Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites
Figures
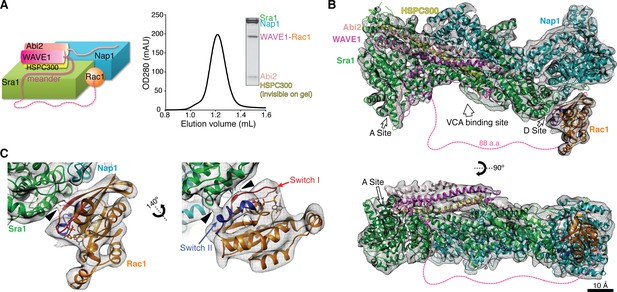
Cryo-EM structure of the WRC-Rac1 complex.
(A) Schematic representation (left) and size-exclusion chromatography profile (right) of the ΔWRC230-Rac1 complex, with Coomassie blue-stained SDS-PAGE gel of the peak fraction shown. Dashed line indicates the tethering sequence between WAVE1 meander region and Rac1. (B) Overall EM density and the fitted model of the ΔWRC230-Rac1 complex. WAVE1(130–147, 161–230) and (GGS)6 leading to Rac1 have no EM density and are shown as dotted lines. (C) Close-up views of Rac1 fitted into the extra density not accounted for by the WRC. Black arrow heads indicate close contacts between Rac1 and the WRC.
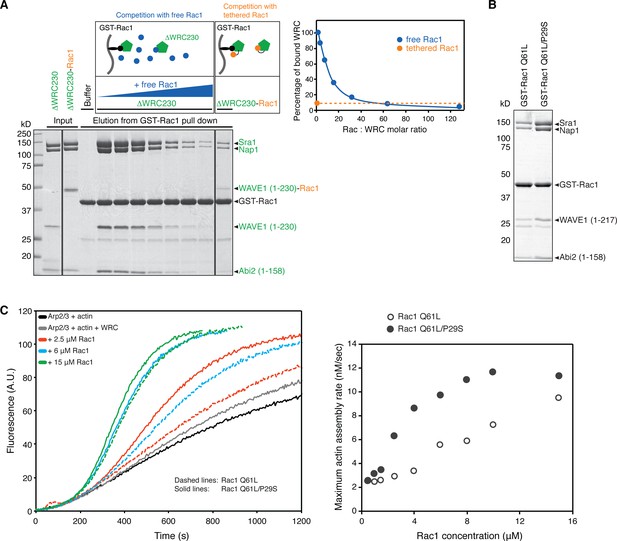
A covalent ΔWRC230-Rac1 complex.
(A) Coomassie blue-stained SDS-PAGE gel of a competition pull-down assay (schematic shown above the gel). GST-Rac1 was used to pull down ΔWRC230 with various ratios of free Rac1 as a competitor (indicated by the blue triangle above the gel), or to pull down ΔWRC230-Rac1 with the tethered Rac1 as an intrinsic competitor (right-most gel lane). Quantification of the WRC bands on the gel is plotted on the right, with the dashed orange line indicating the concentration of free Rac1 (blue curve) needed to achieve the same level of competition as the tethered Rac1 (orange circle). All Rac1 used here is residues 1–177 and has the Q61L/P29S mutations. (B) Coomassie blue-stained SDS-PAGE gel showing GST-Rac1 (Q61L or Q61L/P29S as indicated; both loaded with GMPPNP) pull-down results for ΔWRC217. (C) Actin assembly assays using Rac1 (1-188) with the indicated mutations. Reactions contain 4 µM actin (5% pyrene labeled), 10 nM Arp2/3 complex, 100 nM WRC217VCA, and the indicated amounts of Rac1 loaded with GMPPNP. On the left are representative actin assembly curves. On the right are the derived maximum actin assembly rates at different Rac1 concentrations (Doolittle et al., 2013).
