Stress responsive miR-31 is a major modulator of mouse intestinal stem cells during regeneration and tumorigenesis
Figures
MiR-31 promotes turnover of intestinal epithelial cells.
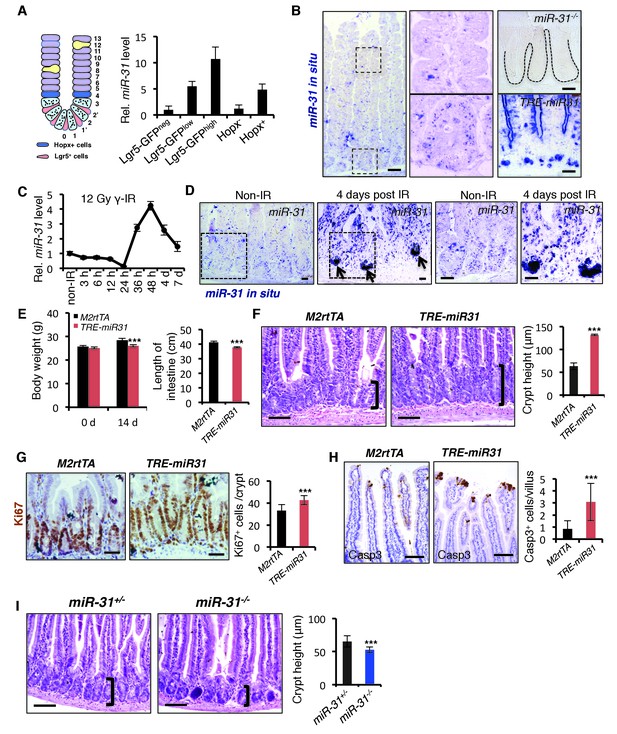
(A) Schematic picture of intestinal crypt showing Lgr5+ CBCs and Hopx+ cells. qRT-PCR for miR-31 in Lgr5-GFPhigh, Lgr5-GFPlow, Lgr5-GFPneg, Hopx- and Hopx+ sorted intestinal epithelial cells. n = 4 biological replicates. (B) In situ hybridization for miR-31 in the intestinal epithelium. Left panel, representative low magnification image (Scale bar: 200 μm); Middle panels, high magnification images indicated by dashed boxes in left panel; Right panels (Scale bar: 50 μm), miR-31 KO intestinal section used as a negative control (Top) and TRE-miR31 (miR-31 overexpressing) intestinal section used as a positive control (Bottom). (C) qRT-PCR for miR-31 in the intestinal epithelium after exposure to 12 Gy γ-IR at indicated time points. n = 3 biological replicates. (D) In situ hybridization for miR-31 in intestines without γ -IR treatment (non-IR), and intestines 4 days after 12 Gy γ-IR. Arrows, miR-31 positive regenerative foci. Dashes boxes indicate the high magnification images in right panels. Scale bar: 50 μm. (E) Quantification of body weight from M2rtTA and TRE-miR31 mice at the age of 8 weeks before and after Dox treatment for 2 weeks. Quantification of intestine length from M2rtTA and TRE-miR31 mice following 2 week Dox induction. n = 6 biological replicates. ***p<0.001. (F) Representative histologic images showing extension of crypt height in jejunum from TRE-miR31 mice, and quantification of crypt height from M2rtTA and TRE-miR31 intestine. Both M2rtTA and TRE-miR31 mice were treated with Dox for 2 weeks. n = 3 biological replicates. Scale bar: 50 μm. ***p<0.001. (G) Immunohistochemistry for Ki67 and quantification of Ki67+ cells per crypt in M2rtTA andTRE-miR31 jejunum, showing an expanded proliferative zone in TRE-miR31 mice following 2 weeks of Dox induction. n = 3 biological replicates. Scale bar: 50 μm. ***p<0.001. (H) Immunohistochemistry for cleaved-Caspase 3 (Casp3) and quantification of Casp3+ cells in the top of intestinal villi from M2rtTA andTRE-miR31 mice following 2 weeks of Dox induction. n = 3 biological replicates. 60 villi were quantified in each mouse. Scale bar: 100 μm. ***p<0.001. (I) Representative histologic images and quantification of crypt height in intestines from miR-31+/− and miR-31−/− mice at 2 months of age. Brackets mark crypts. Scale bar: 100 μm. n = 3 biological replicates. ***p<0.001.
-
Figure 1—source data 1
Source data for Figure 1C,E,F,G,H and I.
- https://doi.org/10.7554/eLife.29538.012
-
Figure 1—source data 2
Source data for Figure 1—figure supplements 1–3.
- https://doi.org/10.7554/eLife.29538.013
-
Figure 1—source data 3
Source data for Figure 1—figure supplements 4–7.
- https://doi.org/10.7554/eLife.29538.014
Generation of inducible TRE-miR-31 transgenic mice, constitutive miR-31 KO and conditional miR-31 KO mice.
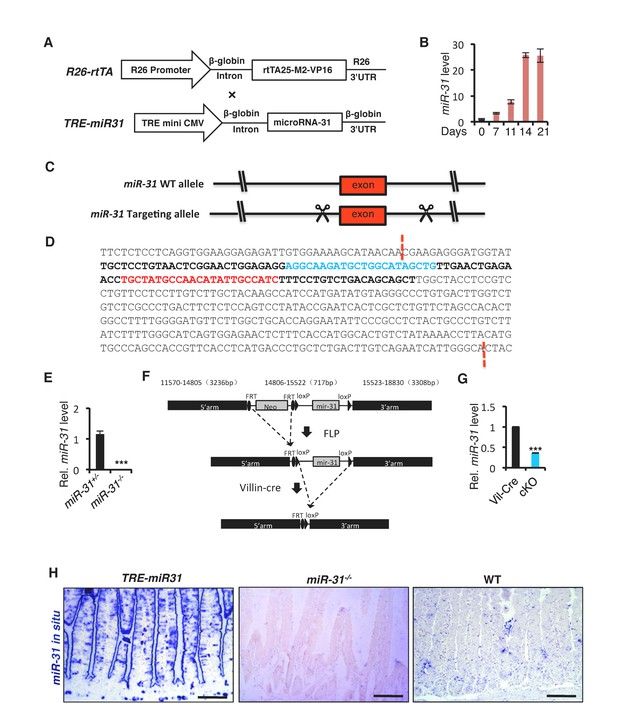
(A) Schematic maps of constructs used to generate Rosa26-rtTA;TRE-miR31 (TRE-miR31) double transgenic mice. (B) qRT-PCR analysis for miR-31 in intestinal tissues from M2rtTA and TRE-miR31 mice at indicated timepoints following Dox treatment. (C) Strategy to generate miR-31 KO mice using Crispr/Cas9 technique. (D) 402 bp DNA fragment containing miR-31 indicated by dash lines was deleted in the KO allele. The miR-31 exon indicated by bold; mature miR-31 indicated by blue, miR-31* indicated by red. (E) qRT-PCR analysis for miR-31 in intestinal tissues from miR-31+/− and miR-31−/− mice. ***p<0.001. (F) Strategy to generate miR-31 conditional null allele. (G) qRT-PCR for miR-31 in intestine from Vil-Cre and Vil-Cre/miR-31fl/fl (cKO) mice. ***p<0.001. (H) In situ hybridization for miR-31 in TRE-miR31, miR-31−/− and WT intestines. TRE-miR31 mice were pretreated with Dox for 2 weeks. Scale bar: 100 μm. Note: The schematic depiction of the general strategy for generating miR-31 mutant mice (Figure 1—figure supplement 1C and D) was also used in another unrelated study on the role of miR-31 in mammary stem cells and breast cancer and the manuscript is currently under consideration in Nature Communications.
MiR-31 induction promotes crypt expansion.
