An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence
Figures
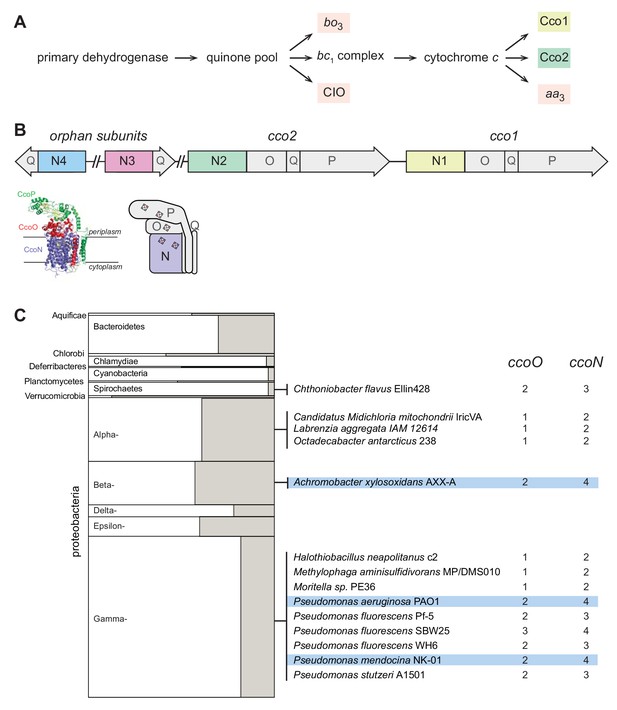
The respiratory chain and arrangement of cco genes and protein products in P. aeruginosa, and the phylogenetic distribution of orphan ccoN genes.
(A) Branched electron transport chain in P. aeruginosa, containing five canonical terminal oxidases. (B) Organization of cco genes in the P. aeruginosa genome. The cartoon of the Cco complex is based on the Cco structure from P. stutzeri (PDB: 3mk7) (Buschmann et al., 2010). (C) Left: graphical representation of the portion of genomes in each bacterial phylum that contain ccoO and N homologs. The clades Chrysiogenetes, Gemmatimonadetes, and Zetaproteobacteria were omitted because they each contain only one species with ccoO and N homologs. The height of each rectangle indicates the total number of genomes included in the analysis. The width of each shaded rectangle represents the portion of genomes that contain ccoN homologs. Middle: genomes that contain more ccoN than ccoO homologs (indicating the presence of orphan ccoN genes) are listed. Right: numbers of ccoO and ccoN homologs in each genome. Blue highlights genomes containing more than one orphan ccoN homolog.
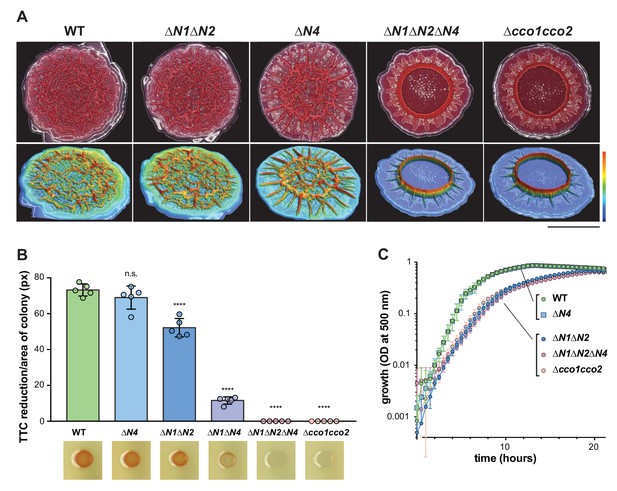
CcoN4-containing heterocomplexes make biofilm-specific contributions to morphogenesis and respiration.
(A) Top: Five-day-old colony biofilms of PA14 WT and cco mutant strains. Biofilm morphologies are representative of more than 10 biological replicates. Images were generated using a digital microscope. Scale bar is 1 cm. Bottom: 3D surface images of the biofilms shown in the top panel. Images were generated using a wide-area 3D measurement system. Height scale bar: bottom (blue) to top (red) is 0–0.7 mm for WT, ∆N1∆N2, and ∆N4; 0–1.5 mm for ∆N1∆N2∆N4 and ∆cco1cco2. (B) TTC reduction by WT and cco mutant colonies after 1 day of growth. Upon reduction, TTC undergoes an irreversible color change from colorless to red. Bars represent the average, and error bars represent the standard deviation, of individually-plotted biological replicates (n = 5). p-Values were calculated using unpaired, two-tailed t tests comparing each mutant to WT (****p≤0.0001). (C) Mean growth of PA14 WT and cco mutant strains in MOPS defined medium with 20 mM succinate. Error bars represent the standard deviation of biological triplicates.
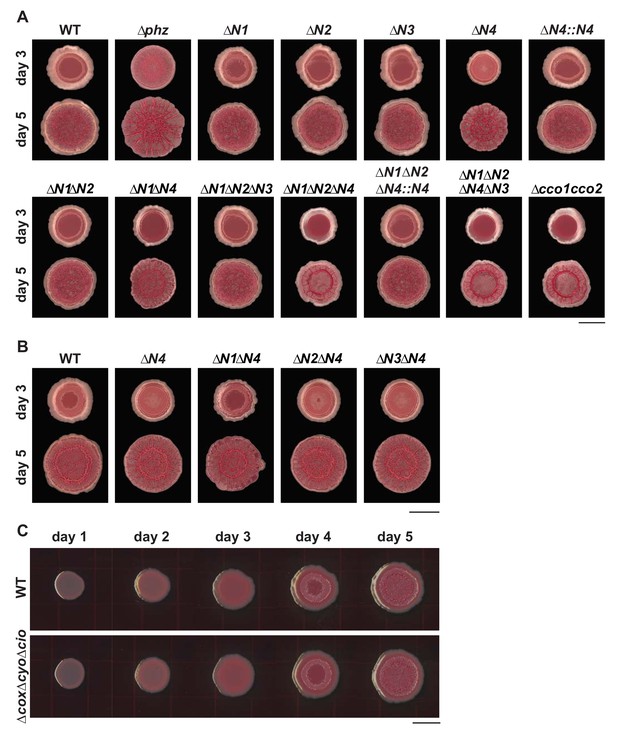
Effects of individual and combined cco gene deletions on colony biofilm morphogenesis.