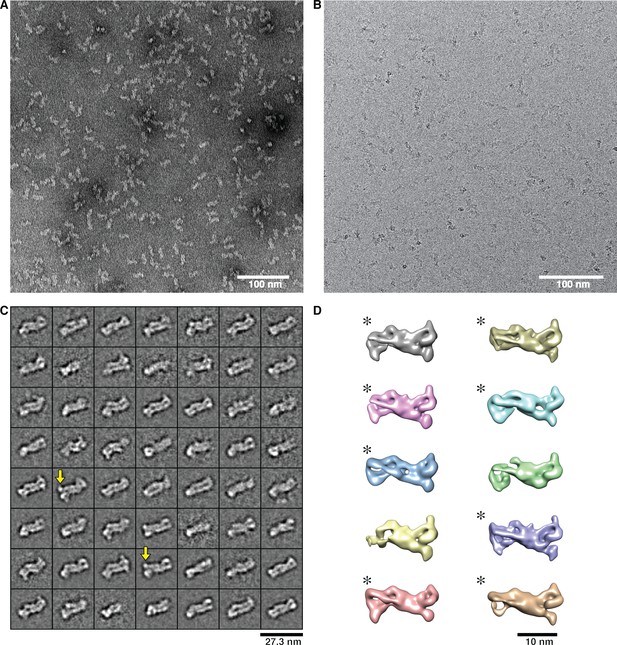
Representative EM images as well as class averages and 3D maps of the ΔWRC230-Rac1 complex.
(A) EM image of negatively stained complex. (B) Cryo-EM image of vitrified complex. (C) The 56 averages obtained by subjecting 11,874 particle images to the iterative stable alignment and clustering (ISAC) procedure (Yang et al., 2012). Yellow arrows indicate extra density in addition to the WRC. (D) After 3D classification of 39,145 particles, 8 classes (indicated by asterisks) showing extra density for Rac1 were combined for subsequent refinement.
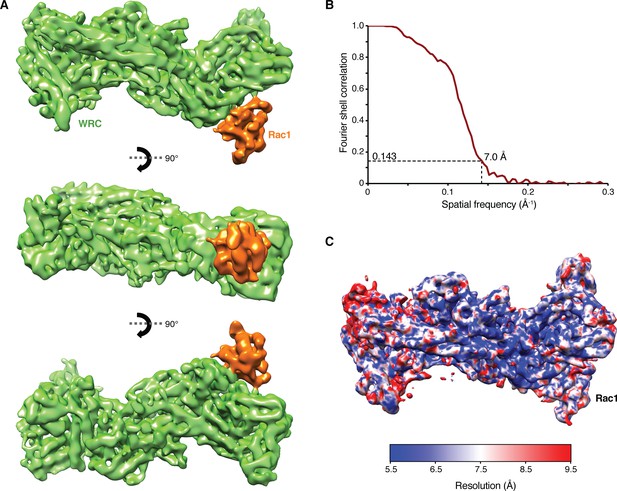
Cryo-EM density map for the ΔWRC230-Rac1 complex.
(A) Different views of the final density map. (B) Fourier shell correlation (FSC) curve obtained from separate refinement of two halves of the whole data set. Using FSC = 0.143 as a cut-off, the density map has a resolution of 7.0 Å. (C) Local resolution of the density map calculated with ResMap (Kucukelbir et al., 2014).
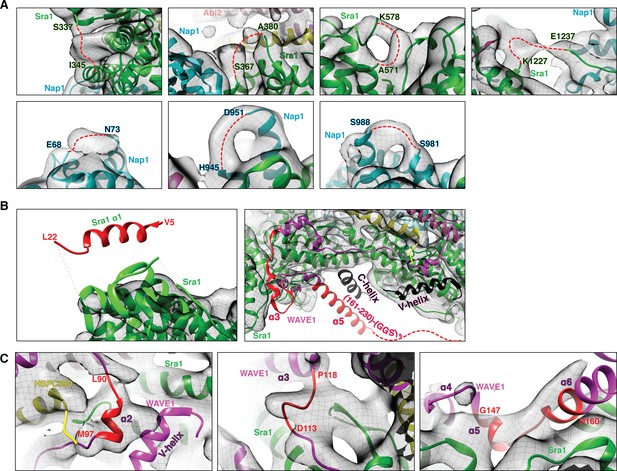
Fitting of the WRC crystal structure to the EM density.
