The CDK-PLK1 axis targets the DNA damage checkpoint sensor protein RAD9 to promote cell proliferation and tolerance to genotoxic stress
Figures
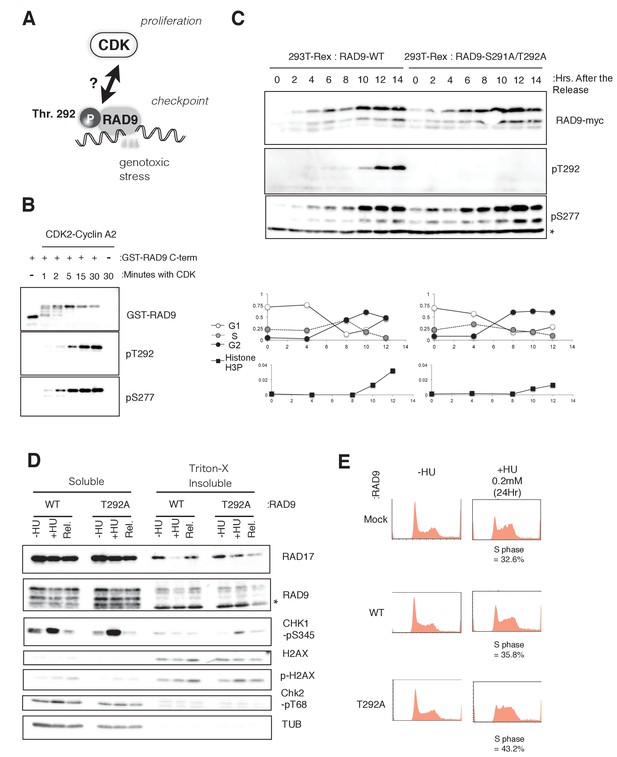
CDK phosphorylates threonine 292 of RAD9.
(A) Schematic of the aim of this manuscript. (B) The recombinant GST-tagged C-terminal (a.a. 266–391) portion of RAD9 was mixed with the purified active CDK2-CyclinA2 complex. Western blotting was performed using the α-RAD9 antibody and the α-pT292 (pT292) and α-phospho-Ser277 (pS277) RAD9 antibodies. (C) Top: The thymidine block and release experiment was performed on HEK293A-T-REx cell lines stably expressing RAD9-WT-mH or RAD9-S291A/T292A-mH. Doxycyclin (0.5 µg/ml) was added during the second thymidine block. The western blotting analysis of cell lysates obtained at the indicated time points is shown, and α-myc, α-pT292 and α-pS277 antibodies were used. The asterisk (*) shows a nonspecific signal. Bottom: The cell cycle profiles were quantified by a flow cytometer analysis. The G1, S, and G2 phases were quantified via propidium iodide staining, and the M phase cells were quantified using an antibody against the phospho-serine 10 of histone H3. (D) A chromatin fractionation assay (Ohashi et al., 2014; Zou et al., 2002) was performed in HEK293A-T-REx cell lines stably expressing RAD9-WT-mH (WT) or RAD9-S291A/T292A-mH (T292A). Cells were grown in media containing 1.5 mM hydroxyurea for 16 hr (+HU) and then released from the hydroxyurea arrest for 1 hr (Rel.). A western blotting analysis is shown, and α-RAD17, α-myc (RAD9), α-phospho-serine 345 of CHK1 (CHK1-pS345), α-phospho-serine 139 of Histone H2AX (p-H2AX), α-H2AX, α-phospho-threonine 68 of CHK2 (CHK2-pT68), and α-tubulin were used. (E) The flow cytometry analysis was performed with U2OS T-REx cells stably expressing RAD9-WT-mH (WT) or RAD9-S291A/T292A-mH (T292A), and the host U2OS T-REx cells (Mock). The cells were grown in media containing 0.2 mM hydroxyurea for 24 hr. The populations of cells showing the S phase peaks were quantified and indicated below the flow cytometer profiles. See also Figure 1—figure supplement 1. CDK phosphorylates threonine 292, and construction of RAD9-WT or -S291A/T292A(T292A) expressing stable cell lines.
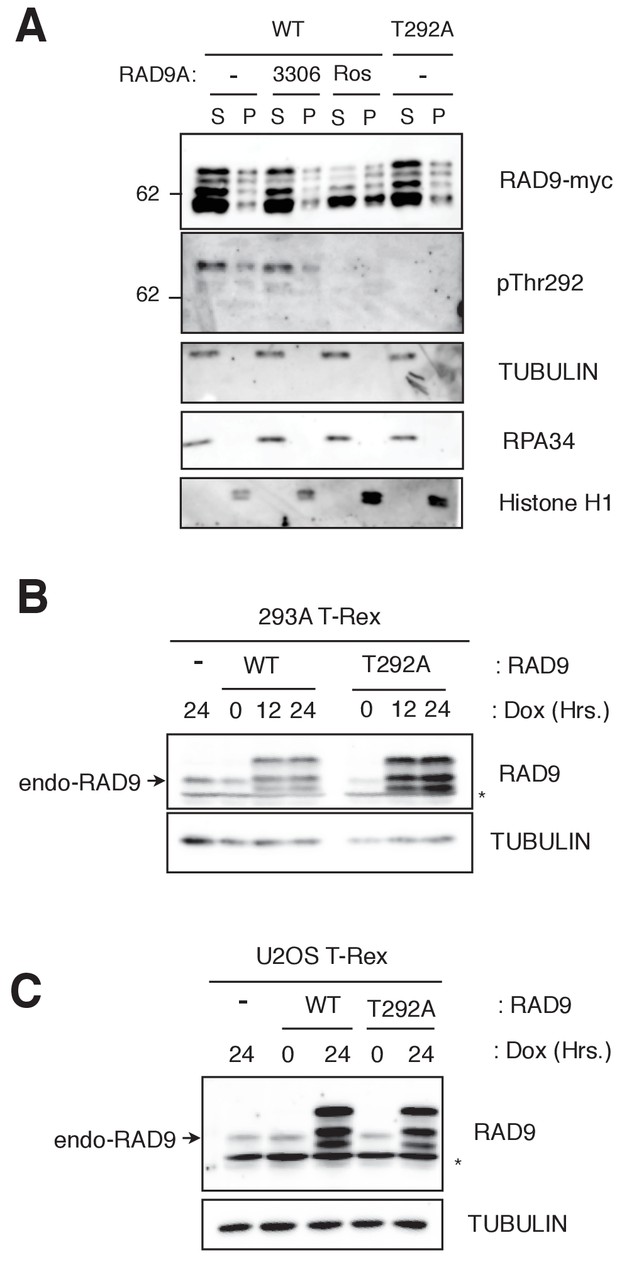
CDK phosphorylates threonine 292, and construction of the RAD9-WT and -S291A/T292A(T292A) expressing stable cell lines.