(A) Morphologies of WT, ∆phz, and cco single, combinatorial, and ccoN4 complementation strains after 3 and 5 days of incubation. Images shown are representative of at least 5 biological replicates and were generated using a digital microscope. Scale bar is 1 cm. (B) Development of WT, ∆N4 and N subunit double mutants containing ∆N4. Images shown are representative of at least 3 biological replicates and were generated using a digital microscope. Scale bar is 1 cm. (C) Development of WT and the triple mutant ∆cox∆cyo∆cio in which only the cbb3-type terminal oxidases are present. Images were generated using a flatbed scanner and are representative of at least 3 biological replicates. Scale bar is 1 cm.
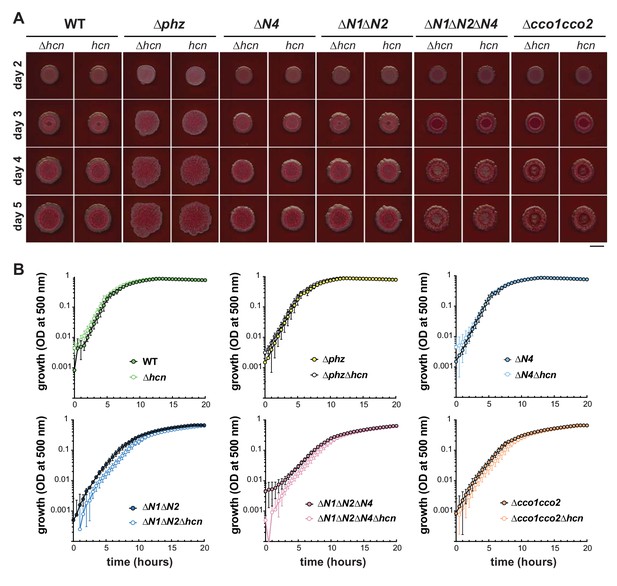
PA14 WT, ∆phz, and cco mutant growth phenotypes are unaffected by endogenous cyanide production.
(A) Colony development over 4 days for ∆phz, ∆hcnABC, and cco combinatorial mutants. Images were generated using a flatbed scanner and are representative of at least 3 biological replicates. Scale bar is 1 cm. (B) Growth of ∆phz, ∆hcnABC, and cco combinatorial mutants in MOPS defined medium with 20 mM succinate. Error bars represent the standard deviation of biological triplicates and are not shown in cases where they would be obscured by the point marker.
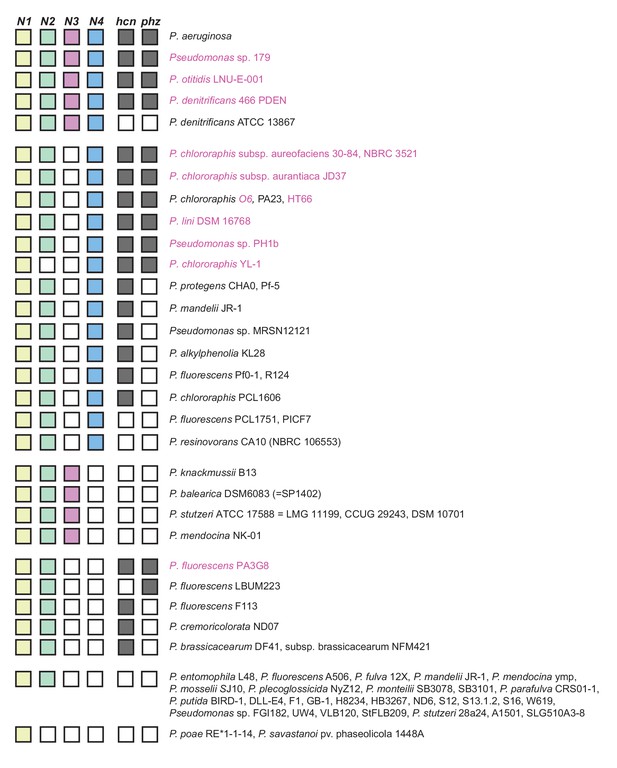
Pseudomonads with CcoN homologs.
We examined genomes available in the Pseudomonas Genome Database (Winsor et al., 2016) for CcoN homologs by performing a protein BLAST search on CcoN1 from P. aeruginosa PA14. All hits from full genomes, excluding other P. aeruginosa strains, were aligned using ClustalW and a tree was built using the geneious tree builder (Geneious 10 (Kearse et al., 2012)). We also included draft genomes that contained genes involved in phenazine biosynthesis (highlighted in purple). The tree revealed four clusters, each being more closely related to one of the four N subunits from PA14, which allowed us to annotate the N subunits accordingly. We next probed all genomes with N subunits for the presence of genes involved in cyanide synthesis (hcnABC) and phenazine biosynthesis (phzABCDEFG). We did not find a clear correlation between the presence of CcoN4 and Hcn proteins (Hirai et al., 2016). We note that with the exception of two P. fluorescens strains, those containing phzABCDEFG operons also contained ccoN4.
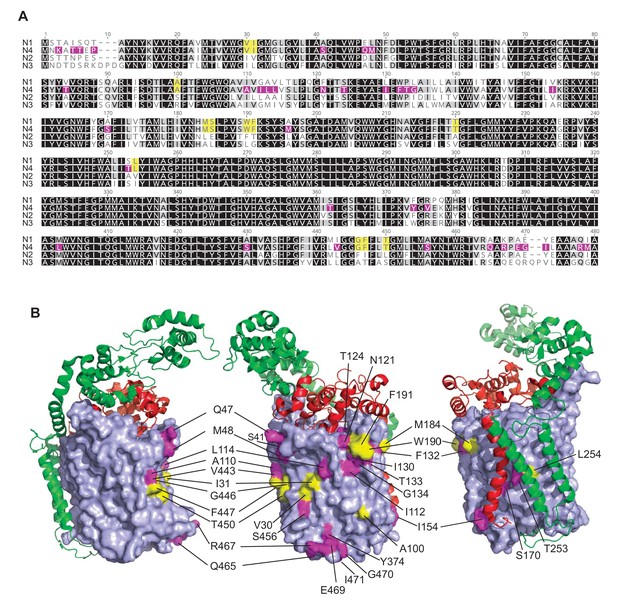
Comparison of the PA14 CcoN subunit sequences and analysis of the predicted structure of CcoN4.