(A) EM density for surface loops (represented by red dashed lines) missing in the mini-WRC crystal structure. (B) Elements observed in the crystal structure but lacking densities in the EM reconstruction, with elements present in the ΔWRC230-Rac1 construct in red and elements absent from the ΔWRC230-Rac1 construct in black. (C) Several structural elements (red) in the meander sequence have EM densities suggestive of different orientations.
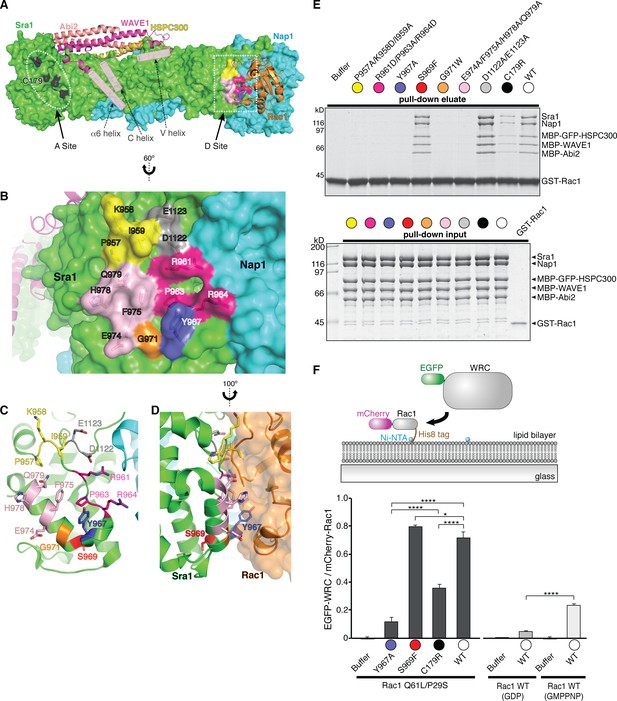
Rac1 binds to two distinct sites on the WRC.
(A) Overall structure of the ΔWRC230-Rac1 complex showing the locations of the A site and D site. Sra1 and Nap1 are shown in surface representation; WAVE1, Abi2, HSPC300 and Rac1 are shown in ribbon representation. Semitransparent pink cylinders are used to refer to the positions of WAVE1 α6 helix (without observable EM density) and the V and C helices (only present in the mini-WRC). Residues in the A Site whose mutation was previously shown to impair Rac1 binding are black. Surface patches in the D Site mutated here are colored as in (B–F). (B) Close-up view of the D Site with semitransparent surface and key side chains shown under the surface. Rac1 was removed for clarity. (C–D) top and side views of the D Site, showing the location of the buried residue S969 (red). (E) Coomassie blue-stained SDS-PAGE gel showing GST-Rac1-GMPPNP (WT) pull-down assay of WRCs bearing the indicated mutations in Sra1 at the Rac1-binding sites. Top panel shows elution of material retained on the beads, bottom panel shows input control. See non-equilibrium GST pull down assay section of Experimental Methods for details of constructs. (F) Top: schematic of membrane recruitment assay using supported lipid bilayers. WRC construct was same as in panel E. Bottom: quantification of the EGFP-WRC/mCherry-Rac1 signals for each reaction used 10 separate TIRF images, with five each collected from two independent experiments. Error bars represent standard error, and p values were calculated by pairwise comparison using Tukey’s test (*p<0.05, and ****p<0.00005).