(A) HEK293A cells were transfected with a plasmid that expresses RAD9-WT-mH (WT) or RAD9-T292A-mH (T292A) under the CMV promoter, cultured in the presence or absence of RO3306 (5 µM, indicated as 3306) or Roscovitin (20 µM, indicated as Ros), and subjected to a fractionation assay. Western blotting analyses using α-myc (RAD9-myc), α-pThr292 (pThr292), α-tubulin (TUBULIN), α-RPA32 (RPA) and α-Histone H1 (Histone H1) are shown. (B), (C) Stable cell lines expressing RAD9-WT-mH (WT) or RAD9-S291A/T292A-mH (T292A) were constructed in HEK293A-T-REx (B) or U2OS T-REx (C) cells. A western blotting analysis was performed to compare the expression levels of endogenous RAD9 and the expressed myc-tagged RAD9. RAD9 was expressed under CMV promoter control via the tetO system. α-RAD9 (Bethyl) was used for the western blot analysis. Doxycyclin (Dox) was added to induce the promoter. Asterisks (*) are nonspecific signals. For the controls, host cell lines (-) were used.
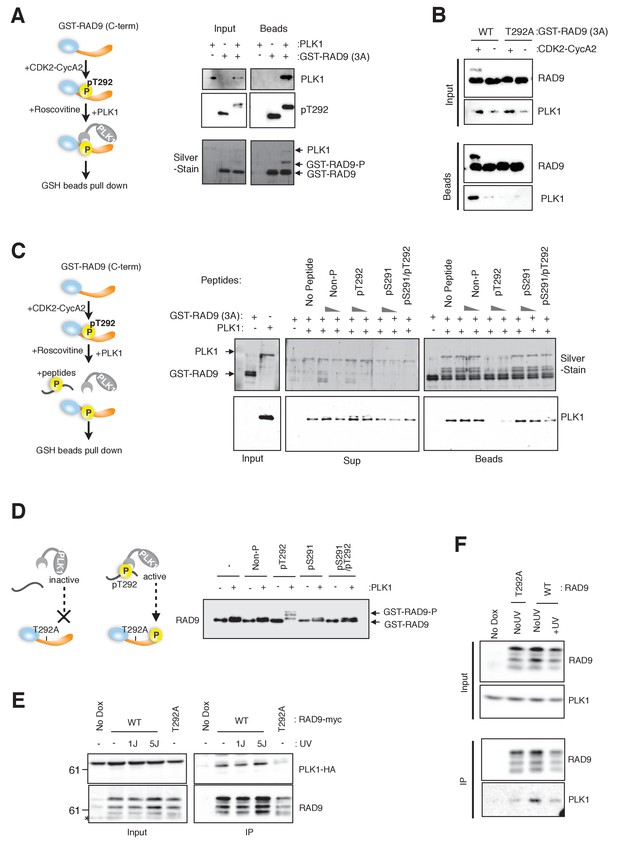
CDK-dependent phosphorylation of threonine 292 of RAD9 accommodates and activates PLK1.
(A) The RAD9 C-terminus co-precipitates with PLK1. GST-RAD9 (3A: S277A, S328A, S336G) (30 pmol) was pre-incubated with CDK2-Cyclin A2 (0.5 pmol) in 15 µl of kinase buffer at 30 ˚C for 30 min, followed by an incubation with 4 pmol of PLK1 (30 ˚C for 5 min followed by 4 ˚C for 30 min). The reaction mixture was captured with GSH-beads for 30 min at 4 ˚C. A schematic of the experiment is shown (left). A western blot analysis was performed using α-PLK1 (PLK1) and α-pT292 (pT292) antibodies, and a silver-stained gel is also shown (right). (B) CDK and Thr292 of RAD9 are responsible for the PLK1-RAD9 interaction. A GSH-bead pull down assay was performed as described in (A), except that the reactions without CDK2-CyclinA2 or GST-RAD9-T292A (T292A) mutant protein were added to the experiment. A western blot analysis was performed using α-PLK1 and α-RAD9 antibodies. (C) The Thr292-phosphorylated peptide can compete with the RAD9-PLK1 interaction. PLK1 (4 pmol) was pre-incubated with phospho- or non-phospho-peptides (8 nmol or 0.8 nmol, Non-P: non-phosphorylated peptide, pT292: a peptide phosphorylated on Thr292, pS291: a peptide phosphorylated on Ser291, pS291/pT292: a peptide phosphorylated on both Ser291 and Thr292; for the pS291/pT292 peptide, only 8 nmol was tested) for 30 min at 4 ˚C, and then mixed with GST-RAD9A (3A) (30 pmol), which was phosphorylated by CDK2-CyclinA2 (0.5 pmol) for 30 min at 30 ˚C in a 20 µl reaction. The reaction mixture was incubated with GSH-beads for 20 min at 4 ˚C. A schematic drawing of the experiment is shown (left). A western blot analysis was performed using α-PLK1, and a silver stained gel is also shown (right). (D) The T292 phosphorylated peptide can promote PLK1-dependent phosphorylation of RAD9. GST-RAD9-T292A (3A) (10 pmol) was incubated with PLK1 (0.3 pmol) and non-phospho- or phospho-peptides (0.8 nmol: same peptides used in (C)), in a 20 µl reaction for 20 min at 30 ˚C. A schematic drawing of the experiment is shown (left). A western blot using the α-RAD9 antibody is also shown (right). (E), (F) RAD9 co-immunoprecipitates with PLK1. Lysates prepared from 293A-T-REx cells stably expressing WT or T292A-mutated RAD9-mH (RAD9-S391A/T292A expressing cells were used for T292A) were subjected to immunoprecipitation, using α-myc antibody-coated agarose beads. Western blots were performed using α-RAD9 and α-PLK1 antibodies on the input (Input) and immunoprecipitates (IP). Ectopically expressed PLK1-HA was used in (E) and endogenously expressed PLK1 was detected in (F). See also Figure 2—figure supplement 1. PLK1 interacts with and phosphorylates RAD9.