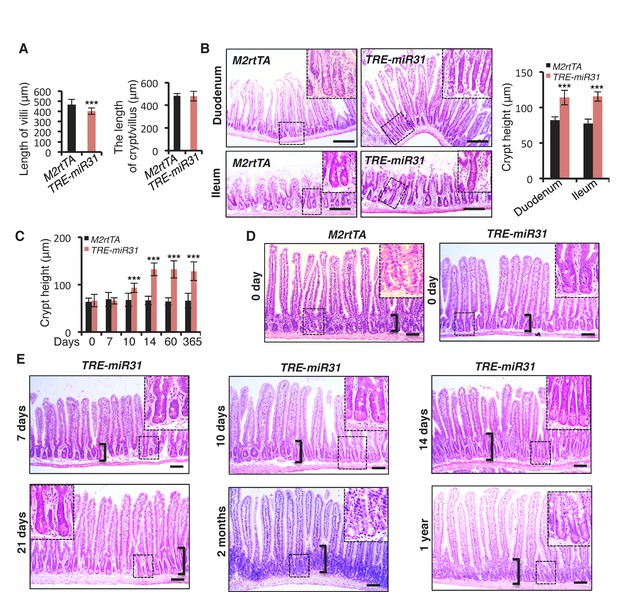
(A) Quantification of length of villi and cypt/villus in M2rtTA and TRE-miR31 mice following 2 weeks of Dox induction. n = 3 biological replicates. ***p<0.001. (B) Representative histologic images of duodenum and ileum from M2rtTA and TRE-miR31 mice following 2 weeks of Dox induction, and quantification of crypt height. Dashed boxes indicate high magnification images of crypts. Scale bar: 100 μm. ***p<0.001. (C) Quantification of crypt height from M2rtTA and TRE-miR31 intestine at indicated time points. Continuous Dox treatment was administered on M2rtTA and TRE-miR31 mice for 0 day, 7 days, 10 days, 14 days, 2 months and 1 year. n = 3 biological replicates at each time points. ***p<0.001. (D) Representative histologic images of jejunum from M2rtTA and TRE-miR31 mice without Dox treatment. Brackets mark crypts. Dashed boxes indicate high magnification images of crypts. Scale bar: 50 μm. (E) Representative histologic images of jejunum from TRE-miR31 mice following 7, 10, 14, 21 days, 2 months and 1 year of Dox induction. Brackets mark crypts. Dashed boxes indicate high magnification images of crypts. Scale bar: 50 μm.
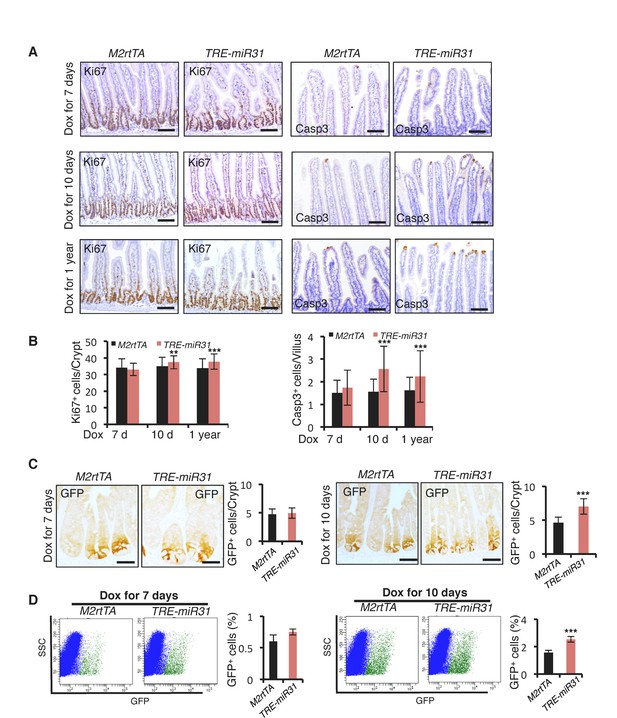
MiR-31 induction promotes cell proliferation in crypts, and apoptosis at the top of villi.
(A) Immunohistochemistry for Ki67 and Cleaved Caspase 3 in jejunum from M2rtTA and TRE-miR31 mice following 7, 10 and 365 days of Dox induction. Scale bar: 100 μm. (B) Quantification of Ki67+ cells in crypts of M2rtTA and TRE-miR31 mice, and Caspase 3+ cells at the tip of villi. **p<0.01; ***p<0.001. (C) Immunohistochemistry for GFP (Lgr5) and quantification of GFP+ cells in crypts from M2rtTA and TRE-miR31 mice following 7 and 10 days of Dox induction. ***p<0.001. (D) Representative FACS profiles and quantification of GFP positive intestinal epithelial cells (Lgr5-GFP+ cells) from an Lgr5-eGFP-CreERT reporter mice crossed with M2rtTA and TRE-miR31 mice. M2rtTA and TRE-miR31 mice were pre-treated with Dox for 7 and 10 days. n = 3 biological replicates. ***p<0.001.
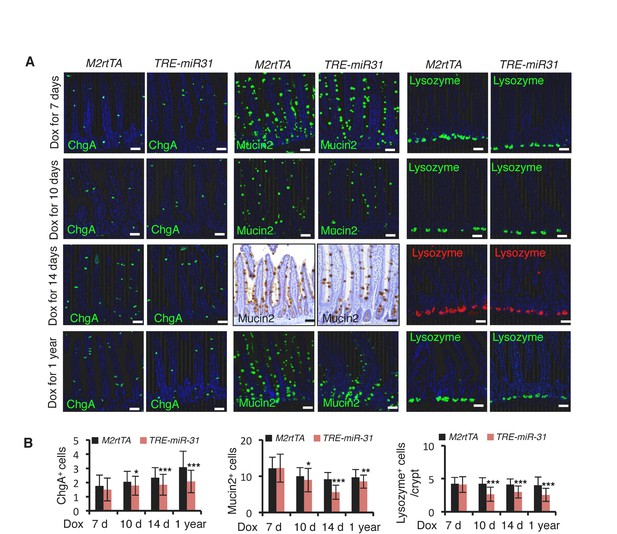
MiR-31 induction impairs cell differentiation.
(A) Immunostaining for ChgA, Mucin2 and Lysozyme in jejunum from M2rtTA and TRE-miR31 mice following 7, 10, 14 days and 1 year of Dox induction. n = 3 biological replicates at each time points. Scale bar: 50 μm. (B) Quantification of ChgA+ cells/crypt-villus, Mucin2+ cells/crypt-villus and Lysozyme+ cells/crypt in Panel A. *p<0.05; **p<0.01, ***p<0.001.
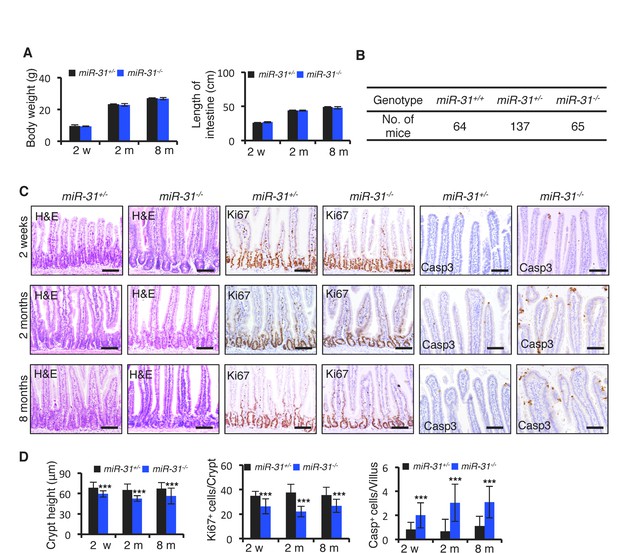
Loss of miR-31 led to shortened crypt.
(A) Quantification of body weight and intestinal length from miR-31+/− (Control) and miR-31−/− mice at 2 weeks, and 2 and 8 months of ages. n = 6 biological replicates for 2 weeks and 2 months; n = 4 biological replicates for 8 months. (B) Quantification of mouse number of miR-31+/+, miR-31+/− and miR-31−/− genotypes. (C) H&E staining and immunohistochemistry for Ki67 and cleaved Caspase 3 (Casp3) in jejunum of miR-31+/− and miR-31−/− mice at 2 weeks, 2 and 8 months of ages. Scale bar: 100 μm. (D) Quantification of crypt height, Ki67+ and Casp3+ cells at different timepoints in Panel C.
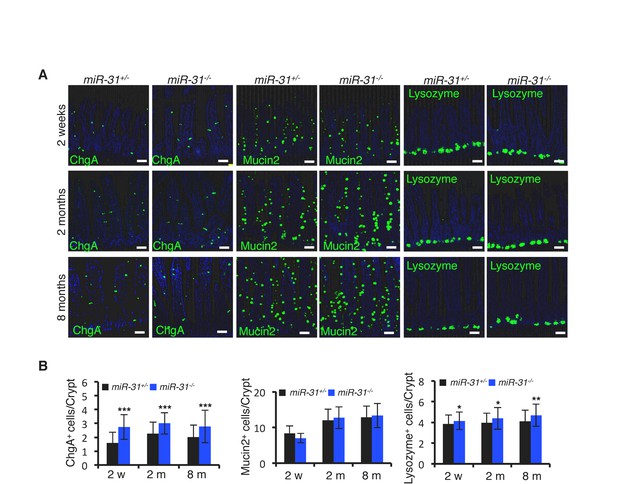
Loss of miR-31 does not affect cell differentiation.
(A) Immunofluorescence for ChgA, Mucin2 and Lysozyme in jejunum from miR-31+/− and miR-31−/− mice at indicated timepoints. Scale bar: 50 μm. (B) Quantification of ChgA+, Mucin2+ and Lysozyme+ cells in Panel A.