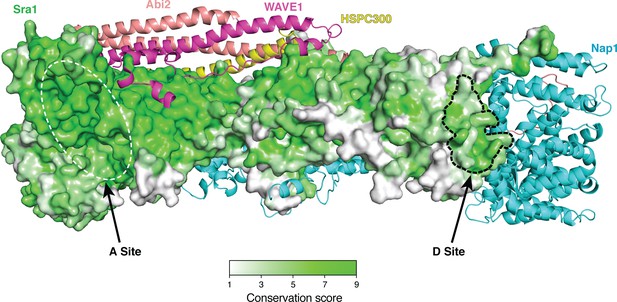
Surface conservation of the Sra1 subunit in the ΔWRC230-Rac1 complex (Rac1 removed for clarity), with a gradient of color from green to white representing the most conserved surface residues (ConSurf score = 9) to the least conserved residues (ConSurf score = 1) (Ashkenazy et al., 2010).
https://doi.org/10.7554/eLife.29795.009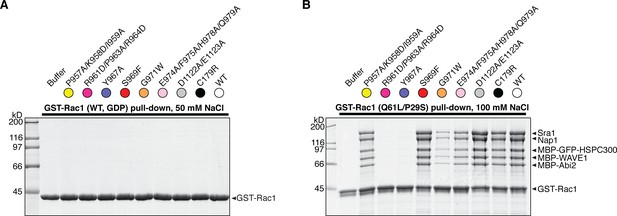
GST-Rac1 pull-down of different WRC mutants.
Coomassie blue-stained SDS-PAGE gels are shown. Each mutant is color-coded on top of the gel as in Figure 2E. (A) Pull-down using wild-type GST-Rac1 loaded with GDP under identical conditions as in Figure 2E. (B) Pull-down using GST-Rac1 (Q61L/P29S) and higher salt concentration (100 mM NaCl). Experiments were performed in parallel with those in main Figure 3, and the same input control gel (Figure 3E, lower panel) applies here.
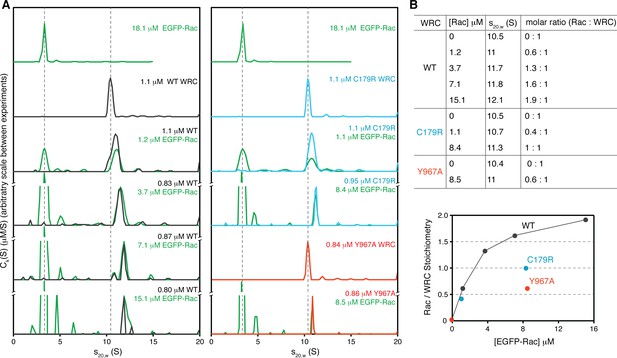
Stoichiometry of the Rac1/WRC complex determined by analytical ultracentrifugation.
(A) Sedimentation profiles (ck(s) distributions) of indicated samples at various concentrations (green for EGFP-Rac1 (Q61L/P29S), black for WT ΔWRC230, blue for C179R, and red for Y967A). Dashed lines are used as a reference to indicate the sedimentation coefficients of the WRC alone and EGFP-Rac alone, respectively. (B) Sedimentation coefficient and stoichiometry of Rac1/WRC in the WRC peak.
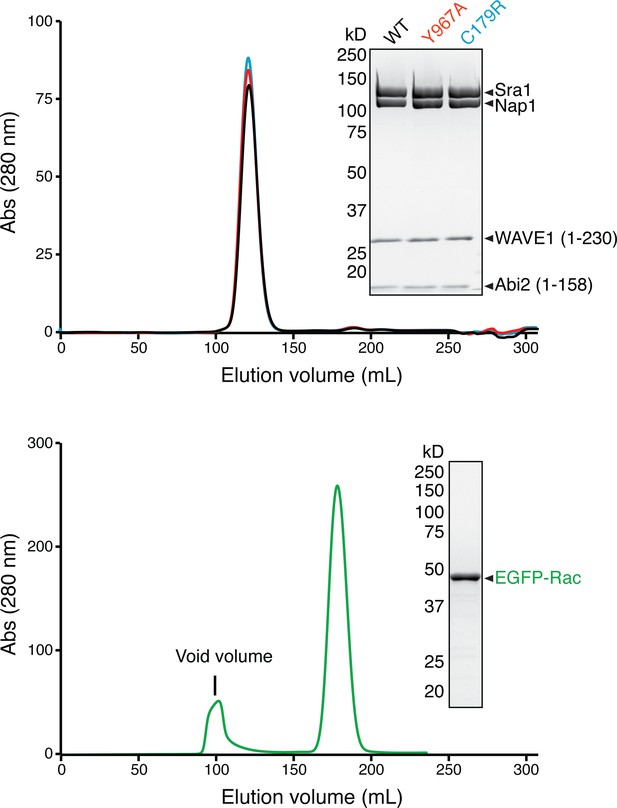
Gel filtration chromatography profiles and Coomassie blue-stained SDS-PAGE gels of the ΔWRC230 and the EGFP-Rac1 samples used in the AUC experiments.