(A) Amino acid alignment (ClustalW) of the four CcoN subunits encoded by the PA14 genome. Residues conserved among all four N subunits are highlighted in black; residues conserved among any three of the four N subunits in gray; residues shared exclusively between CcoN1 and CcoN4 in yellow; and residues unique to CcoN4 in purple. (B) Predicted structure of CcoN4 from P. aeruginosa PA14, obtained by threading the PA14 sequence through the reported structure for the CcoN subunit of P. stutzeri (PDB: 5DJQ; Buschmann et al., 2010) using SWISS-MODELL (Biasini et al., 2014). Surface-exposed residues that are shared exclusively between CcoN1 and CcoN4 are shown in yellow, while residues that are unique to CcoN4 are shown in magenta. Ribbon structures of the CcoO and CcoP subunits from P. stutzeri are shown in red and green, respectively. Structures were generated using PyMol (Schrödinger, LLC, 2015).
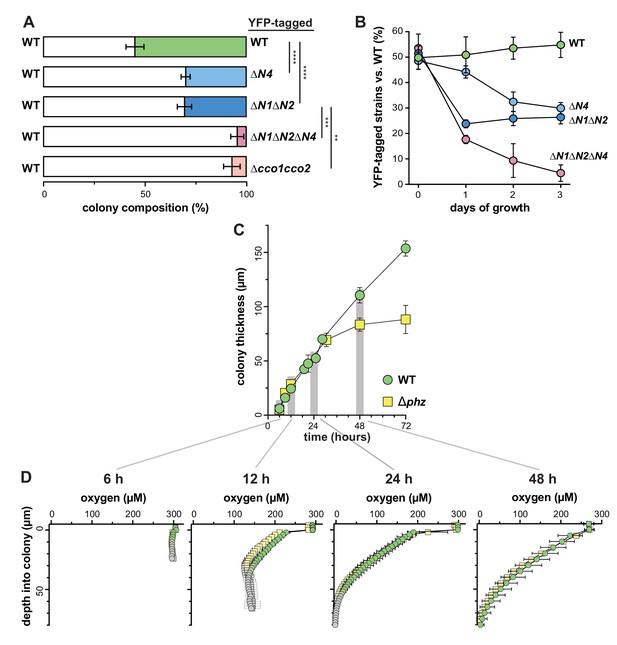
CcoN4 confers a competitive advantage in biofilms, particularly when O2 becomes limiting.
(A) Relative fitness of various YFP-labeled cco mutants when co-cultured with WT in mixed-strain biofilms for 3 days. Error bars represent the standard deviation of biological triplicates. p-Values were calculated using unpaired, two-tailed t tests (**p≤0.01; ***p≤0.001; ****p≤0.0001). (B) Time course showing relative fitness, over a period of 3 days, of various cco mutants when co-cultured with WT in mixed-strain biofilms. Results are shown for experiments in which the WT was co-cultured with various ‘labeled’ strains, that is, those that were engineered to constitutively express YFP. (See Figure 3—figure supplement 1 for results from experiments in which the labeled WT was co-cultured with unlabeled mutants.) Error bars represent the standard deviation of biological triplicates. (C) Change in thickness over 3 days of development for colony biofilms of WT and ∆phz as assessed by thin sectioning and DIC microscopy. After the onset of wrinkling, thickness was determined for the base (i.e. the ‘valley’ between wrinkles). Error bars represent the standard deviation of biological triplicates. (D) O2 profiles of colonies at selected timepoints within the first 3 days of biofilm development. Gray point markers indicate measurements made in the agar directly below the colony. Error bars denote standard deviation of biological triplicates.
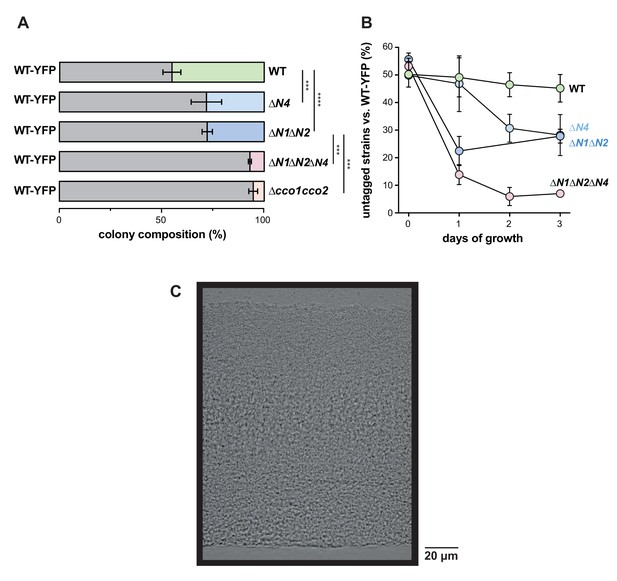
CcoN4 is necessary for optimal fitness in biofilms, particularly when O2 becomes limiting.
(A) Relative fitness of YFP-labeled WT when co-cultured with various cco mutant strains in mixed-strain biofilms for 3 days. Error bars represent the standard deviation of biological triplicates. p-Values were calculated using unpaired, two-tailed t tests (***p≤0.001; ****p≤0.0001). (B) Time course showing relative fitness, over a period of 3 days, of YFP-labeled WT when co-cultured with various cco mutant strains in mixed-strain biofilms. Error bars represent the standard deviation of biological triplicates. (C) DIC image of a 3-day-old WT biofilm, which is representative of at least 10 biological replicates.