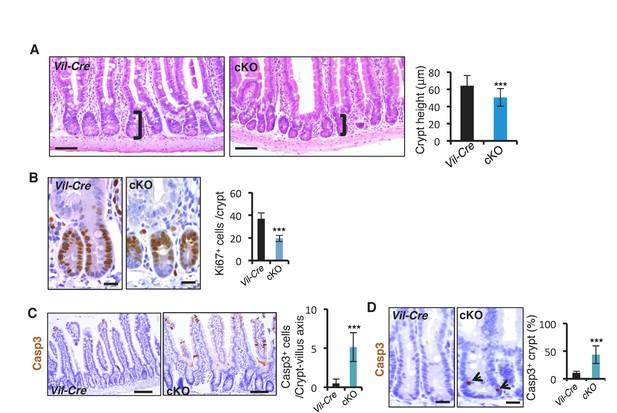
Conditional deletion of miR-31 resulted in shortened crypt, reduced proliferation and enhanced apoptosis.
(A) Representative histologic images of jejunum from Vil-Cre and miR-31 cKO mice, and quantification of crypt height. Brackets mark crypts. Scale bar: 50 μm. n = 4 biological replicates. ***p<0.001. (B) Immunohistochemistry for Ki67 in Vil-Cre and miR-31 cKO intestines. Quantification of Ki67+ cells per crypt. n = 3 biological replicates. Scale bar: 25 μm. (C) Immunohistochemistry for cleaved Caspase 3 (Casp3) in Vil-Cre and miR-31 cKO intestines. Quantification of Casp3+ cells per crypt-villus axis (excluding the tip of villus). n = 3 biological replicates. Scale bar: 100 μm. (D) High magnification images of immunohistochemistry for cleaved Caspase 3 (Casp3) in Vil-Cre and miR-31 cKO intestines. Quantification of percentage of Casp3+ crypts. n = 3 biological replicates. Scale bar: 25 μm.
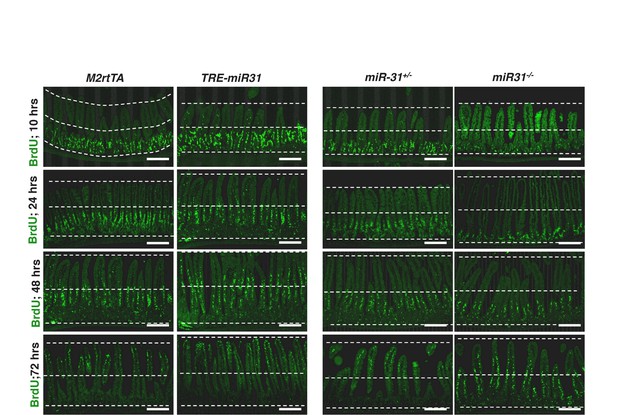
MiR-31 promotes cell turnover from crypt to villi.
Immunofluorescence for BrdU in M2rtTA, TRE-miR31, miR-31+/− and miR-31−/− intestinal crypts at indicated time points post 1 dose of BrdU pulse. The dashed lines marked the top of villi, middle line of intestine, and the bottom of crypt, respectively. Scale bar: 100 μm. n = 3 biological replicates at each time points.
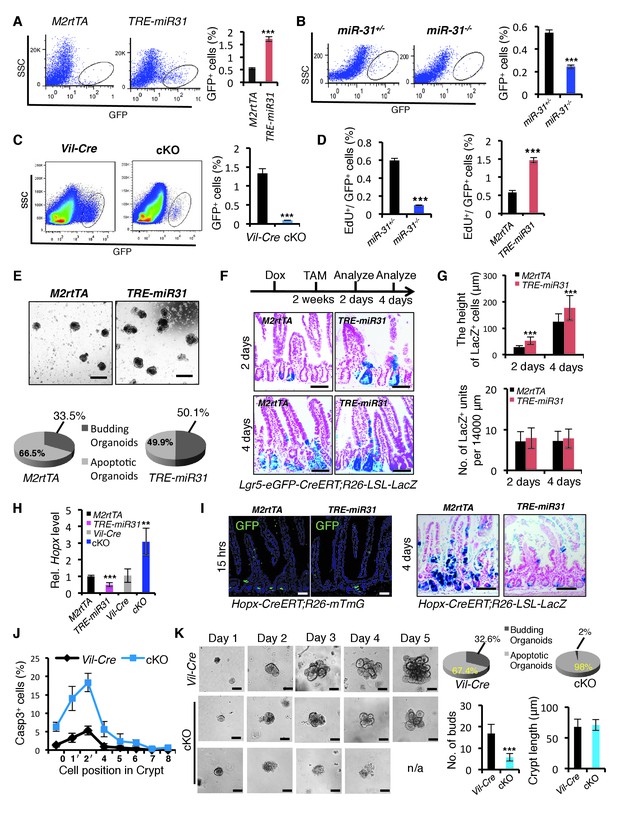
MiR-31 promotes expansion of Lgr5+ CBC stem cells.
(A) Representative FACS profiles and quantification of GFP positive intestinal epithelial cells (Lgr5-GFP+ cells) from an Lgr5-eGFP-CreERT reporter mice crossed with M2rtTA (control) and TRE-miR31 mice. M2rtTA (control) and TRE-miR31 mice were pre-treated with Dox for two weeks. n = 4 biological replicates. ***p<0.001. (B, C) FACS profiles and quantification of Lgr5-GFP+ cells from an Lgr5-eGFP-CreER reporter mice crossed with miR-31+/− (control) and miR-31−/− mice (B), or Vil-Cre (Villin-Cre) and cKO (Vil-Cre;miR-31fl/fl) mice (C). n = 4 biological replicates. ***p<0.001. (D) Assessment of 1.5-hour-pulse EdU incorporation in Lgr5+ CBC cells in M2rtTA, and TRE-miR31 mice following 2 weeks of Dox treatment, and in miR-31+/− and miR-31−/− intestine. ***p<0.001. (E) Crypts purified from M2rtTA and TRE-miR31 mice grown in organoid cultures with Dox. Representative gross images of budding organoids, and quantification of budding and apoptotic organoids at day 7. Scale bar: 500 μm. n = 5 technical replicates. (F) X-gal staining showing lineage tracing events from Lgr5+ ISCs. Lgr5-eGFP-CreERT;R26-LSL-LacZ;TRE-miR31 mice and its control counterpart were pretreated with Dox for 2 weeks, injected with a single dose tamoxifen, and analyzed 2 and 4 days after injection. Scale bar: 100 μm. n = 3 biological replicates. (G) Quantification of the length of LacZ+ cells and LacZ+ units in Panel F. ***p<0.001. (H) qRT-PCR analysis for Hopx in intestines from M2rtTA, TRE-miR31, Vil-Cre and cKO mice. n = 3 biological replicates. **p<0.01; ***p<0.001. (I) Lineage tracing events from Hopx+ ISCs. Hopx-CreERT;mTmG;TRE-miR31 mice and their control counterparts were pretreated with Dox for 2 weeks, injected with a single dose of tamoxifen, and analyzed 15 hr after injection. Hopx-CreERT;R26-LSL-LacZ;TRE-miR31 and their control counterparts were analyzed 4 days after inject with the same treatment. Scale bar: 100 μm. n = 3 biological replicates. (J) Quantification of Cleaved Caspase3+ cells at indicated positions in the intestinal crypts of Vil-Cre and miR-31 cKO mice in Figure 1—figure supplement 7D. n = 3 biological replicates, 50 crypts per sample. (K) Crypts purified from Vil-Cre and miR-31 cKO mice grown in organoid cultures at indicated time points. Quantification of budding organoids and apoptotic organoids, budding number and crypt length. n = 3 biological replicates. ***p<0.001.
-
Figure 2—source data 1
Source data for Figure 2.
- https://doi.org/10.7554/eLife.29538.017
-
Figure 2—source data 2
Source data for Figure 2—figure supplement 1.