https://doi.org/10.7554/eLife.29795.012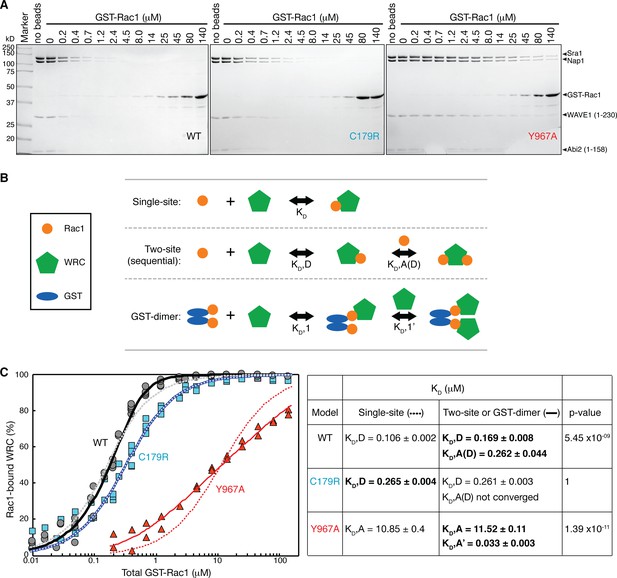
Quantification of the binding affinity by equilibrium pull-down assay.
(A). Representative Coomassie-blue-stained SDS-PAGE gels of the supernatant samples in equilibrium pull down experiments involving GST-Rac1 (Q61L/P29S) binding to ΔWRC230. (B) Schematic representation of the three successful equilibrium models used to fit the binding isotherms. (C) Binding isotherms fit to the indicated models. For WT and C179R ΔWRC230, results for single-site (dotted curves) and two-site (solid curves) models are shown. For Y967A WRC, results for single-site (dotted curves) and GST-dimer models (solid curves) are shown. The data were pooled from multiple independent experiments (5 experiments for WT and C179R, and 2 experiments for Y967A) covering various concentration ranges. Derived dissociation constants are shown in the right table, with KD,D representing the dissociation constant for the D Site, KD,A for the A Site, KD,A(D) for the A Site when the D Site is occupied by Rac1, and KD,A’ for the A Site of the second WRC in the GST-dimer model. Standard errors of the fit for the derived KD values are shown. p values were obtained by F-test; the most appropriate dissociation constants based on the p values are shown in bold. See Figure 5—figure supplement 2 and Methods for additional details.
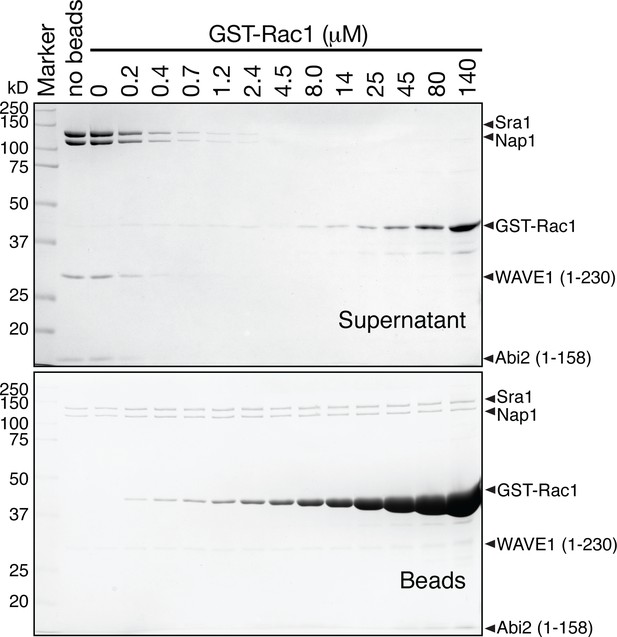
Representative Coomassie blue-stained SDS-PAGE gels of the supernatant (top gel) and the beads samples (bottom gel) in an equilibrium pull-down assay involving GST-Rac1 (Q61L/P29S) binding to ΔWRC230.