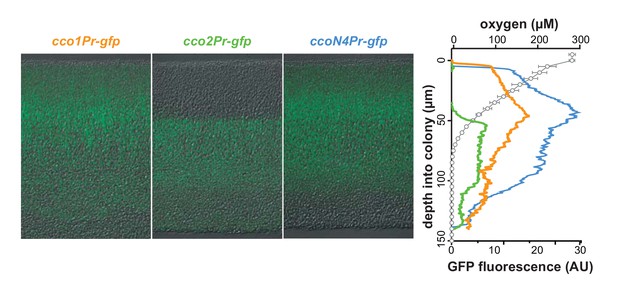
cco genes are differentially expressed over biofilm depth.
Left: Representative images of thin sections prepared from WT biofilms grown for 3 days. Each biofilm is expressing a translational GFP reporter under the control of the cco1, cco2, or ccoN4Q4 promoter. Reporter fluorescence is shown in green and overlain on respective DIC images. Right: Fluorescence values corresponding to images on the left. Fluorescence values for a strain containing the gfp gene without a promoter (the empty MCS control) have been subtracted from each respective plot. O2 concentration over depth (open circles) from 3-day-old WT biofilms is also shown. Error bars represent the standard deviation of biological triplicates and are not shown in cases where they would be obscured by the point markers. y-axis in the right panel provides a scale bar for the left panel. Reporter fluorescence images and values are representative of 4 biological replicates.
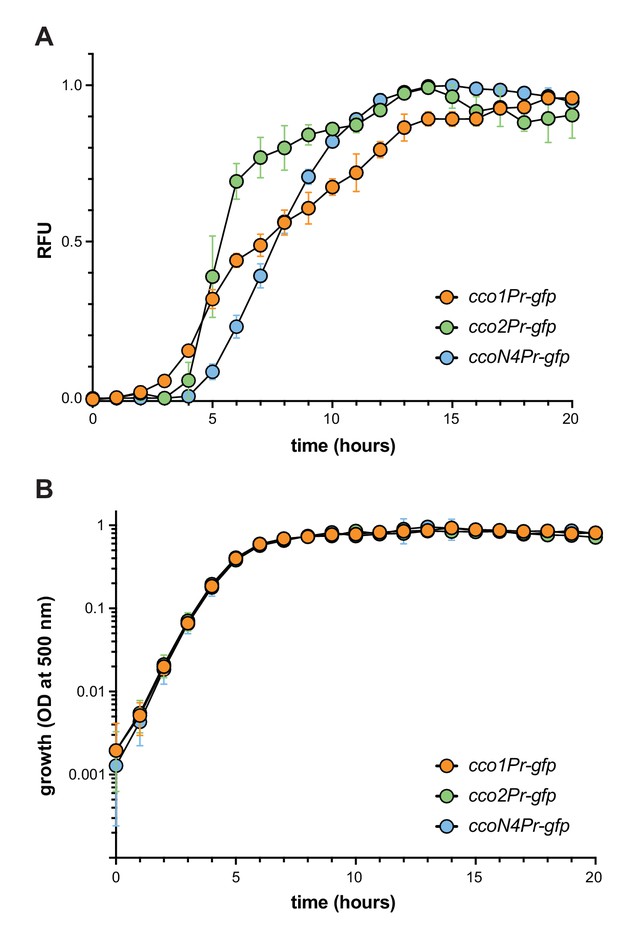
Expression of cco reporters in shaken liquid cultures.
(A) Fluorescence of translational reporter strains, engineered to express GFP under the control of the cco1, cco2, or ccoN4Q4 promoter during growth in 1% tryptone. Fluorescence values for a strain containing the gfp gene without a promoter (the MCS control) were treated as background and subtracted from each growth curve. (B) Liquid-culture growth of translational reporter strains in 1% tryptone. Error bars in (A) and (B) represent the standard deviation of biological triplicates and are not drawn in cases where they would be obscured by point markers.
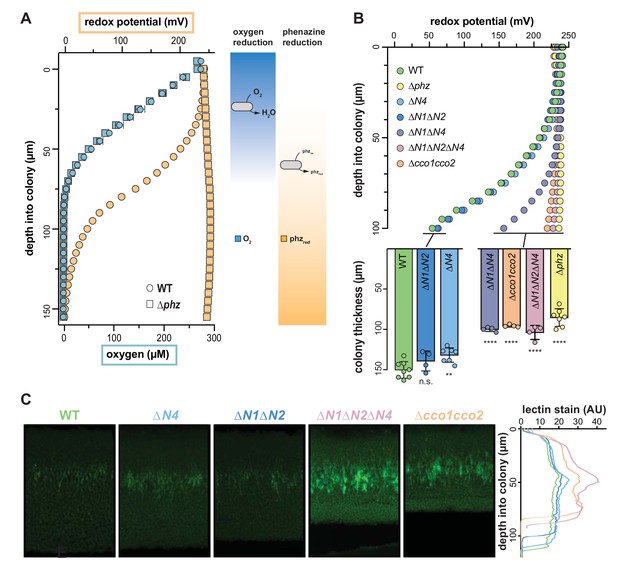
Characterization of chemical gradients and matrix distribution in PA14 WT and mutant colony biofilms.
(A) Left: Change in O2 concentration (blue) and redox potential (orange) with depth for WT and ∆phz biofilms grown for two days. WT biofilms are ~150 µm thick while ∆phz biofilms are ~80 µm thick. For O2 profiles, error bars represent the standard deviation of biological triplicates. For redox profiles, data are representative of at least 5 biological replicates. Right: model depicting the distribution of O2 and reduced vs. oxidized phenazines in biofilms. (B) Top: Change in redox potential with depth for WT and various mutant biofilms grown for 2 days. Data are representative of at least 5 biological replicates. Bottom: Thickness of 3-day-old colony biofilms of the indicated strains. Bars represent the average of the plotted data points (each point representing a biological replicate, n ≥ 4), and error bars represent the standard deviation. p-Values were calculated using unpaired, two-tailed t tests comparing each mutant to WT (n.s., not significant; **p≤0.01; ****p≤0.0001). (C) Left: Representative thin sections of WT and cco mutant biofilms, stained with lectin and imaged by fluorescence microscopy. Biofilms were grown for 2 days before sampling. Right: Relative quantification of lectin stain signal intensity. Coloration of strain names in the left panel provides a key for the plotted data, and the y-axis in the right panel provides a scale bar for the left panel. Lectin-staining images and values are representative of 4 biological replicates.