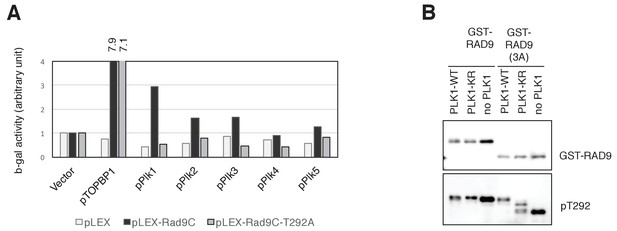
PLK1 interacts with and phosphorylates RAD9.
(A) Yeast two hybrid assays were performed using the wild type (WT) or T292A-mutated (T292A) C-terminus of pLEX-RAD9 (pLEX-RAD9C) as the bait, and the PBD of PLK1-5, which was cloned into pGADT7, as the prey. pACT2-TopBP1 (a kind gift from Dr. Seiji Tanaka (Kochi University of Technology)) was used as a positive control. Results from a liquid ONPG (2-Nitrophenyl-β-D-galactopyranoside) assay are shown. (B) Two different constructs of the GST-tagged C-terminus of RAD9 (GST-RAD9: wild type and GST-RAD9 (3A): S277A, S328A, S336G) were used for the in vitro kinase assay, using the wild type (PLK1-wt) or K81R (PLK1-KR) mutant of recombinant PLK1. Prior to the PLK1 assay, the GST-RAD9 proteins were phosphorylated by CDK-CyclinA2. Western blotting analyses using α-RAD9 (GST-RAD9) and α-pT292 (pT292) are shown.
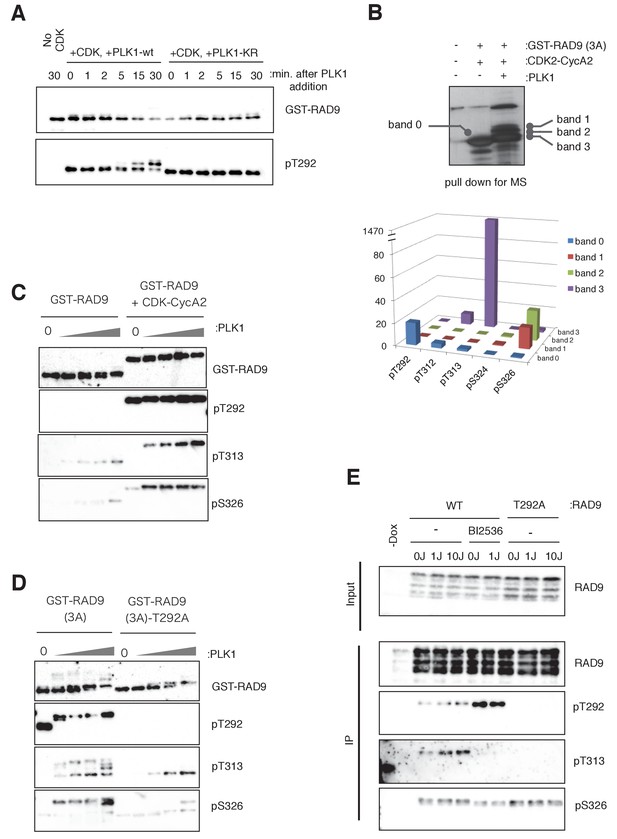
PLK1 phosphorylates RAD9 on Thr 313 and Ser 326 in vivo and in vitro.
(A) PLK1 phosphorylates CDK-phosphorylated RAD9. PLK1 or PLK1-KR (K82R) (0.3 pmol) was incubated with GST-RAD9 (3A) (10 pmol; phosphorylated with the CDK complex (0.5 pmol) for 30 min at 30 ˚C), at 30 ˚C for the indicated time points in the presence of roscovitine (10 µM). Samples were obtained and western blot analyses using α-RAD9 (GST-RAD9) and α-pT292 (pT292) antibodies were performed. (B) Top: Samples for mass spectrometry were prepared by an in vitro CDK-PLK1 kinase assay. GST-RAD9 (3A) (5 µg) was incubated with CDK2-Cyclin A2 (0.5 µg), followed by an incubation with PLK1 (5 µg) in a 100 µl reaction. Phosphorylated GST-RAD9 was pulled down using GSH-agarose beads. A 10% portion of the captured materials was subjected to SDS-PAGE and silver staining. Bottom: Mass spectrometry analysis of the PLK1-phosphorylated GST-RAD9 (3A), excised from the bands corresponding to band 0 to band 3 at the top. The total counts of the reliable MS/MS spectra (confidence ≥95%) corresponding to the peptides originating from GST-RAD9 (3A), in which Thr292 (pT292), Thr312 (pT312), Thr313 (pT313), Ser324 (pS324), or Ser326 (pS326), was phosphorylated. (C), (D) PLK1 phosphorylates Thr313 (pT313) and Ser326 (pS326) of RAD9, when RAD9 was pre-phosphorylated by CDK on Thr 292 (pT292). GST-RAD9 (10 pmol) was incubated with or without CDK2-CyclinA2 (0.5 pmol) prior to incubations with different amounts of PLK1 (0, 0.3, 0.8, 2.6, 8 pmol). The western blot is shown in (C). GST-RAD9 (3A) (10 pmol) or GST-RAD9 (3A)-T292A (10 pmol) was incubated with different amounts of PLK1 (0, 0.3, 0.8, 2.6, 8 pmol). The western blot is shown in (D). α-RAD9 (GST-RAD9), α-pT292 (pT292), α-pT313 (pT313), and α-pS326 (pS326) antibodies were used for the western blot analyses in (C) and (D). When the GST-RAD9 was incubated with PLK1, roscovitine (10 µM) was added to inhibit the remaining CDK. (E) 293A T-REx cells stably expressing RAD9-WT-mH (WT) or RAD9-S291A/T292A-mH (T292A) were collected, two hours after UV-irradiation (1 J/m2, 10 J/m2). To inhibit the cellular PLK1, BI2536 (2 µM) was added for 15 min, prior to the harvest. Cell lysates were subjected to immunoprecipitation using α-myc antibody-coated agarose beads. Western blotting analyses of the input (Input) and immunoprecipitates (IP) were performed, using α-RAD9 (RAD9), α-pT292 (pT292), α-pT313 (pT313), and α-pS326 (pS326) antibodies. See also Figure 3—figure supplement 1. PLK1 phosphorylates RAD9 on Thr 313 and Ser 326, Figure 3—figure supplement 2. PLK1 phosphorylates RAD9 on Thr 313 and Ser 326 in vivo and Figure 3—figure supplement 3. The phosphorylation status of RAD9 under various genotoxic stresses.
