modifications in naïve reprogramming.
(A) Alluvial plots showing the global dynamics of genes covered by different chromatin states during naïve reprogramming. Each line represents a gene. Red bar represents genes with promoter that covered only by H3K4me2. Yellow bar represents genes with promoter that covered only by H3K4me3. Blue bar represents genes with promoter that covered by both H3K4me2 and H3K4me3. Grey bar represents genes with promoter that covered by neither H3K4me2 nor H3K4me3. (B) Enrichment of p-value of representative GO term of the selected clusters in Figure 5C. (C) Average H3K9me3 profiles around integrants of SVA family during naïve reprogramming.
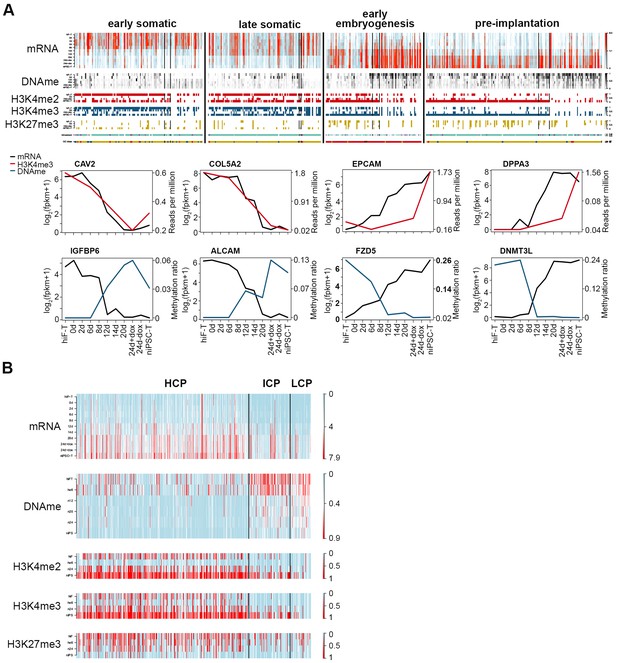
Integrative analysis of transcriptional and epigenetic dynamics during naïve reprogramming.
(A) Transcriptional dynamics of genes with different patterns are closely correlated with epigenetic modifications at promoter regions during naïve reprogramming. Black lines in the heatmaps separate genes regulated by histone modifications (upper panel, left part), DNA methylation (upper panel, right part) or both (upper panel, middle part). Line plots (lower panels) show the representative genes in each category that are regulated by H3K4me3 modification or DNA methylation. (B) Up-regulated genes with different CG ratios in their promoters exhibit different kinetics with regard to transcription, DNA methylation and H3K4me2/H3K4me3/H3K27me3 coverage during naïve reprogramming.
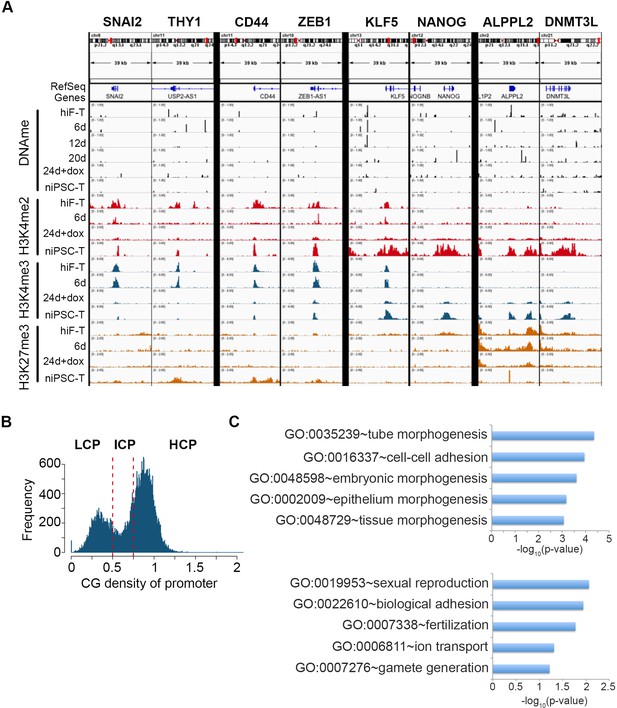
Epigenetic changes of representative genes during naïve reprogramming path.