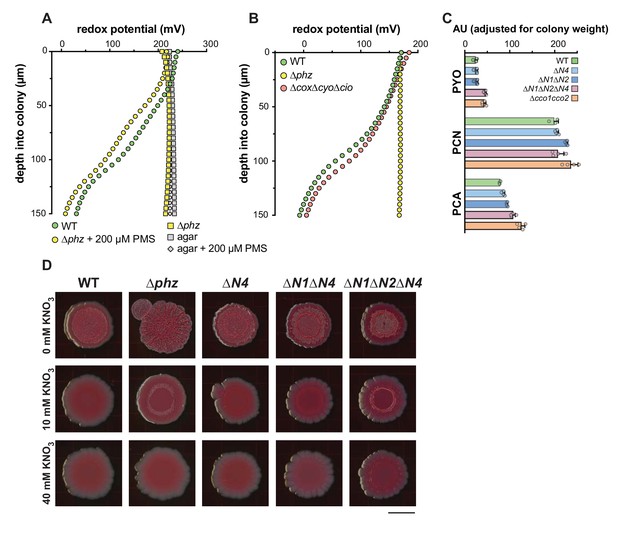
Use of a redox microelectrode to measure phenazine reduction in colony biofilms.
(A) Change in redox potential over depth for 2-day-old biofilms of PA14 WT, ∆phz, and ∆phz grown on 200 µM phenazine methosulfate (PMS). Data are representative of at least 3 biological replicates. To ensure that addition of PMS did not alter the baseline redox potential, a measurement was also taken of agar only. (B) Change in redox potential with depth for WT, ∆phz, and ∆cox∆cyo∆cio biofilms grown for 2 days. Data are representative of at least 2 biological replicates. (C) Levels of phenazines extracted from the agar medium underneath the colony and separated by HPLC, adjusted for biomass, for PA14 WT and various cco mutant biofilms grown for 2 days. Data represent the area under each peak in absorbance units for the phenazines indicated, and error bars represent standard deviation of at least 3 biological replicates. The phenazines pyocyanin (PYO), phenazine-1-carboxamide (PCN), and phenazine-1-carboxylic acid (PCA) were quantified. (D) Colony biofilm morphologies on day 4 of development for WT and various cco mutant biofilms grown on colony morphology plates containing 0, 10, and 40 mM potassium nitrate. Images were generated using a flatbed scanner and are representative of at least 3 biological replicates. Scale bar is 1 cm.
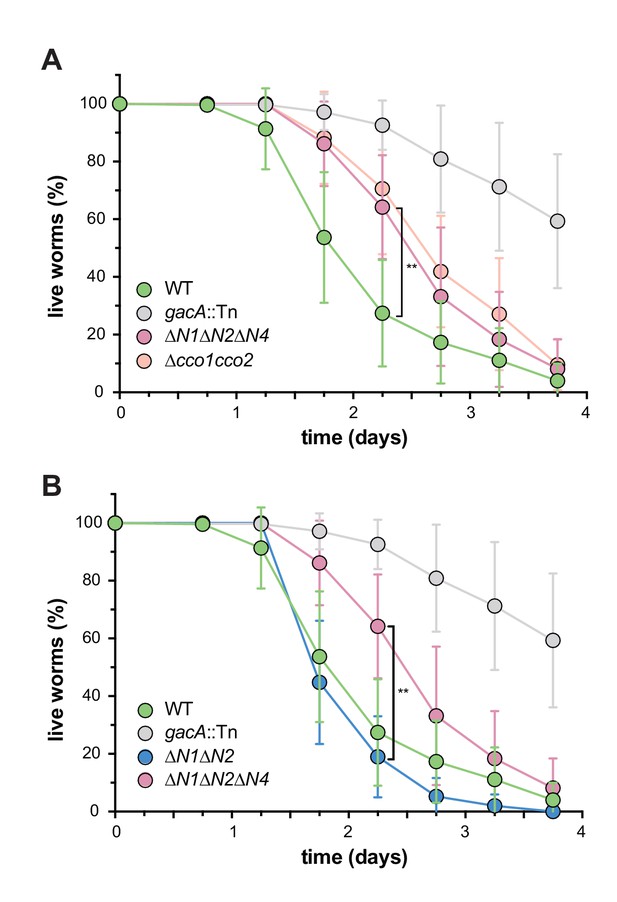
CcoN4-containing isoform(s) make unique contributions to PA14 virulence.