- https://doi.org/10.7554/eLife.29538.018
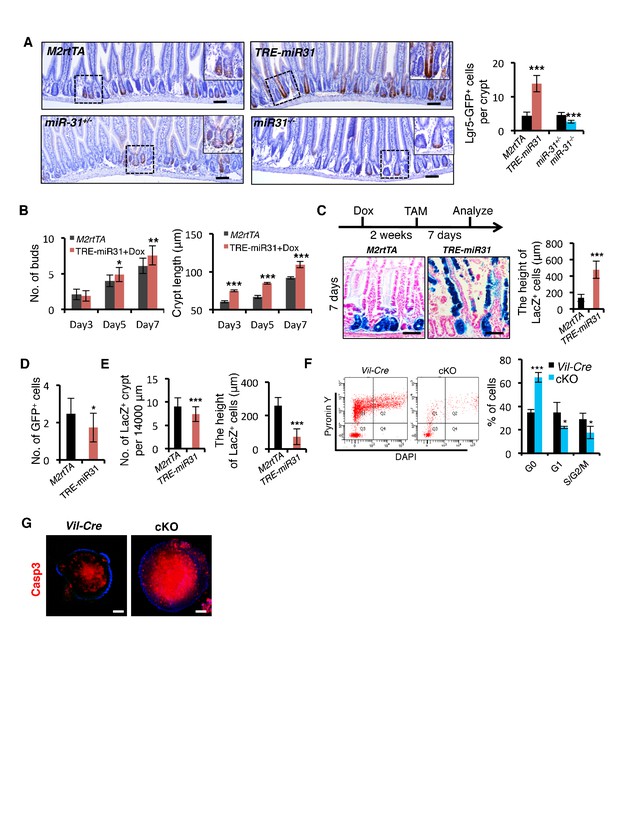
MiR-31 promotes ISC expansion.
(A) Immunohistochemistry for GFP (Lgr5-GFP) in intestines from Lgr5-eGFP-CreERT reporter mice crossed with M2rtTA (control), TRE-miR31, miR-31+/− (control) and miR-31−/− mice. Scale bar: 100 μm. The dashed boxes indicate the high magnification images of insets. Quantification of Lgr5-GFP positive intestinal epithelial cells (Lgr5-GFP+ cells). n = 4 biological replicates. 50 crypts were quantified at each single mouse. ***p<0.001. (B) Quantification of budding number and crypt length in organoids at indicated time points in Figure 2E. n = 5 technical replicates. *p<0.05; **p<0.01; ***p<0.001. (C) X-gal staining showing lineage tracing events from Lgr5+ ISCs and quantification of the length of LacZ+ cells. Lgr5-eGFP-CreERT;R26-LSL-LacZ;TRE-miR31 mice and its control counterpart were pretreated with Dox for 2 weeks, injected with a single dose tamoxifen, and analyzed 7 days after injection. Scale bar: 100 μm. n = 3 biological replicates. ***p<0.001. (D) Quantification on number of GFP+ cells per crypt 15 hrs after Tamoxifen injection in Figure 2I. *p<0.05. (E) Statistical analysis on Hopx lineage tracing 4 days after Tamoxifen injection in Figure 2I. ***p<0.001. (F) Analysis of cell cycle distribution of FACS-purified Lgr5-GFP+ cells using Pyronin Y and DAPI staining from Vil-Cre and miR-31 cKO mice. n = 3 biological replicates. *p<0.05; ***p<0.001. (G) Immunofluorescence for cleaved Caspase 3 (Casp3) in cultured organoids from Vil-Cre and cKO crypts in Figure 2K. Scale bar: 50 μm.
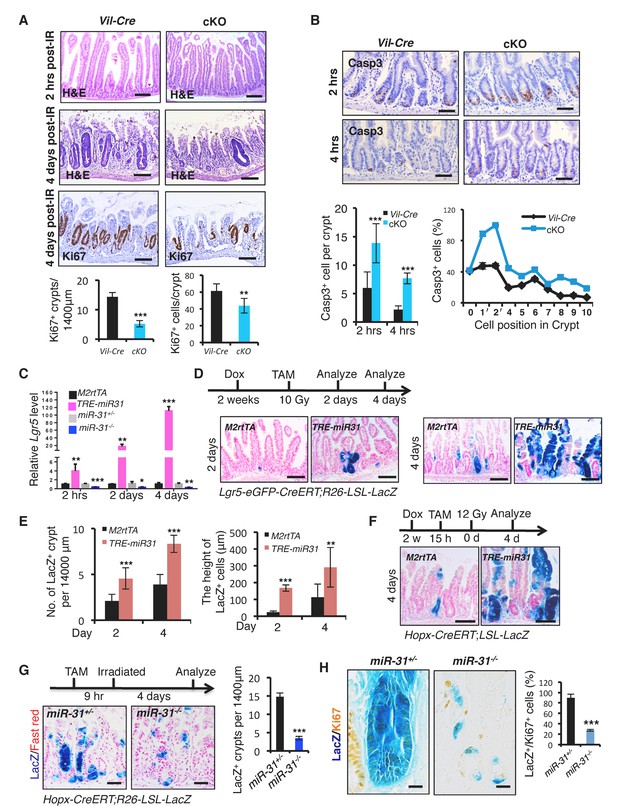
Loss of miR-31 abrogates epithelial regeneration following irradiation.
(A) Representative images of H&E and/or Ki67 immunohistochemistry from jejunum of irradiated Vil-Cre and cKO mice 2 hrs and 4 days post 12 Gy γ-IR. Quantification of Ki67+ regenerative foci per 1400 μm and No. of Ki67+ cells per regenerative focus. Top panel: n = 6 biological replicates; Scale bar: 200 μm. Middle and bottom panels: n = 5 biological replicates; Scale bar: 50 μm. **p<0.01; ***p<0.001. (B) Immunohistochemistry for Casp3, quantification of the number of Casp3+ cells in intestinal crypts of Vil-Cre and cKO mice 2 and 4 hrs post 12 Gy γ-IR. Quantification of Casp3+ cells at indicated positions in intestinal crypts of Vil-Cre and cKO mice 2 hrs post γ-IR. Scale bar: 50 μm. n = 3 biological replicates, and 50 crypts were quantified in each single mouse. ***p<0.001. (C) qRT-PCR analysis for Lgr5 in intestines from M2rtTA, TRE-miR31, miR-31+/− and miR-31−/− mice 2 hrs, 2 and 4 days post 12 Gy irradiation. M2rtTA and TRE-miR31 mice were pre-treated with Dox for two weeks. n = 3 biological replicates at each time points. *p<0.05; **p<0.01; ***p<0.001. (D) Schematic of Lgr5-eGFP-CreERT;R26-LSL-LacZ lineage tracing experiment after irradiation. X-gal staining showing lineage tracing events from Lgr5+ ISCs. Lgr5-eGFP-CreERT;R26-LSL-LacZ;TRE-miR31 mice and their control counterparts were pretreated with Dox for 2 weeks, injected with a single dose tamoxifen and then immediately exposed to 10 Gy γ-IR, and analyzed 2 and 4 days after γ-IR. Scale bar: 100 μm. n = 3 biological replicates at each time points. (E) Quantification of LacZ+ units and the length of LacZ+ cells in Panel D. (F) Schematic of Hopx-CreERT;R26-LSL-LacZ lineage tracing experiment. Hopx-CreERT;R26-LSL-LacZ;TRE-miR31 and their control counterparts were pretreated with Dox for 2 weeks, then injected with a single dose of tamoxifen, and then irradiated 15 hrs after injection and analyzed 4 days after irradiation. Representative images of LacZ staining in M2rtTA and TRE-miR31 intestine 4 days post 12 Gy γ-IR. Scale bar: 50 μm. Statistics of LacZ+ regenerative foci were shown in Figure 3—figure supplement 1E. n = 3 biological replicates. (G) Schematic of Hopx-CreERT;R26-LSL-LacZ lineage tracing experiment. Representative images of LacZ staining in miR-31+/− and miR-31−/− intestine 4 days post 12 Gy γ-IR. Scale bar: 50 μm. Statistics of LacZ+ regenerative foci. n = 3 biological replicates. (H) Representative images of LacZ (blue) and Ki67 (yellow) immunostaining in miR-31+/− and miR-31−/− intestinal crypts, and statistics of percentage of LacZ+/Ki67+cells in regenerative foci. Scale bar: 25 μm. n = 3 biological replicates. ***p<0.001.
-
Figure 3—source data 1
Source data for Figure 3.
- https://doi.org/10.7554/eLife.29538.021
-
Figure 3—source data 2
Source data for Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.29538.022
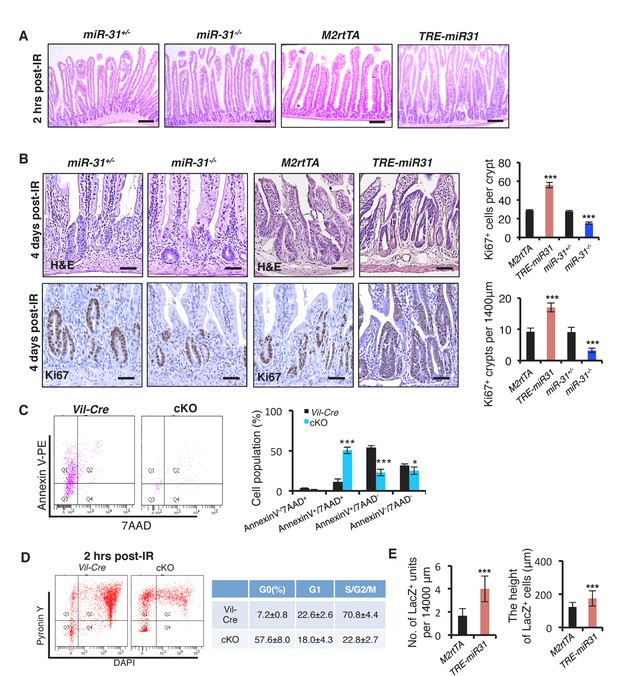
MiR-31 is required for intestinal epithelial regeneration in response to γ-IR.