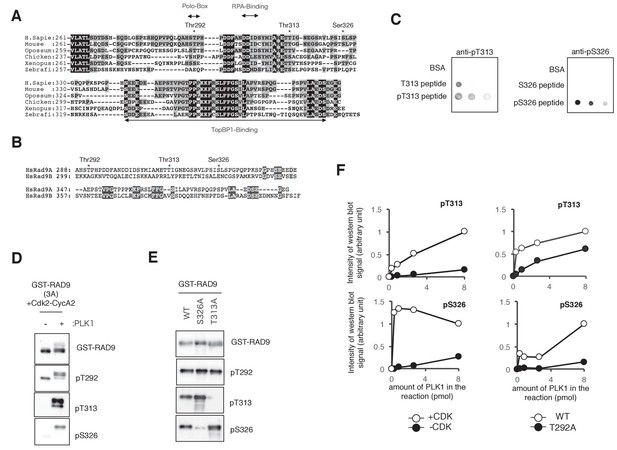
PLK1 phosphorylates RAD9 on Thr 313 and Ser 326.
(A) An alignment of the RAD9 C-terminal region is shown. RAD9 (RAD9A) proteins from Human (H. sapiens), Mouse, Opossum, Chicken, Frog (Xenopus) and Zebrafish were aligned. Identical amino acids for all of the species are shown in black boxes with white letters. Similar amino acids are shown in grey boxes. (B) Comparison between Human RAD9A (HsRAD9A) and RAD9B (HsRAD9B). Amino acids in common are indicated in black boxes with white letters. (C) Dot blot analysis to check the specificity of the phospho-peptide antibodies against Thr 313 (T313) or Ser 326 (S326). Dilution series (1 µg, 0.3 µg and 0.1 µg) of each peptide were spotted onto nitrocellulose membranes. (D) The GST-RAD9 (3A) protein, which was pre-phosphorylated by CDK2-CyclinA2, was subjected to the PLK1 kinase assay. Western blots using α-RAD9, α-pT292, α-pT313 and α-pS326 antibodies are shown. GST-RAD9 (3A) was phosphorylated by CDK2-CyclinA2 prior to the PLK1 kinase assay. (E) GST-RAD9 proteins (WT, T313A, and S326A) were subjected to the PLK1 kinase assay following phosphorylation by CDK2-CyclinA2. Western blots using α-RAD9, α-pT292, α-pT313 and α-pS326 antibodies are shown. (F) The western blot signals in Figure 3C were quantified and are shown in (left), and the signals in Figure 3D are shown in (right).
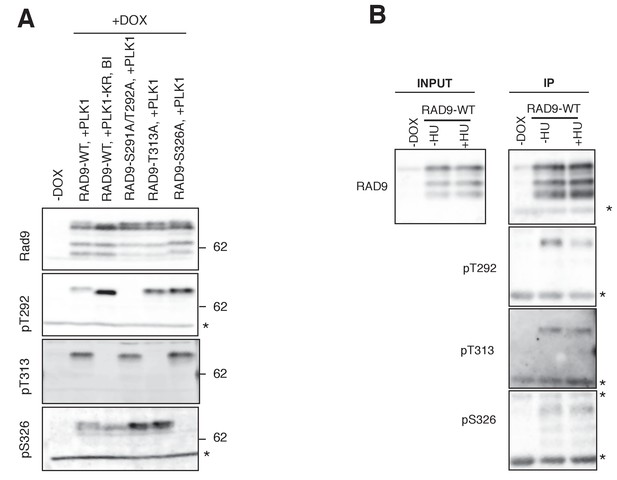
PLK1 phosphorylates RAD9 on Thr 313 and Ser 326 in vivo.
(A) 293A T-REx cells, stably expressing the wild type or S291A/T292A-, T313A- or S326A- mutated RAD9-mH from the FRT locus, were transfected with CMV-PLK1 or CMV-PLK1-KR. Calyculin A (0.5 µM, Santa Cruz) was applied for 15 min before the harvest of the cells. The cells transfected with PLK1-KR were also treated with BI2536 (BI: 2 µM) for 15 min before the harvest. Cell lysates were subjected to immunoprecipitation, using α-myc antibody-coated agarose beads. Western blotting analyses were performed using α-RAD9, α-pT292, α-pT313, and α-pS326 antibodies. The asterisk (*) shows a nonspecific signal. (B) 293A T-REx cells stably expressing wild type RAD9-mH were subjected to a thymidine block and release experiment. Cells were harvested 22 hr after the release from the thymidine block, and hydroxyurea (0.2 mM) was added to the media for 10 hr prior to the harvest. The asterisk (*) shows a nonspecific signal.
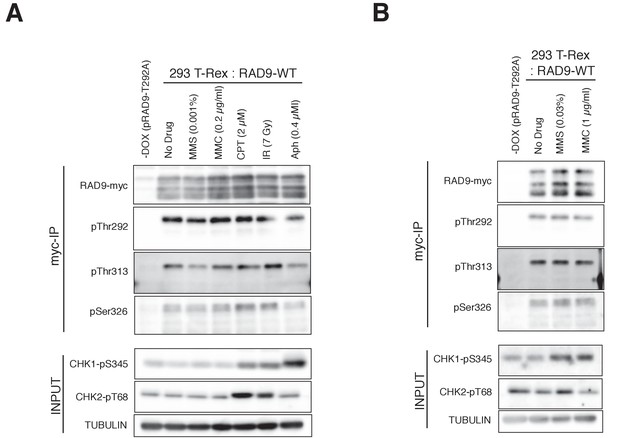
The phosphorylation status of RAD9 under various genotoxic stresses.
(A), (B) Cell lysates were subjected to immunoprecipitation, using α-myc antibody-coated agarose beads. Western blotting analyses of the immunoprecipitates (myc-IP) were performed using α-myc (RAD9-myc), α-pThr292 (pThr292), α-pThr313 (pThr313) and α-pSer326 (pSer326) antibodies, to monitor the phosphorylation status of RAD9. The input lysate (INPUT) was subjected to western blot analyses using α-CHK1-pS345 (CHK1-pS345), α-CHK2-pT68 (CHK2-pT68) and α-Tubulin (TUBULIN), to monitor the checkpoint response. Cells were treated with MMS (0.001%), MMC (0.2 µg/ml), CPT (2 µM) or Aphidicolin (Aph: 0.4 µM) for 1 hr before the harvest, and the cells were subjected to 7 Gy of gamma-irradiation (IR) 10 min before the harvest in (A). The amounts of MMS (0.03%) or MMC (1 µg/ml) used to treat the cells were increased in (B), as the checkpoint responses in (A) were rather moderate for MMS and MMC.