Slow-killing kinetics of WT, gacA, and various cco mutant strains in the nematode Caenorhabditis elegans. Nearly 100% of the C. elegans population exposed to WT PA14 is killed after 4 days of exposure to the bacterium, while a mutant lacking GacA, a regulator that controls expression of virulence genes in P. aeruginosa, shows decreased killing, with ~50% of worms alive 4 days post-exposure. (A) ∆N1∆N2∆N4 and ∆cco1cco2 show comparably attenuated pathogenicity relative to WT. Error bars represent the standard deviation of at least 6 biological replicates. At 2.25 days post-exposure, significantly less C. elegans were killed by ∆N1∆N2∆N4 than by WT (unpaired two-tailed t test; p=0.0022). (B) ∆N1∆N2 displays only slightly reduced pathogenicity when compared to WT. At 2.25 days post-exposure, significantly more C. elegans were killed by ∆N1∆N2 than by ∆N1∆N2∆N4 (unpaired two-tailed t test; p=0.003). Error bars represent the standard deviation of at least 4 biological replicates, each with a starting sample size of 30–35 worms per replicate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (UCBPP-PA14 Pseudomonas aeruginosa) | wild type (WT) | PMID: 7604262 | ||
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆phz | PMID: 16879411 | LD24 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1; ∆N1 | this study | LD1784 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN2; ∆N2 | this study | LD1614 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN3; ∆N3 | this study | LD1620 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN4; ∆N4 | this study | LD2833 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2; ∆N1∆N2 | this study | LD1888 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN4; ∆N1∆N4 | this study | LD1951 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN2∆ccoN4; ∆N2∆N4 | this study | LD1692 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN3∆ccoN4; ∆N3∆N4 | this study | LD1649 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆ccoN3; ∆N1∆N2∆N3 | this study | LD1977 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆ccoN4; ∆N1∆N2∆N4 | this study | LD1976 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆ccoN4 ∆ccoN3; ∆N1∆N2∆N4∆N3 | this study | LD2020 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆cco1cco2 | this study | LD1933 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆cox∆cyo∆cio | this study | LD2587 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆hcn | this study | LD2827 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆phz∆hcn | this study | LD2828 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN4∆hcn; ∆N4∆hcn | this study | LD2829 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆hcn; ∆N1∆N2∆hcn | this study | LD2830 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆ccoN4 ∆hcn; ∆N1∆N2∆N4∆hcn | this study | LD2831 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆cco1cco2∆hcn | this study | LD2832 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | gacA::Tn | PMID: 16477005 | LD1560 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN4::ccoN4; ∆N4::N4 | this study | LD1867 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆ccoN4:: ccoN4; ∆N1∆N2∆N4::N4 | this study | LD2576 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | MCS-gfp | this study | LD2820 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | Pcco1-gfp; cco1Pr-gfp | this study | LD2784 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | Pcco2-gfp; cco2Pr-gfp | this study | LD2786 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | PccoN4-gfp; ccoN4Pr-gfp | this study | LD2788 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | PA14-yfp | this study | LD2780 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2-yfp; ∆N1∆N2-yfp | this study | LD2013 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN4-yfp; ∆N4-yfp | this study | LD2834 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆ccoN1∆ccoN2∆cco N4-yfp; ∆N1∆N2∆N4-yfp | this study | LD2136 | |
| strain, strain background (UCBPP-PA14 P. aeruginosa) | ∆cco1cco2-yfp | this study | LD2012 | |
| strain, strain background (Escherichia coli) | UQ950 | other | From D. Lies, Caltech; LD44 | |
| strain, strain background (E. coli) | BW29427 | other | From W. Metcalf, University of Illinois; LD661 | |
| strain, strain background (E. coli) | β2155 | PMID: 8990308 | LD69 | |
| strain, strain background (E. coli) | S17-1 | doi:10.1038/nbt1183-784 | LD2901 | |
| strain, strain background (Saccharomyces cerevisiae) | InvSc1 | Invitrogen | LD676 | |
| recombinant DNA reagent | pMQ30 (plasmid) | PMID: 16820502 | For generation of deletion constructs listed above; further information can be found in the Materials and Methods section. | |
| recombinant DNA reagent | pAKN69 (plasmid) | PMID: 15186351 | For generation of strains constitutively expression eyfp; further information can be found in the Materials and Methods section. | |
| recombinant DNA reagent | pLD2722 (plasmid) | this study | For generation of gfp reporter constructs; further information can be found in the Materials and Methods section. | |
| recombinant DNA reagent | pFLP2 (plasmid) | PMID: 9661666 | For generation of gfp reporter constructs; further information can be found in the Materials and Methods section. | |
| software, algorithm | EggNOG Database | PMID: 26582926 | http://eggnogdb.embl.de/#/app/home | |
| software, algorithm | SensorTrace Profiling | Unisense | For data acquisition for redox and oxygen microprofiling; further information can be found in the Materials and Methods section. | |