(A) Representative images of H&E from jejunum of irradiated M2rtTA, TRE-miR31, miR-31+/− and miR-31−/− mice 2 hrs post 12 Gy irradiation. n = 3 biological replicates. Scale bar: 200 μm. (B) Histology and immunohistochemistry for Ki67 from jejunum of irradiated miR-31+/−, miR-31−/−, M2rtTA and TRE-miR31 mice 4 days post 12 Gy irradiation. Quantification of Ki67+ regenerative foci and No. of Ki67+ cells per regenerative focus. Scale bar: 50 μm. n = 4 biological replicates. ***p<0.001. (C) The FACS profile and quantification of Annexin V−7AAD−, Annexin V-7AAD+, Annexin V+7 AAD−, Annexin V+7AAD+ cells in Lgr5-GFP+ cells from Vil-Cre and cKO mice. n = 3 biological replicates. *p<0.05; ***p<0.001. (D) Flow cytometry analysis of cell cycle distribution of FACS-purified Lgr5-GFP+ cells using PyroninY and DAPI staining from Vil-Cre and miR-31 cKO mice 2 hrs post γ-IR. PyroninYlowDAPIlow: G0; PyroninYhighDAPIlow: G1; PyroninYhighDAPIhigh: S/G2/M. n = 3 biological replicates. (E) Quantification of LacZ+ units and the length of LacZ+ cells in Figure 3F. n = 3 biological replicates.
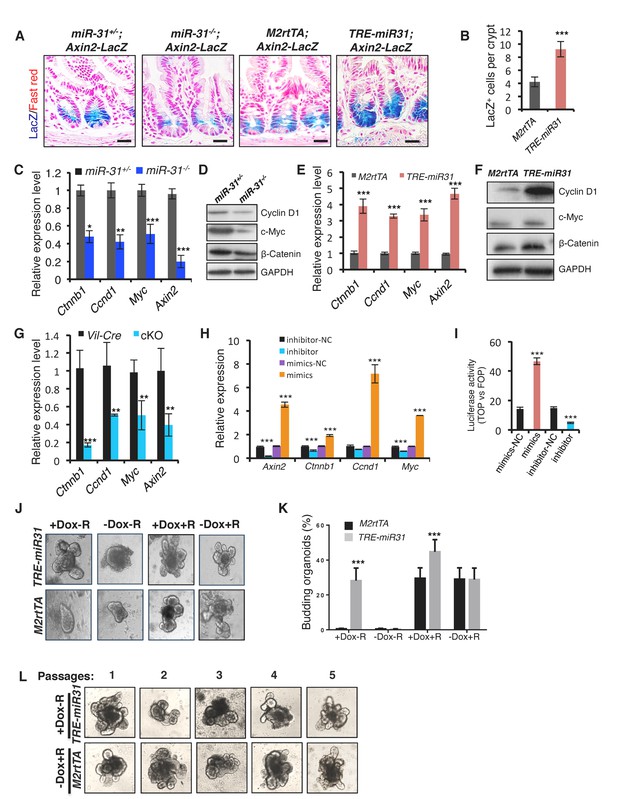
MiR-31 activates Wnt pathway activity.
(A) Wnt activity was evaluated by Axin2-LacZ reporter activity in M2rtTA and TRE-miR31 intestine following 2 week Dox induction, and in miR-31+/− and miR-31−/− intestine. Blue, LacZ signals. n = 3 biological replicates. Scale bar: 25 μm. (B) Quantification of LacZ+ cells per crypt in M2rtTA and TRE-miR31 mice. ***p<0.001. (C) qRT-PCR analysis for Ctnnb1 (encoding β-Catenin), Ccnd1 (encoding Cyclin D1), Myc, and Axin2 in miR-31+/− and miR-31−/− intestine. *p<0.05; **p<0.01; ***p<0.001. (D) Western blotting for Cyclin D1, c-Myc and β-Catenin in miR-31+/− and miR-31−/− intestine. GAPDH was used as a loading control. (E) qRT-PCR for Ccnd1, Myc, Axin2 and Ctnnb1 in intestine from M2rtTA and TRE-miR31 mice following 2 weeks of Dox induction. ***p<0.001. (F) Western blotting for Cyclin D1, c-Myc, and β-Catenin in intestine from M2rtTA and TRE-miR31 mice following 2 weeks of Dox induction. (G) qRT-PCR for Ctnnb1, Ccnd1, Myc, and Axin2 in intestine from Vil-Cre and cKO mice. n = 4 biological replicates. **p<0.01; ***p<0.001. (H) qRT-PCR for Axin2, Ccnd1, Myc, and Ctnnb1 in HCT116 colon cancer cells treated with miR-31 inhibitor and negative control (NC, Scramble RNA), as well as miR-31 mimics and negative control (NC, Scramble RNA) for 24 hrs. ***p<0.001. (I) Luciferase activity of TOPflash versus FOPflash in HCT116 cells treated with miR-31 inhibitor and negative control (NC, Scramble RNA), as well as miR-31 mimics and negative control (NC, Scramble RNA) for 24 hrs. n = 3 technical replicates. ***p<0.001. (J) Representative images of organoids cultures from purified M2rtTA and TRE-miR31 crypts at indicated conditions. R; R-Spondin. n = 3 biological replicates. (K) Quantification of budding organoids in Panel J. ***p<0.001. (L) Representative images of organoids cultures from purified M2rtTA and TRE-miR31 crypts at serial passages. M2rtTA organoids were cultured with R-Spondin; TRE-miR31 organoids were cultured with Dox and without R-Spondin. n = 4 biological replicates.
-
Figure 4—source data 1
Source data for Figure 4.
- https://doi.org/10.7554/eLife.29538.025
-
Figure 4—source data 2
Source data for Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.29538.026

MiR-31 activates Wnt signaling pathway.
(A) Immunohistochemistry for β-Catenin in jejunum from miR-31+/− and miR-31−/− mice at 2 and 4 months of ages. Quantification of nuclear β-Catenin positive cells in miR-31+/− and miR-31−/− crypts. Scale bar: 25 μm. *p<0.05; **p<0.01. (B) Immunohistochemistry for β-Catenin in jejunum from M2rtTA and TRE-miR31 mice following 7 days, 14 days and 2 months of Dox induction. Quantification of nuclear β-Catenin positive cells in M2rtTA and TRE-miR31 crypts. Scale bar: 25 μm. ***p<0.001. (C) Western blotting for β-Catenin in HCT116 cells under treatment of control (mimics-NC) and miR-31 mimics, or control (inhibitor-NC) and anti-miR31. β-Tubulin was used as a loading control.
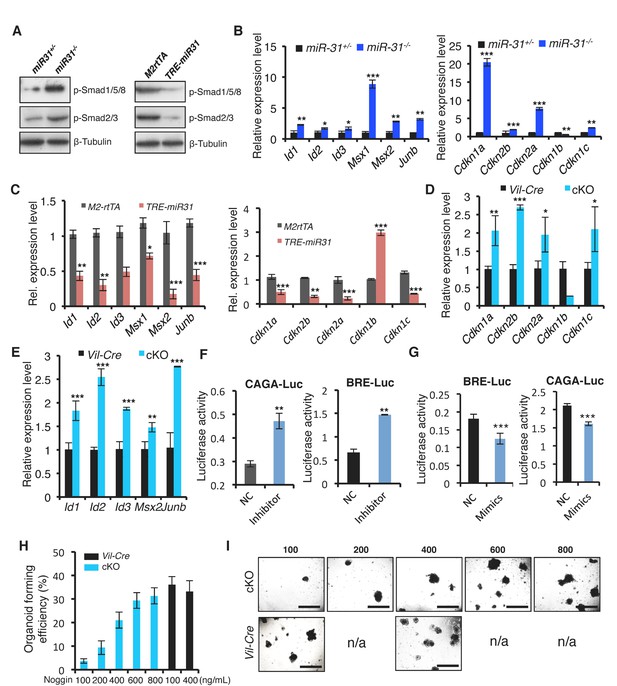
MiR-31 represses BMP/TGFβ signaling pathways.