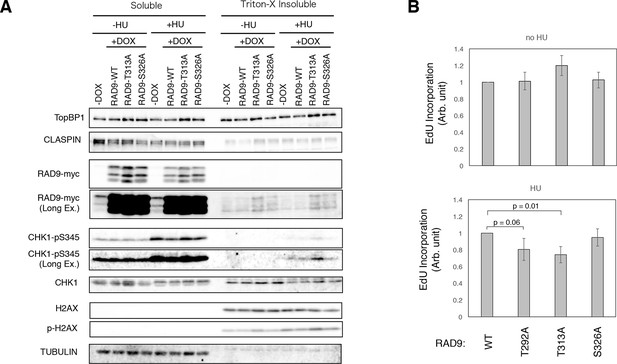
Enhanced checkpoint signaling when PLK1 fails to phosphorylate RAD9.
(A) A chromatin fractionation assay (see Materials and methods) was performed in HEK293A-T-REx cell lines stably expressing RAD9-WT-mH (RAD9-WT), RAD9- T313A-mH (RAD9-T313A) or RAD9- S326A-mH (RAD9-S326A). Cells were grown in media containing 0.2 mM hydroxyurea for 24 hr (+HU). A western blotting analysis is shown. α-TopBP1, α-CLASPIN, α-myc (RAD9-myc), α-phospho-serine 345 of CHK1 (CHK1-pS345), α-CHK1, α-phospho-serine 139 of Histone H2AX (p-H2AX), α-H2AX, and α-tubulin were used. (B) The incorporation of ethynyl-deoxy-uridine (EdU) was quantified in U2OS cells stably expressing RAD9-WT-mH (WT), RAD9-S291A/T292A-mH (T292A), RAD9-T313A-mH (T313A) or RAD9-S326A-mH (S326A) from the FRT locus (see Materials and methods). The cells were incubated with 5 µM of EdU before the harvest (30 min for cells without hydroxyurea treatment, 1 hr for cells with 0.2 mM hydroxyurea). The samples were stained with Alexa Fluor 488 azide using the Click reaction, and subjected to the flow cytometry analysis. An example of the data is shown in Figure 4—figure supplement 3. The incorporated signals were quantified, and the mean values from three independent experiments are plotted. See also Figure 4—figure supplement 1. U2OS stable cell line that expresses RAD9-mycHis, Figure 4—figure supplement 2. DNA damage signaling was increased when PLK1 failed to phosphorylate RAD9, Figure 4—figure supplement 3. dNTP incorporation was decreased in HU treated cells where PLK1-dependent phosphorylation on RAD9 was defective, Figure 4—figure supplement 4. The analysis using the CMV-RAD9-expressing U2OS cell line under endogenous RAD9 knock down, Figure 4—figure supplement 5. Decreased dNTP incorporation was observed in telomerase-positive HEK293A cells that expressed CDK-PLK1-dependent phosphorylation defective RAD9 and Figure 4—figure supplement 6. EdU incorporation assay under Aphidicolin- and MMC-induced stress.
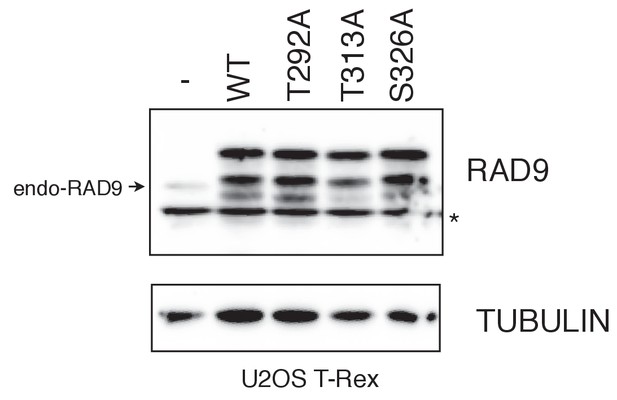
U2OS stable cell line that expresses RAD9-mycHis.
Western blotting analyses were performed to monitor the expression levels of myc-tagged RAD9 (RAD9-WT-mH (WT), RAD9-S291A/T292A (T292A), and RAD9-T313A-mH (T313A). RAD9-S326A (S326A)) was expressed from the FRT locus, under the CMV promoter controlled via the tetO system in U2OS cells. Doxycyclin was added for 24 hr to induce the promoter. For the control, the host U2OS T-REx cells (-) were used. The asterisk (*) shows a nonspecific signal.
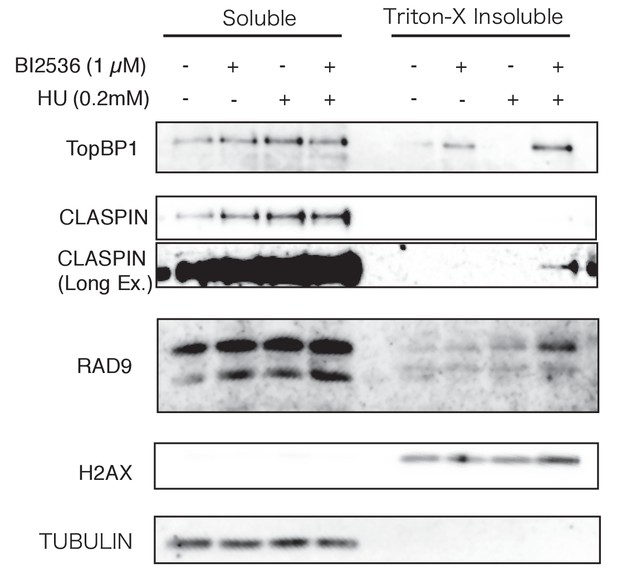
DNA damage signaling was increased when PLK1 failed to phosphorylate RAD9.
U2OS cells were treated with hydroxyurea (0.2 mM, 24 hr) and/or BI2536 (1 µM, 15 min) before the harvest. A chromatin fractionation assay was performed. Western blotting analyses were performed using α-TopBP1 (TopBP1), α-CLASPIN (CLASPIN), α-RAD9 (RAD9), α-H2AX (H2AX) and α-tubulin (TUBULIN) antibodies.