| other | Agar | Teknova | For colony morphology assays; further information can be found in the Materials and Methods section. | |
| other | Lectin | Vector Laboratories | For visualization of matrix; further information can be found in the Materials and Methods section. |
Strains used in this study.
https://doi.org/10.7554/eLife.30205.016| Strain | Number | Description | Source |
|---|---|---|---|
| Pseudomonas aeruginosa strains | |||
| UCBPP-PA14 | Clinical isolate UCBPP-PA14. | Rahme et al. (1995) | |
| PA14 ∆phz | LD24 | PA14 with deletions in phzA1-G1 and phzA2-G2 operons. | Dietrich et al., 2006a |
| PA14 ∆ccoN1 | LD1784 | PA14 with deletion in PA14_44370. | this study |
| PA14 ∆ccoN2 | LD1614 | PA14 with deletion in PA14_44340. | this study |
| PA14 ∆ccoN3 | LD1620 | PA14 with deletion in PA14_40510. | this study |
| PA14 ∆ccoN4 | LD2833 | PA14 with deletion in PA14_10500. | this study |
| PA14 ∆ccoN1 ∆ccoN2 | LD1888 | PA14 with deletions in PA14_44370 and PA14_44340. Made by mating pLD1610 into LD1784. | this study |
| PA14 ∆ccoN1 ∆ccoN4 | LD1951 | PA14 with deletions in PA14_44370 and PA14_10500. Made by mating pLD1264 into LD1784. | this study |
| PA14 ∆ccoN2 ∆ccoN4 | LD1692 | PA14 with deletions in PA14_44340 and PA14_10500. Made by mating pLD1264 into LD1614. | this study |
| PA14 ∆ccoN3 ∆ccoN4 | LD1649 | PA14 with deletions in PA14_40510 and PA14_10500. Made by mating pLD1264 into LD1620. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN3 | LD1977 | PA14 with deletions in PA14_443470, PA14_44340, and PA14_40510. Made by mating pLD1616 into LD1888. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4 | LD1976 | PA14 with deletions in PA14_443470, PA14_44340, and PA14_10500. Made by mating pLD1264 into LD1888. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4 ∆ccoN3 | LD2020 | PA14 with deletions in PA14_443470, PA14_44340, PA14_10500, and PA14_40510. Made by mating pLD1264 into LD1977. | this study |
| PA14 ∆cco1cco2 | LD1933 | PA14 with both cco operons (PA14_44340-PA14_44400) deleted simultaneously. | this study |
| PA14 ∆cox ∆cyo ∆cio | LD2587 | PA14 with deletions in PA14_01290–01320 (cox/aa3 operon), PA14_47150–47210 (cyo/bo3 operon), and PA14_13030–13040 (cio operon). Made by mating pLD1966, pLD1967, and pLD2044, in that order, to PA14. | this study |
| PA14 ∆hcn | LD2827 | PA14 with deletion in hcnABC operon (PA14_36310–36330). | this study |
| PA14 ∆phz ∆hcn | LD2828 | PA14 with deletions in phzA1-G1, phzA2-G2, and hcnABC operons. Made by mating pLD2791 into LD24. | this study |
| PA14 ∆ccoN4 ∆hcn | LD2829 | PA14 with deletions in PA14_10500 and hcnABC operon. Made by mating pLD2791 into LD2833. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆hcn | LD2830 | PA14 with deletions in PA14_44370, PA14_44340, and hcnABC operon. Made by mating pLD2791 into LD1888. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4 ∆hcn | LD2831 | PA14 with deletions in PA14_44370, PA14_44340, PA14_10500 and hcnABC operon. Made by mating pLD2791 into LD1976. | this study |
| Pseudomonas aeruginosa strains | |||
| PA14 ∆cco1cco2 ∆hcn | LD2832 | PA14 with deletions in cco1, cco2, and hcnABC operons. Made by mating pLD2791 into LD1933. | this study |
| PA14 gacA::Tn | LD1560 | MAR2xT7 transposon insertion into PA14_30650. | Liberati et al. (2006) |
| PA14 ∆ccoN4::ccoN4 | LD1867 | PA14 ∆ccoN4 strain with wild-type ccoN4 complemented back into the site of deletion. Made by mating pLD1853 into LD2833. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4::ccoN4 | LD2576 | PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4 strain with wild-type ccoN4 complemented back into the site of deletion. Made by mating pLD1853 into LD1976. | this study |
| PA14 MCS-gfp | LD2820 | PA14 without a promoter driving gfp expression. | this study |
| PA14 Pcco-1-gfp | LD2784 | PA14 with promoter of cco1 operon driving gfp expression. | this study |
| PA14 Pcco-2-gfp | LD2786 | PA14 with promoter of cco2 operon driving gfp expression. | this study |
| PA14 PccoN4-gfp | LD2788 | PA14 with promoter of ccoN4Q4 operon driving gfp expression. | this study |
| PA14-yfp | LD2780 | WT PA14 constitutively expressing eyfp. | this study |
| PA14 ∆ccoN1 ∆ccoN2-yfp | LD2013 | PA14 ∆ccoN1 ∆ccoN2 constitutively expressing eyfp. Made by mating pAKN69 into LD1888. | this study |
| PA14 ∆ccoN4-yfp | LD2834 | PA14 ∆ccoN4 constitutively expressing eyfp. Made by mating pAKN69 into LD2833. | this study |
| PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4-yfp | LD2136 | PA14 ∆ccoN1 ∆ccoN2 ∆ccoN4 constitutively expressing eyfp. Made by mating pAKN69 into LD1976. | this study |
| PA14 ∆cco1cco2-yfp | LD2012 | PA14 ∆cco1cco2 constitutively expressing eyfp. Made by mating pAKN69 into LD1933. | this study |
| Escherichia coli strains | |||
| UQ950 | LD44 | E. coli DH5 λpir strain for cloning. F-∆(argF- lac) 169φ80 dlacZ58(∆M15) glnV44(AS) rfbD1 gyrA96(NaIR) recA1 endA1 spoT thi-1 hsdR17 deoR λpir+ | D. Lies, Caltech |
| BW29427 | LD661 | Donor strain for conjugation. thrB1004 pro thi rpsL hsdS lacZ ∆M15RP4-1360 ∆(araBAD)567 ∆dapA1314::[erm pir(wt)] | W. Metcalf, University of Illinois |
| β2155 | LD69 | Helper strain. thrB1004 pro thi strA hsdsS lacZ∆M15 (F’lacZ∆M15 lacIq traD36 proA + proB + ) ∆dapA::erm (Ermr)pir::RP4 [::kan (Kmr) from SM10] | Dehio and Meyer (1997) |
| S17-1 | LD2901 | StrR, TpR, F− RP4-2-Tc::Mu aphA::Tn7 recA λpir lysogen | Simon et al. (1983) |
| Saccharomyces cerevisiae strains | |||
| InvSc1 | LD676 | MATa/MATalpha leu2/leu2 trp1-289/trp1-289 ura3−52/ura3-52 his3-∆1/his3-∆1 | Invitrogen |
Primers used in this study.