(A) Western blotting for p-Smad1/5/8 and p-Smad2/3 in miR-31+/−, miR-31−/−, M2rtTA and TRE-miR31 intestine. Both M2rtTA and TRE-miR31 mice were treated with DOX for 2 weeks. β-Tubulin was used as a loading control. (B) qRT-PCR analysis for BMP downstream genes, Id1, Id2, Id3, Msx-1, Msx-2 and Junb, and TGFβ downstream genes, Cdkn1c (p57), Cdkn1a (p21), Cdkn2a (p16), Cdkn2b (p15) and Cdkn1b (p27) in miR-31+/− and miR-31−/− intestine. *p<0.05; **p<0.01; ***p<0.001. (C) qRT-PCR analysis for BMP downstream genes, Id1, Id2, Id3, Msx-1, Msx-2 and Junb, and TGFβ downstream genes, Cdkn1c, Cdkn1a, Cdkn2a, Cdkn2b and Cdkn1b in M2rtTA and TRE-miR31 intestine following 2 weeks of Dox induction. **p<0.01; ***p<0.001. (D) qRT-PCR analysis for TGFβ downstream genes, Cdkn1c, Cdkn1a, Cdkn2a, Cdkn2b and Cdkn1b in intestine from Vil-Cre and cKO mice. *p<0.05; **p<0.01; ***p<0.001. (E) qRT-PCR analysis for BMP downstream genes, Id1, Id2, Id3, Msx2 and Junb in Vil-Cre and cKO intestine. **p<0.01; ***p<0.001. (F and G) HEK293T cells were transfected with CAGA- or BRE- luciferase reporter vector, combined with scramble RNA (negative control, NC) or anti-miR-31 (miR-31 inhibitors) (F), or scramble RNA (negative control, NC) and miR-31 mimics (G) for 24 hrs and then harvested for luciferase activity determination. n = 3 technical replicates. **p<0.01; ***p<0.001. (H) Quantification of organoid forming efficiency (budding organoids per 100 crypts) after Vil-Cre or cKO crypts cultured with noggin at indicated concentrations for 4 days. n = 3 technical replicates. (I) Representative images of organoids from Vil-Cre and cKO crypts cultured with noggin at indicated concentrations (100, 200, 400, 600 and 800 ng/mL) for 4 Days in Panel H.
-
Figure 5—source data 1
Source data for Figure 5.
- https://doi.org/10.7554/eLife.29538.029
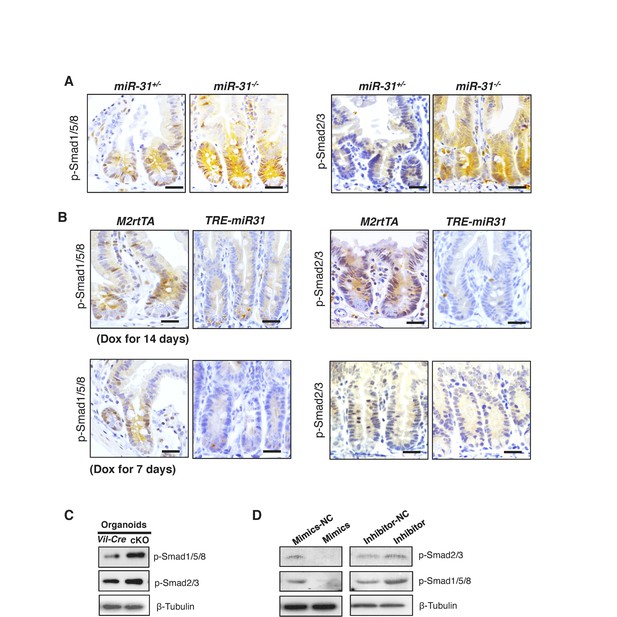
MiR-31 represses BMP and TGFβ signaling pathways.
(A) Immunohistochemistry for p-Smad1/5/8 and p-Smad2/3 in jejunum from miR-31+/− and miR-31−/− mice at 2 months of age. Scale bar: 25 μm. (B) Immunohistochemistry for p-Smad1/5/8 and p-Smad2/3 in jejunum from M2rtTA and TRE-miR31 mice following 7 days and 14 days of Dox induction. Scale bar: 25 μm. (C) Western blotting for p-Smad1/5/8 and p-Smad2/3 in cultured organoids from Vil-Cre and miR-31 cKO crypts. n = 4 biological replicates. β-Tubulin was used as a loading control. (D) Western blotting for p-Smad2/3 and p-Smad1/5/8 in HCT116 cells under treatment of control (mimics-NC) and miR-31 mimics, or control (inhibitor-NC) and anti-miR31. β-Tubulin was used as a loading control, which is identical to Figure 4—figure supplement 1C.
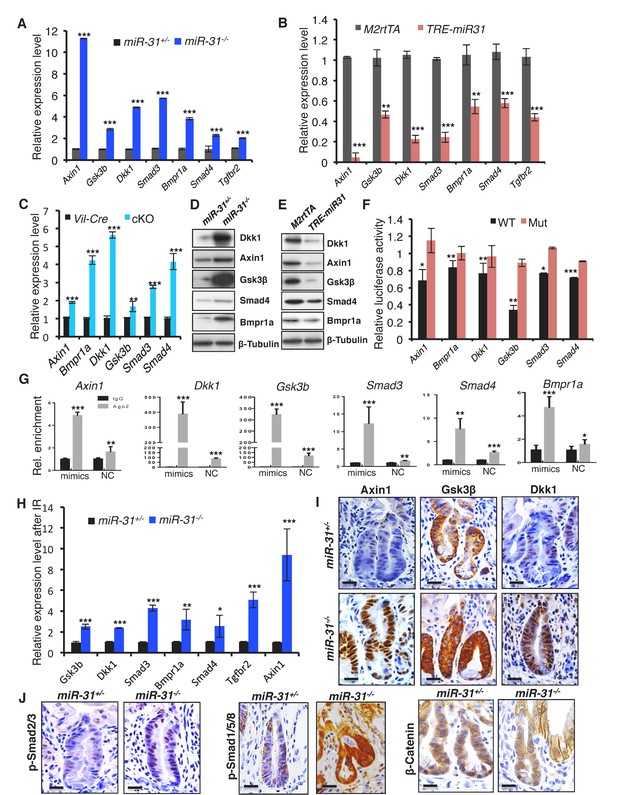
Identification of miR-31 target genes in intestinal epithelium.
(A, B) qRT-PCR analysis for Axin1, Gsk3b, Dkk1, Smad3, Bmpr1a, Smad4 and Tgfbr2 in miR-31+/− and miR-31−/− intestine (A), as well as M2rtTA and TRE-miR31 intestine following 2 weeks of Dox induction (B). **p<0.01; ***p<0.001. (C) qRT-PCR analysis for Axin1, Bmpr1a, Dkk1, Gsk3b, Smad3, and Smad4 in Vil-Cre and cKO intestine. **p<0.01; ***p<0.001. (D) Western blotting for Axin1, Gsk3β, Dkk1, Smad4, and Bmpr1a in miR-31+/− and miR-31−/− intestine. β-Tubulin was used as a loading control, which is identical with Figure 5A. n = 3 biological replicates. (E) Western blotting for Axin1, Gsk3β, Dkk1, Bmpr1a and Smad4 in M2rtTA and TRE-miR31 intestine following 2 weeks of Dox induction. β-Tubulin was used as a loading control. n = 3 biological replicates. (F) Ratio of luciferase activity of miR-31 mimics versus scramble RNA in wild type and mutant 3’UTR constructs based on 3 independent experiments. *p<0.05; **p<0.01; ***p<0.001. (G) RNA crosslinking, immunoprecipitation, and qRT-PCR (CLIP-PCR) assay for Dkk1, Axin1, Gsk3b, Smad3, Smad4 and Bmpr1a upon Ago2 antibody immunoprecipitates in response to miR-31 mimics and scramble RNA (NC). IgG was used as a negative control. (H) qRT-PCR analysis for Axin1, Gsk3b, Dkk1, Smad3, Bmpr1a, Smad4 andTgfbr2 in miR-31+/− and miR-31−/− intestine 4 days post 12 Gy γ-IR. n = 3 biological replicates. *p<0.05; **p<0.01; ***p<0.001. (I) Immunohistochemistry for Axin1, Gsk3β and Dkk1 in miR-31+/− and miR-31−/− intestinal crypts 4 days post 12 Gy γ-IR. Scale bar: 25 μm. (J) Immunohistochemistry for p-Smad2/3, p-Smad1/5/8 and β-Catenin in miR-31+/− and miR-31−/− intestinal crypts 4 days post 12 Gy γ-IR. Scale bar: 25 μm.
-
Figure 6—source data 1
Source data for Figure 6.
- https://doi.org/10.7554/eLife.29538.034

Identification of miR-31 target genes.