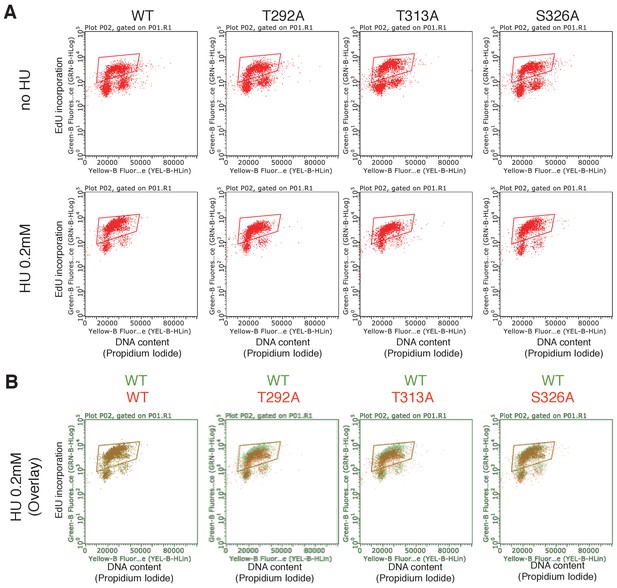
dNTP incorporation was decreased in HU treated cells in which PLK1-dependent phosphorylation of RAD9 was defective.
(A) An EdU incorporation assay was performed. An example of the raw data of the dNTP incorporation presented in Figure 4B is shown. The fluorescent value (Y-axis) of the enclosed area in the scatter plots was quantified. (B) The scatter plots of HU treated cells (HU 0.2 mM) shown in (A) were modified. The plots from WT-RAD9 expressing cells are shown in green and the scatter plots from WT, T292A, T313A and T326A- RAD9 expressing cells are shown in red and overlaid for the direct comparison.
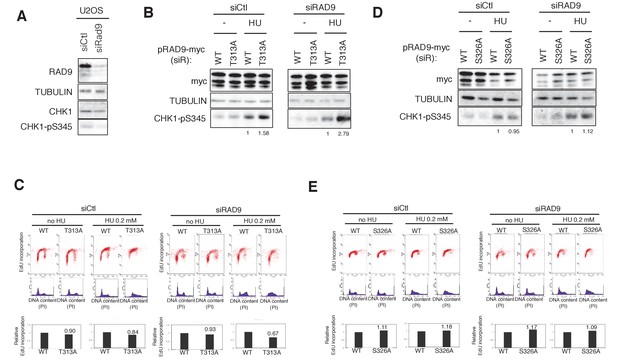
The analysis using CMV-RAD9-expressing U2OS cell line under endogenous RAD9 knock down.
(A) U2OS T-REx cells were subjected to siRNA treatment against RAD9 (siRAD9: s11719 (Ambion)). Cells were harvested 48 hr after the siRNA treatments. The antibodies used were α-RAD9 (RAD9), α-Tubulin (TUBULIN), α-CHK1 (CHK1) and α-CHK1-pS345 (CHK1-pS345). (B), (D) Western blot analysis of cell lysates from U2OS T-REx cells that expressed siRNA-resistant (siR) RAD9s. At 48 hr after the siRNA transfection, the cells were subjected to the HU treatment for 24 hr, and then harvested for the preparation of lysates. The antibodies used were α-RAD9 (RAD9), α-Tubulin (TUBULIN) and α-CHK1-pS345 (CHK1-pS345). The effects of the T313A-mutated RAD9 are shown in (B) and those of the S326A-mutated RAD9 are shown in (D). (C), (E) An EdU incorporation assay was performed. Scatter plots showing EdU incorporation and histograms showing DNA content (PI: propidium iodide staining) are presented. Quantification of EdU incorporation is shown below. Relative amount of phosphorylation of CHK1-pS345 was measured by the Licor system and was shown in (B) and (D). Relative values against WT were calculated for T313A in (C) and for S326A in (E).
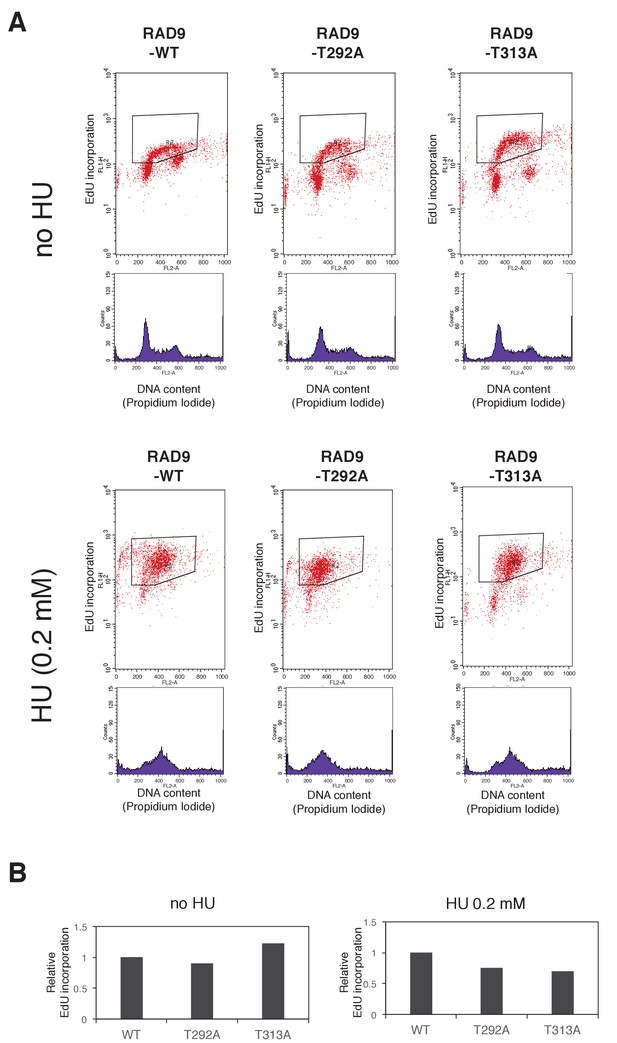
Decreased dNTP incorporation was observed in telomerase-positive HEK293A cells that expressed CDK-PLK1-dependent phosphorylation defective RAD9.
(A) An EdU incorporation assay was performed. Scatter plots showing EdU incorporation and histograms showing DNA content (propidium iodide staining) are presented. Cells (RAD9-WT, RAD9-T292A, RAD9-T313A stably expressing 293A-Trex cells) were incubated with (HU (0.2 mM)) or without (no HU) hydroxyurea for 24 hr before the harvest. Before the harvest, EdU was added for 1 hr (HU (0.2 mM) samples) or 30 min (no HU samples). The fluorescent value (Y-axis) of the enclosed area in the scatter plots was quantified. (B) A quantification of EdU incorporation (example shown in black-enclosed area in (A)) is shown. Average values of two experiments are indicated. Relative values against WT were calculated for T313A.