The top gel is identical to the top gel in Figure 5A. Each reaction contained 30 μL of beads and 70 μL of supernatant. Samples for the top gel (supernatant) were prepared by mixing 40 μL of the supernatant with 8 μL of a 6X SDS PAGE loading buffer, with 15 μL of the samples (thus 12.5 μL of the supernatant) loaded to each lane. Samples for the bottom gel (beads) were prepared by adding 60 μL of a 2X SDS PAGE loading buffer to the remaining 60 μL of reaction (containing 30 μL of supernatant and 30 μL of beads), with 2.5 μL of the mixture (thus 0.625 μL of the supernatant and 0.625 μL of beads) loaded in each lane.
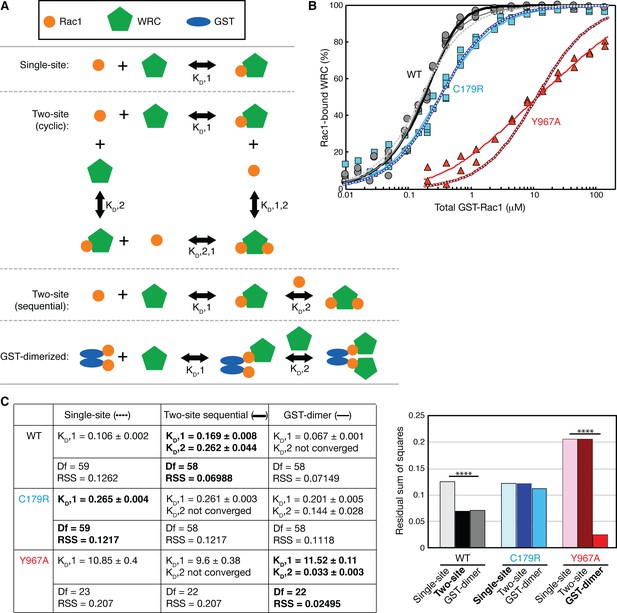
Quantification of the binding affinity by equilibrium pull-down assay involving GST-Rac1 (Q61L/P29S) bindingΔWRC230.
(A) Schematic representation of all equilibrium models used to fit the binding isotherms. (B) Binding isotherms fit to the indicated models, with dotted curves for single-site, thick solid curves for two-site sequential, and thin solid lines for GST-dimerization models. The data were pooled from multiple independent experiments (5 experiments for WT and C179R, and 2 experiments for Y967A) covering various concentration ranges. (C) The derived dissociation constant KD (μM, shown with standard errors of the fit) and the fitting results used for F-test (Df for degree of freedom, RSS for residual sum of squares) are shown in the left table. F-test results are shown on the right with column colors matched to the curve colors in (B) (**** for p<0.0000001). The results for the best fits based on the p values are shown in bold.