(A) MiR-31 binding sites in 3’UTR of these putative target genes. (B) Mutant binding sites of genes, Axin1, Bmpr1a, Dkk1, Gsk3b, Smad3 and Smad4.
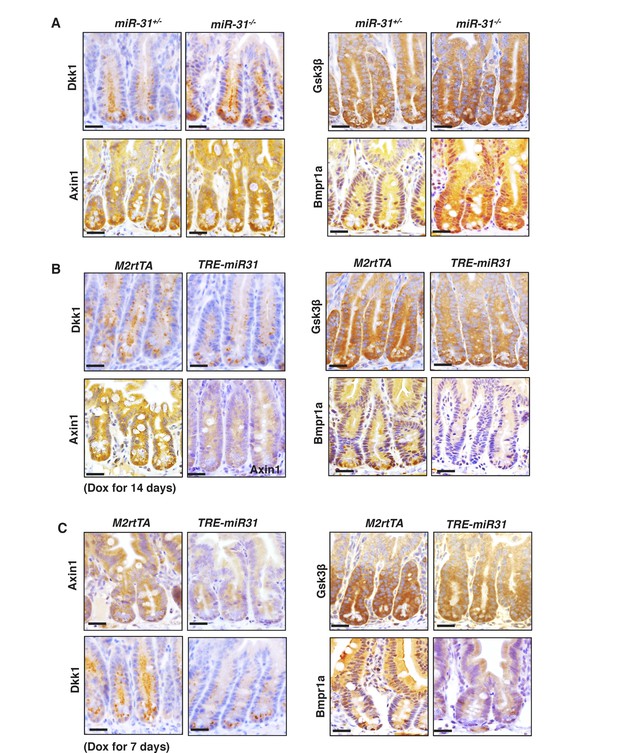
Identification of miR-31 target genes.
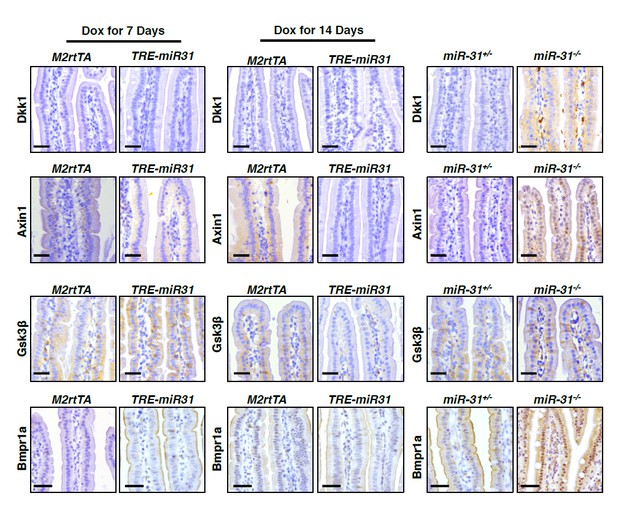
(A) Immunohistochemistry for Dkk1, Gsk3β, Axin1 and Bmpr1a in jejunum from miR-31+/− and miR-31−/− mice at 2 months of age. Scale bar: 25 μm. (B) Immunohistochemistry for Dkk1, Gsk3β, Axin1 and Bmpr1a in jejunum from M2rtTA and TRE-miR31 mice following 7 days and 14 days of Dox induction. Scale bar: 25 μm.

Identification of miR-31 target genes.
(A) Western blotting for Gsk3β, Dkk1, Axin1, Smad4 and Bmpr1a in cultured organoids from Vil-Cre and miR-31 cKO crypts. n = 4 technical replicates. β-Tubulin was used as a loading control. (B) Western blotting for Dkk1, Axin1, Gsk3β, Smad4 and Bmpr1a in HCT116 cells under treatment of control (mimics-NC) and miR-31 mimics, or control (inhibitor-NC) and anti-miR-31. β-Tubulin was used as a loading control, which is identical to Figure 4—figure supplement 1C and Figure 5—figure supplement 1D.
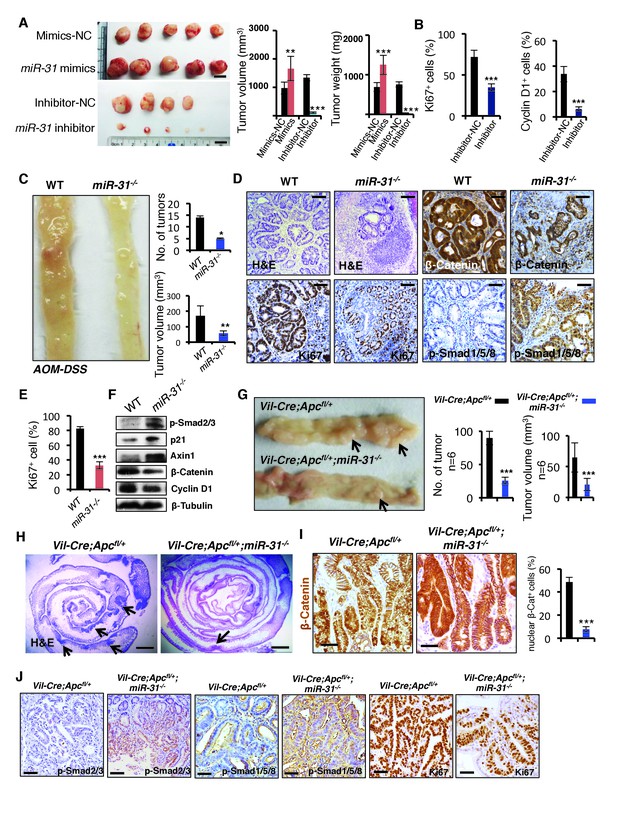
MiR-31 promotes tumor growth in vivo.
(A) Gross appearance of tumors of HCT116 colorectal cancer cell xenograft 30 days post transplantation. HCT116 colorectal cancer cells were transfected with mimics-NC or miR-31 mimics, and inhibitor-NC or anti-miR-31 (inhibitor) for 36 hrs before xenograft. NC-mimics, n = 5; miR-31 mimics, n = 5; NC-inhibitor, n = 4; anti-miR-31, n = 5. Quantification of tumor volume and tumor weight at indicated conditions. **p<0.01; ***p<0.001. Scale bar: 1 cm. (B) Quantification of Ki67+ and Cyclin D1+ cells in NC-inhibitor and miR-31 inhibitor treated tumors in Figure 7—figure supplement 1B. ***p<0.001. (C) Representative photograph of distal colon resected from WT and miR-31−/− mice at the end of AOM-DSS protocol. Frequency and tumor size of inflammation-driven colorectal adenomas in mice treated with the AOM-DSS protocol, with or without miR-31 deletion. n = 6 mice per group, *p<0.05; **p<0.01. (D) H&E, and immunohistochemistry for Ki67, β-Catenin and p-Smad1/5/8 in adenomas of WT and miR-31−/− mice resulting from AOM-DSS treatment. Scale bar: 100 μm. (E) Quantification of Ki67+ cells in Panel D. ***p<0.001. (F) Western blotting for p-Smad2/3, p21, Axin1, β-Catenin, Cyclin D1 in adenomas of WT and miR-31−/− mice resulting from AOM-DSS treatment. β-Tubulin was used as a loading control. (G) Representative photograph of intestine resected from Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− mice at 6 months of age. Arrows point to tumors. Quantification of tumor number and tumor volume in intestines from these mice. n = 6 biological replicates. ***p<0.001. (H) Representative histology of intestine resected from Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− mice at 6 months of age. Arrows point to tumors. Scale bar: 2.5 mm. (I) Immunohistochemistry for β-Catenin and quantification of nuclear β-Catenin positive cells in Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− tumors. (Black, Vil-Cre;Apcfl/+; Blue, Vil-Cre;Apcfl/+;miR-31−/−). n = 6 biological replicates. Scale bar: 50 μm. ***p<0.001. (J) Immunohistochemistry for p-Smad2/3, p-Smad1/5/8 and Ki67 in Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− tumors. Scale bar: 50 μm.
-
Figure 7—source data 1
Source data for Figure 7.
- https://doi.org/10.7554/eLife.29538.037
-
Figure 7—source data 2
Source data for Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.29538.038

MiR-31 promotes tumor growth.
(A) In vitro MTT proliferation assay of human colorectal cancer cell lines, HCT116, SW480 and LOVO upon transfection of miR-31 inhibitor (anti-miR-31), and inhibitor-NC, as well as miR-31 mimics and mimics-NC. n = 3 technical replicates. (B) H&E, and immunohistochemistry for Ki67, Cyclin D1, β-Catenin, p-Smad1/5/8 and p-Smad2/3 in inhibitor-NC and miR-31 inhibitor treated tumors in Figure 7A. n = 3 biological replicates. Scale bar: 50 μm. (C) Quantification of p-Smad2/3+, p-Smad1/5/8+ and Ki67+ cells in Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− tumors shown in Figure 7J. n = 3 biological replicates. ***p<0.001. (D) Immunohistochemistry for Dkk1, Axin1, Gsk3β, Smad4 and Bmpr1a in Vil-Cre;Apcfl/+ and Vil-Cre;Apcfl/+;miR-31−/− tumors. n = 4 biological replicates. Scale bar: 50 μm.