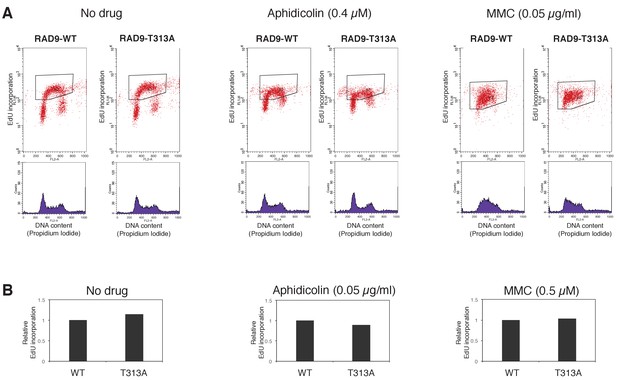
EdU incorporation assay under Aphidicolin- and MMC-induced stress.
(A), (B) An EdU incorporation assay was performed. Scatter plots showing EdU incorporation and histograms showing DNA content (propidium iodide staining) are presented. Cells were incubated without any genotoxic drug (No drug) or with 0.4 µM aphidicolin or 0.05 µg/ml MMC for 24 hr before the harvest. Before the harvest, EdU was added for 1 hr. The fluorescent value (Y-axis) of the enclosed area in the scatter plots was quantified. The cells expressing RAD9-T313A showed a reduced dNTP incorporation rate when treated with aphidicolin. Although the reduction of dNTP incorporation was not observed when the cells were treated with MMC, the accumulation in early S-phase (the histogram plot) was observed instead. (B) A quantification of EdU incorporation (example shown in black-enclosed area in (A)) is shown. Average values of two experiments are indicated. Relative values against WT were calculated for T313A.
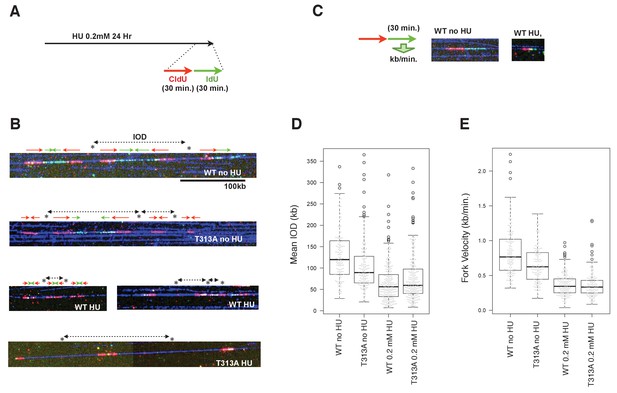
The effect of origin firing and DNA replication fork integrity of PLK1-dependent phosphorylation on RAD9.
(A) A schematic of the time course applied for the combing assay analysis. U2OS cells expressing WT- or T313A-RAD9 from genomic FRT sites were used for the analysis. (B) Examples of DNA fibers seen in the DNA combing assay. The vertical strips seen were either the boundary of the assembled photo frames or noise produced upon scanning of DNA fibers. See the supplement for the surrounding regions (Supplementary file 3). (C) A schematic of the measurement of replication fork velocity. A green signal (IdU labeled DNA) was measured on a unidirectionally moving fork that is associated with a red signal (CldU labeled DNA). (D), (E) Mean Inter Origin Distance (IOD) values were measured and plotted on the graph in (D) (WT no HU: n = 140, T313A no HU: n = 146, WT 0.2 mM HU: n = 231, T313A 0.2 mM HU: n = 229). The velocity of each DNA replication fork was measured and plotted in (E). Boxplots overlaid with beeswarm plots are shown. Boxes indicate the upper and lower quartiles, and the central bar indicates the median value of the measurements. The graphs were made with the R program. The p-values in the IOD analysis were 8 × 10−8 (without HU) and 0.058 (with HU) (WT vs. T313A; the measurement values that were statistically significant (within the top and bottom whisker bars) were applied to calculate the p-values). The p-value in the replication fork velocity analysis was 5.8 × 10−7 (WT vs. T313A, without HU). See also Figure 5—figure supplement 1. The DNA combing assay in aphidicolin-treated cells, Figure 5—figure supplement 2. Enhanced RAD9-CLASPIN complex formation when CDK or PLK1 failed to phosphorylate RAD9.
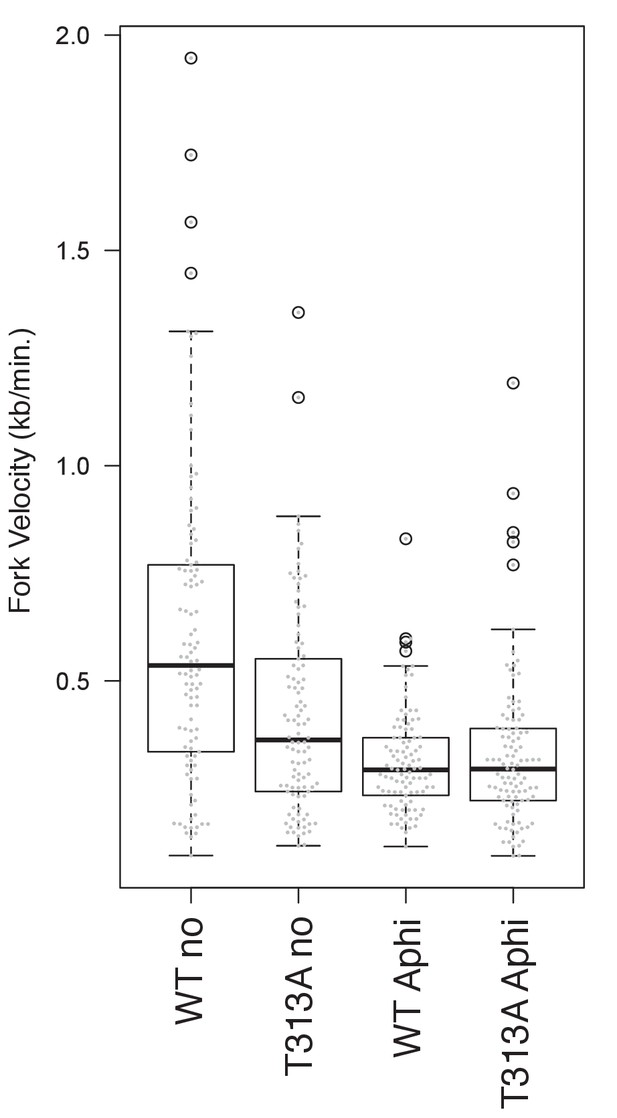
The DNA combing assay in aphidicolin-treated cells.