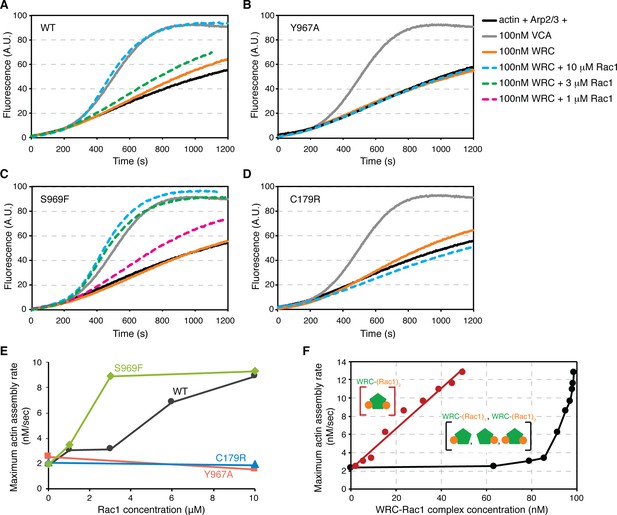
Activation of the WRC requires both Rac1-binding sites.
(A–D) Actin assembly assays of the wild-type WRC230VCA (A), Sra1 Y967A (B), Sra1 S969F (C), and Sra1 C179R (D), with corresponding reaction conditions shown on the right. Reactions contain 4 μM actin (5% pyrene labeled), 10 nM Arp2/3 complex, 100 nM WRC230VCA or VCA, and indicated amount of Rac1 Q61L loaded with GMPPNP. (E) Maximum actin assembly rates at different Rac1 concentrations derived from data in (A–D) (Doolittle et al., 2013). (F) Maximum actin assembly rates at different concentrations of distinct WRC-Rac1 species, obtained from the data shown in Figure 1—figure supplement 1C for WRC activation by Rac1 Q61L/P29S. Concentrations of the indicated species of WRC-Rac1 complexes (shown as cartoons) formed at different Rac1 concentrations in the actin assembly assay were calculated using DynaFit (Kuzmic, 1996) by using a two-site cyclic model shown in Figure 5—figure supplement 2A, using KDD of 0.265 μM and KDA of 11.52 μM (derived from Figure 5, assuming no cooperativity between the A and D Site, since the actin assembly assays were performed in solution). Black data plot the summed concentration of all WRC-Rac1 species, while red data plot only WRC-(Rac1)2. A linear fit of the latter data yields r2 of 0.98.
Gel filtration chromatography profiles of the WRC230VCA samples used in the actin assembly assays.
https://doi.org/10.7554/eLife.29795.017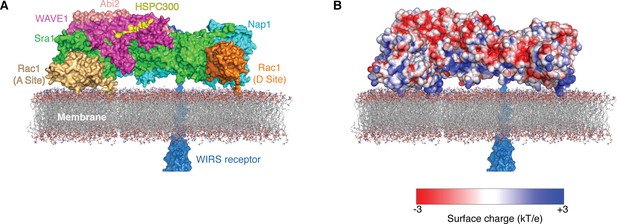
Two Rac1 molecules bind to the WRC on the membrane.
Surface representation (A) and surface charge representation (B) (calculated using APBS in Pymol) (Dolinsky et al., 2007) show how the WRC can be oriented on the membrane to simultaneously interact with multiple signaling molecules, including two Rac1 molecules (gold and orange) anchored on the membrane through prenylation of their C-terminal tails, WIRS-containing surface receptors (dark blue), and acidic phospholipids through electrostatic interaction (represented by membrane). The Rac1 at the D site is taken from the ΔWRC230-Rac1 complex structure determined here; Rac1 at the A site was manually docked in a plausible orientation to contact Sra1 residues 179, 190, 434, 626 and 632 and place the C terminus near the membrane. WIRS-containing receptors could approach the WRC from the back (shown) or the front (not shown).
Additional files
-
Supplementary file 1
Sequence alignments of Sra1 proteins from representative organisms.
Residues in the A Site mutated previously are colored grey. Surface residues shown in Figure 2A–B are highlighted using the same color scheme. For clarity, insertions in three of the sequences were hidden and indicated by amino acids colored in cyan.
- https://doi.org/10.7554/eLife.29795.019
-
Supplementary file 2
Scripts used in the DynaFit program to fit the equilibrium pull-down data.
- https://doi.org/10.7554/eLife.29795.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29795.021