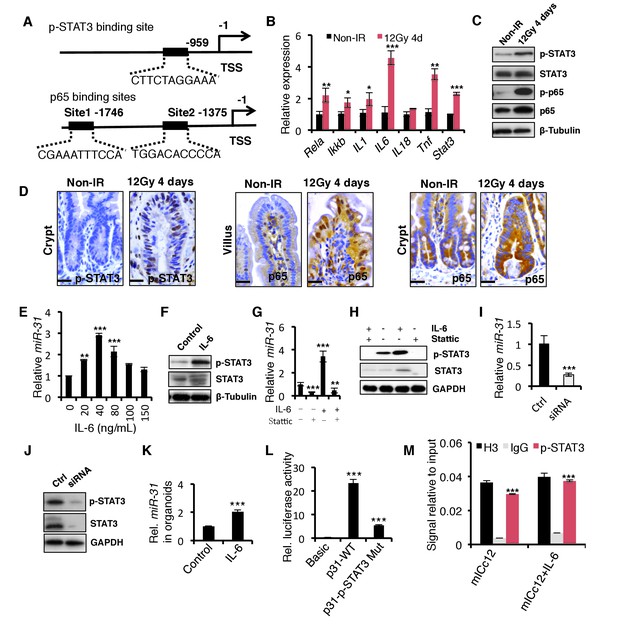
The STAT3 pathway mediates the induction of miR-31 caused by γ-IR.
(A) The schematic diagram showed two potential p65 binding sites and one p-STAT3 binding site in the miR-31 promoter. (B) qRT-PCR analysis for Rela, Ikk-b, IL-1, IL-6, IL-18, Tnf and Stat3 in the intestinal epithelium 4 days after exposure to 12 Gy γ-IR, relative to non-irradiated controls. n = 3 biological replicates. *p<0.05, **p<0.01, ***p<0.001. (C) Western blotting for STAT3, p-STAT3, p65 and p-p65 in the intestinal epithelium 4 days after exposure to 12 Gy γ-IR, relative to non-irradiated controls. n = 3 biological replicates. (D) Immunohistochemistry for p-STAT3 and p65 in control and the intestinal epithelium 4 days after exposure to 12 Gy γ-IR. n = 3 biological replicates. Scale bar: 25 μm. (E) qRT-PCR for miR-31 in mouse intestinal epithelial cell line (mICc12) in response to IL-6 with concentrations of 20, 40, 80, 100 and 150 ng/mL. n = 3 technical replicates. **p<0.01; ***p<0.001. (F) Western blotting for STAT3 and p-STAT3 in mICc12 cells in response to 40 ng/mL IL-6. (G) qRT-PCR analysis for miR-31 in mICc12 cells treated with IL-6 and STAT3 inhibitor, Stattic. **p<0.01; ***p<0.001. (H) Western blotting for p-STAT3 in mICc12 cells treated with IL-6 and Stattic. (I) qRT-PCR analysis for miR-31 in mICc12 cells treated with Stat3 siRNA. ***p<0.001. (J) Western blotting for STAT3 and p-STAT3 in mICc12 cells treated with STAT3 siRNA. (K) qRT-PCR analysis for miR-31 in cultured organoids treated with IL-6. n = 4 technical replicates. ***p<0.001. (L) Luciferase activity in lysates of mICc12 cells transfected with luciferase reporter plasmids of pGL3-basic empty vector (basic), wild type miR-31 promoter or mutant promoter with mutation of p-STAT3 binding sites. ***p<0.001. (M) Chromatin immunoprecipitation (ChIP) assay carried out on mICc12 cells using antibodies against p-STAT3 and Histone 3. The antibody against Histone 3 was used as a positive control. The enrichment of p-STAT3 binding to miR-31 promoter was quantified using qPCR. ***p<0.001.
-
Figure 8—source data 1
Source data for Figure 8.
- https://doi.org/10.7554/eLife.29538.040
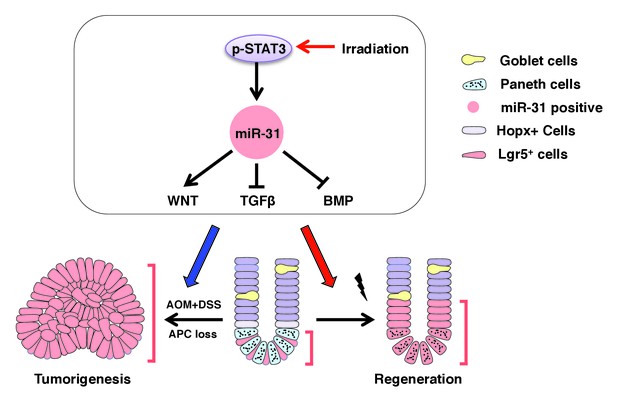
The miR-31 working model in intestinal epithelial regeneration and tumorigenesis.
https://doi.org/10.7554/eLife.29538.041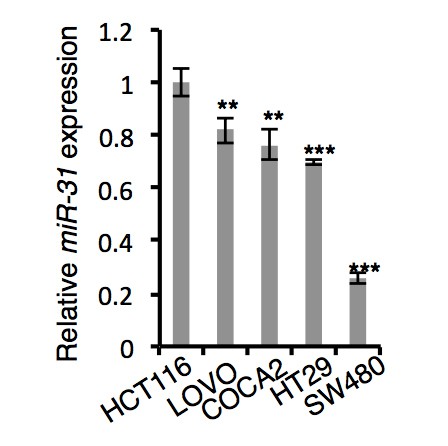
qRT-PCR analysis for miR-31 in HCT116, LOVO, COCA2, HT29 and SW480 colorectal cancer cells.
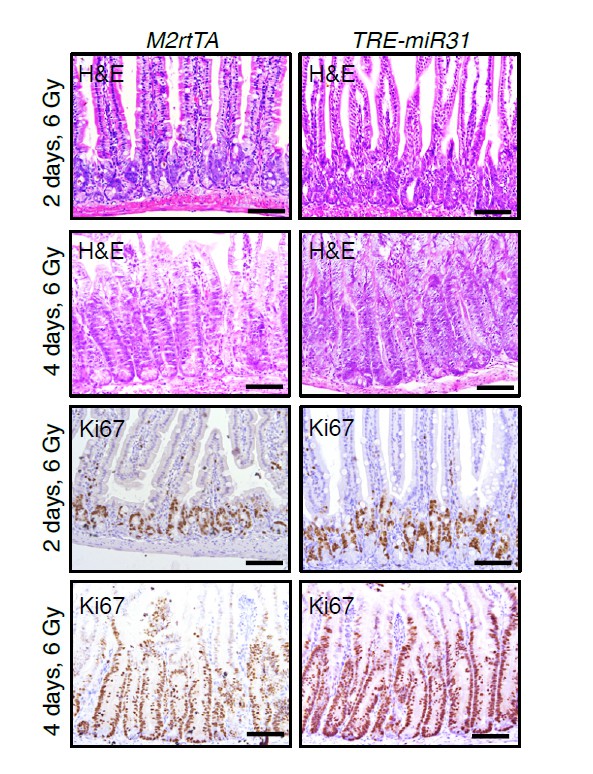
H&E and immunohistochemistry for Ki67 in intestines from M2rtTA control and TRE-miR31 mutant mice 2 and 4 days post 6 Gy irradiation.
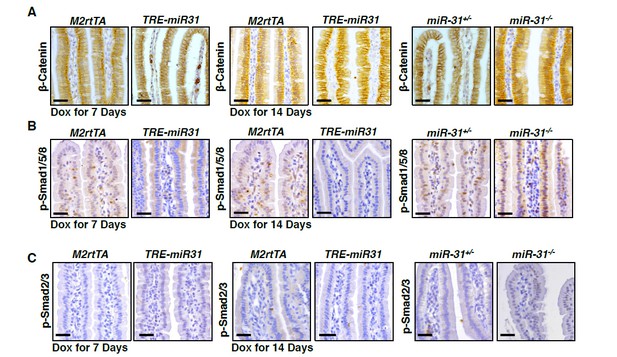
Immunohistochemistry for β-Catenin (A), p-Smad1/5/8 (B), and p-Smad2/3 (C) in intestinal villi from M2rtTA and TRE-miR31 mice, as well as miR-31+/- and miR-31-/- mice.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29538.042