The combing assay was performed. In this assay, the cells (U2OS T-REx cells expressing RAD9-WT or RAD9-T313A) were first labeled with IdU for 30 min, followed by CldU labeling for 30 min at the end of 4 hr of aphidicolin treatment (0.4 µM), before the harvest. Boxplots overlaid with beeswarm plots were constructed with the R program and are shown (Average fork progression rates in untreated cells, WT: 0.60 ± 0.36 kb/minutes, T313A: 0.42 ± 0.24 kb/minutes; in aphidicolin-treated cells, WT Aphi: 0.31 ± 0.12 kb/minutes, T313A Aphi: 0.33 ± 0.18 kb/minutes, p-value in non-treated cells was 2.8 × 10−5).
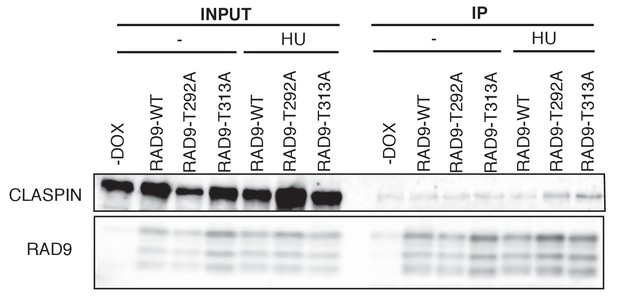
Enhanced RAD9-CLASPIN complex formation when CDK or PLK1 failed to phosphorylate RAD9.
293A T-REx cells were cultured in HU (0.2 mM) containing media for 24 hr, and the cell lysates were subjected to immunoprecipitation using α-myc antibody-coated agarose beads. HEK293A cells stably expressing myc-tagged RAD9s (RAD9-WT-mH (WT), RAD9-S291A/T292A (T292A), RAD9-T313A-mH (T313A)) from the FRT locus were used. Western blotting analyses were performed using α-RAD9 and α-CLASPIN antibodies.
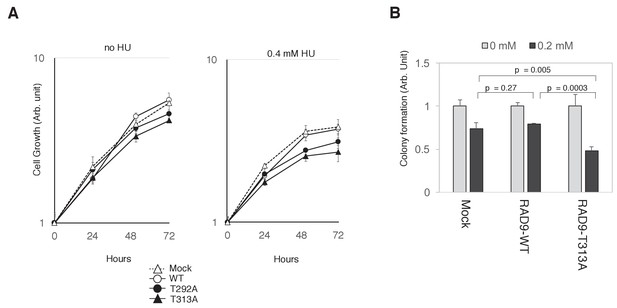
PLK1-dependent phosphorylation of RAD9 is required for proliferation under replicative stress.
(A) Cell growth was monitored using the Presto-Blue Cell viability reagent (see Materials and methods). U2OS cells stably expressing RAD9-WT-mH (WT), RAD9-S291A/T292A-mH (T292A), and RAD9-T313A-mH (T313A) from the FRT locus, and the host U2OS T-REx cells (Mock) were used. (B) The cellular viability in the presence of hydroxyurea was monitored by a colony formation assay (see Materials and methods). U2OS cells stably expressing RAD9-WT-mH (RAD9-WT) and RAD9-T313A-mH (RAD9-T313A) from the FRT locus, and the host U2OS T-REx cells (Mock) were used. See also Figure 6—figure supplement 1. Reduced cell proliferation by expression of non-phosphorylatable RAD9 in telomerase-positive HEK293A cells.
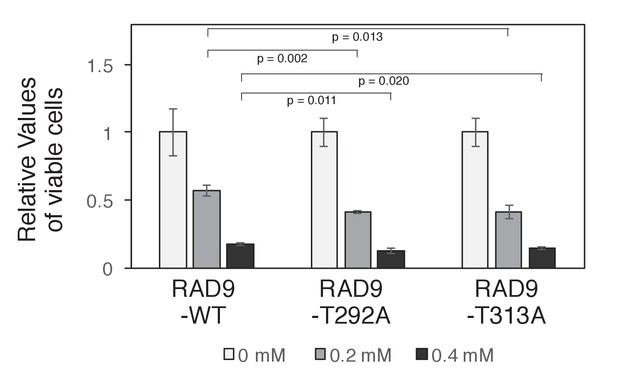
Reduced cell proliferation by expression of non-phosphorylatable RAD9 in telomerase-positive HEK293A cells.
HEK293A T-REx cells expressing RAD9-WT, RAD9-T292A or RAD9-T313A were grown in 96 well plates in the presence of 0.2 mM or 0.4 mM hydroxyurea for 4 days. siRNA (s11719: Ambion) was transfected 48 hr before starting the incubation with HU, to knock down the endogenous RAD9. Viable cells were measured using Presto-Blue (Thermo Fisher), and relative values against that obtained from cells incubated without hydroxyurea are indicated. Averages and standard deviations obtained from the values from the three slots are shown as a graph.
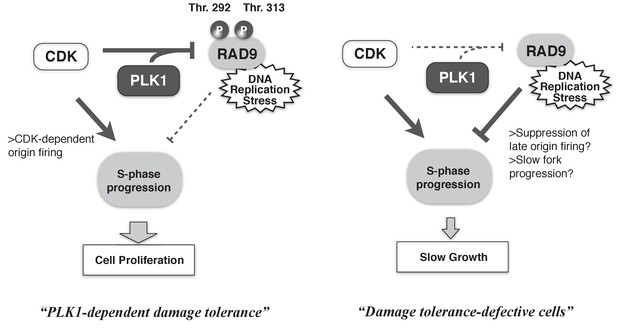
A schematic of PLK1-dependent damage tolerance.
(Left) The collaboration of CDK and PLK1 targets and suppresses the DNA damage detection machinery in the DNA damage checkpoint process, and promotes the S-phase progression to support cell proliferation. (Right) When CDK and PLK1 fail to phosphorylate RAD9 (in the case of the PLK1 inhibitor, RAD9-T292A or -T313A mutant expressing cells), DNA damage checkpoint activation is increased and confers the slow growth.
Additional files
-
Supplementary file 1
Primers used in this study
Primers used in this study was listed.
- https://doi.org/10.7554/eLife.29953.023
-
Supplementary file 2
Statistic data to construct Figures 4B and 6A,B
An excel formatted file was presented.
- https://doi.org/10.7554/eLife.29953.024
-
Supplementary file 3
The expanded photos of DNA fibers used in Figure 5B
The field in white squares are region presented in Figure 5B.
- https://doi.org/10.7554/eLife.29953.025
-
Transparent reporting form
- https://doi.org/10.7554/eLife.29953.026