(A) Base-level visualization of DNA methylation and histone modifications in the promoter regions of representative genes. (B) Classification of genes by the CG ratio in their promoter. (C) Enrichment of p-value of representative GO term of clusters based on different CG ratios in promoters as shown in Figure 6B.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene () | NA | NA | ||
| strain, strain background () | NA | NA | ||
| genetic reagent () | NA | NA | ||
| cell line () | Human embryonic fibroblasts (HEFs); Primary primed iPSC lines; hiF-T cell lines; Secondary primed iPSC lines; Secondary naïve iPSC lines | This paper; Cacchiarelli, D., Trapnell, C., Ziller, M.J., Soumillon, M., Cesana, M., Karnik, R., Donaghey, J., Smith, Z.D., Ratanasirintrawoot, S., Zhang, X., et al. (2015). Cell. 2015 Jul 16;162(2):412–424. doi: 10.1016/j.cell.2015.06.016. Yan, L., Yang, M., Guo, H., Yang, L., Wu, J., Li, R., Liu, P., Lian, Y., Zheng, X., Yan, J., et al. (2013). Nat Struct Mol Biol. 2013 Sep;20(9):1131–9. doi: 10.1038/nsmb.2660. Epub 2013 Aug 11. | ||
| transfected construct () | dox-inducible, polycistronic OKMS lentiviral vector | Addgene 51543. Cacchiarelli, D., Trapnell, C., Ziller, M.J., Soumillon, M., Cesana, M., Karnik, R., Donaghey, J., Smith, Z.D., Ratanasirintrawoot, S., Zhang, X., et al. (2015).Cell. 2015 Jul 16;162(2):412–424. doi: 10.1016/j.cell.2015.06.016. | ||
| biological sample () | hiF-T/0d/2d/6d/8d/12d/14d/20d/ 24d+dox/24d-dox/niPSC-T; Oocyte/Zygote/2 cell/4 cell/8 cell/ Morula/MTE/PTE/EPI/PE/hESC0/hESC10; hiF-T/2d/6d/8d/14d/20d/24d+ dox/24d-dox/piPSC-T; | this paper; | ||
| antibody | anti-SSEA3, SSEA4, TRA-1–60, UTF1, DPPA3, ZSCNA4, MBD3L2 | Millipore MAB4304, Millipore MAB4360, Abcam ab24273, Santa Cruz sc- 67249, Millipore AB4340, Abcam ab174802 | ||
| recombinant DNA reagent | NA | NA | ||
| sequence-based reagent | KAPA Stranded mRNA-Seq Kit; KAPA DNA Library Preparation Kits | KAPA KK8401; KAPA KK8234 | ||
| peptide, recombinant protein | human LIF recombinant protein; bFGF recombinant protein | Peprotech 300–05; Peprotech 450–33 | ||
| commercial assay or kit | bowtie; TopHat; Cufflinks; edgeR; MACS2; | KAPA KK8401; KAPA KK8234 | ||
| chemical compound, drug | Activin A; PD0325901; IM-12; SB590885; WH-4–023; Y-27632; | Peprotech 120–14; Stemgent 04–0012; Enzo BML-WN102; R and D systems 2650; A Chemtek 0104–002013; Stemgent 04–0012; | ||
| software, algorithm | bowtie; TopHat; Cufflinks; edgeR; MACS2; | PMID: 22388286; PMID: 19289445; PMID: 22383036; PMID: 24743990; PMID: 18798982; | RRID:SCR_005476; RRID:SCR_013035; RRID:SCR_014597; RRID:SCR_012802; RRID:SCR_013291 | NA |
| other |
Additional files
-
Source data 1
Source Data for Figures 2–6 and Figure Supplements.
The Source Data files include the source R code for Figures 2–6. The R scripts use the data in the ‘Expression Data’ folder with the relative path. The ‘Expression Data’ folder contains the non-redundant related gene expression, methylation and histone modification tables for the main figures and figure supplements.
- https://doi.org/10.7554/eLife.29518.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29518.017