https://doi.org/10.7554/eLife.30205.017| Primer number | Sequence | used to make plasmid number |
|---|---|---|
| LD717 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCAGGACAAGCAGTGGGAAC | pLD1852 |
| LD718 | aggtgttgtaggccatcagcTGGCGGACCACCTTATAGTT | |
| LD958 | aactataaggtggtccgccaCGGTGGTTTCTTCCTCACC | |
| LD959 | ggaattgtgagcggataacaatttcacacaggaaacagctGGTCCAGCCTTTTTCCTTGT | |
| LD725 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCCCCTCAGAGAAGTCAGTCG | pLD1610 |
| LD726 | aggtgttgtaggccatcaggGGCGGACCACCTTGTAGTTA | |
| LD727 | taactacaaggtggtccgccCCTGATGGCCTACAACACCT | |
| LD728 | ggaattgtgagcggataacaatttcacacaggaaacagctCAGCGGGTTGTCATACTCCT | |
| LD741 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatTCGAGGGCTTCGAGAAGAT | pLD1616 |
| LD742 | aggtgttgtaggccatcagcCAGGGTCATCAGGGTGAACT | |
| LD743 | agttcaccctgatgaccctgGCTGATGGCCTACAACACCT | |
| LD744 | ggaattgtgagcggataacaatttcacacaggaaacagctCGGGTGATGTCGACGTATTC | |
| LD438 | ggaattgtgagcggataacaatttcacacaggaaacagctCCGTTGATTTCCTTCTGCAT | pLD1264 (LD438 - LD441) pLD1853 (LD438 and LD441) |
| LD439 | ctacaaggtggttcgccagtCGCTGACCTACTCCTTCGTC | |
| LD440 | gacgaaggagtaggtcagcgACTGGCGAACCACCTTGTAG | |
| LD441 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCATCGACCTGGAAGTGCTC | |
| LD725 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCCCCTCAGAGAAGTCAGTCG | pLD1929 |
| LD1063 | gttgcccaggtgttcctgtGGCGGACCACCTTGTAGTTA | |
| LD949 | ggaattgtgagcggataacaatttcacacaggaaacagctTGTAGTCGAGGGACTTCTTGC | |
| LD1064 | taactacaaggtggtccgccACAGGAACACCTGGGCAAC | |
| LD2168 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatATGTAGGGATCGAGCGACAG | pLD2791 |
| LD2169 | acacgatatccagcccctctTGGACATCGCGCCGTTCCTC | |
| LD2170 | gaggaacggcgcgatgtccaAGAGGGGCTGGATATCGTGT | |
| LD2171 | ggaattgtgagcggataacaatttcacacaggaaacagctAAGAGGTCATAATCGGCGGT | |
| LD2120 | gattcgacatcactagtACGCCCAGCTCCAACAAA | pLD2777 |
| LD2121 | gattcgatgccctcgaGCTAGGGGTTCCACGGTTAAT | |
| LD2122 | gattcgactgcactagtCATCGACTTGCCGCCCAG | pLD2778 |
| LD2123 | g attcg atg ccctcgaGCTATGGGCTTCCATC CAC | |
| LD2124 | gattcgactgcactagtGGCTACTTCCTCTGGCTGG | pLD2779 |
| LD2125 | gattcgactgcctcgagCTGTACAGTCCCGAAAGAAATGAAC | |
| LD1118 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatTCTTCAGGTTCTCGCGGTAG | pLD1966 |
| LD1119 | aagtgccagtaccaactggcGCAGATCCAGAAGATGGTCA | |
| LD1120 | tgaccatcttctggatctgcGCCAGTTGGTACTGGCACTT | |
| LD1121 | ggaattgtgagcggataacaatttcacacaggaaacagctATCGCGAGACTCATGGTTTT | |
| LD1134 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatCGCTGCTTGTCGATCTGTT | pLD1967 |
| LD1135 | gcgacatgaccctgttcaacCTGACCGGCTACTGGACC | |
| LD1136 | ggtccagtagccggtcagGTTGAACAGGGTCATGTCGC | |
| LD1137 | ggaattgtgagcggataacaatttcacacaggaaacagctCCTCGGCGACCATGAATAC | |
| LD1126 | ccaggcaaattctgttttatcagaccgcttctgcgttctgatTTCAGGTTCTTCGGGTTCTC | pLD2044 |
| LD1187 | aacagcgcgccgaccagcatCTCTTCGTTCGTTTTCAGCC | |
| LD1188 | ggctgaaaacgaacgaagagATGCTGGTCGGCGCGCTGTT | |
| LD1189 | ggaattgtgagcggataacaatttcacacaggaaacagctGCGTTGATGAAGCGGATAAC |
Plasmids used in this study.
https://doi.org/10.7554/eLife.30205.018| Plasmid | Description | Source |
|---|---|---|
| pMQ30 | 7.5 kb mobilizable vector; oriT, sacB, GmR. | Shanks et al. (2006) |
| pAKN69 | Contains mini-Tn7(Gm)PA1/04/03::eyfp fusion. | Lambertsen et al. (2004) |
| pLD2722 | GmR, TetR flanked by Flp recombinase target (FRT) sites to resolve out resistance casettes. | this study |
| pFLP2 | Site-specific excision vector with cI857-controlled FLP recombinase encoding sequence, sacB, ApR. | Hoang et al. (1998) |
| pLD1852 | ∆ccoN1 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD1610 | ∆ccoN2 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD1616 | ∆ccoN3 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD1264 | ∆ccoN4 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD1929 | ∆cco1 cco2 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD2791 | ∆hcn PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. | this study |
| pLD1853 | Full genomic sequence of ccoN4 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain InvSc1. Verified by sequencing. | this study |
| pLD1966 | ∆aa3 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain IncSc1. | this study |
| pLD1967 | ∆bo3 PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain IncSc1. | this study |
| pLD2044 | ∆cio PCR fragment introduced into pMQ30 by gap repair cloning in yeast strain IncSc1. | this study |
| pLD2777 | PCR-amplified cco1 promoter ligated into pSEK103 using SpeI and XhoI. | this study |
| pLD2778 | PCR-amplified cco2 promoter ligated into pSEK103 using SpeI and XhoI. | this study |
| pLD2779 | PCR-amplified ccoN4 promoter ligated into pSEK103 using SpeI and XhoI. | this study |
Statistical analysis.
https://doi.org/10.7554/eLife.30205.019| Figure 2B | Number of values (biological replicates) | mean | median | SD | SEM | Lower 95% confidence interval of mean | Upper 95% confidence interval of mean |
|---|---|---|---|---|---|---|---|
| WT | 5 | 73.22 | 72.94 | 3.387 | 1.515 | 69.02 | 77.43 |
| ∆N4 | 5 | 68.97 | 70.6 | 6.44 | 2.88 | 60.97 | 76.96 |
| ∆N1∆N2 | 5 | 52.18 | 50.46 | 5.142 | 2.3 | 45.79 | 58.56 |
| ∆N1∆N4 | 5 | 11.57 | 12.42 | 2.011 | 0.8991 | 9.074 | 14.07 |
| ∆N1∆N2∆N4 | 5 | 0.001958 | 0.001117 | 0.001696 | 0.0007586 | −0.0001481 | 0.004064 |
| ∆cco1cco2 | 5 | 0.001367 | 0.0008644 | 0.001237 | 0.0005532 | −0.0001686 | 0.002903 |
| t-test | p value | p value summary | |||||
| WT vs. ∆N4 | 0.2273 | ns | |||||
| WT vs. ∆N1∆N2 | <0.0001 | **** | |||||
| WT vs. ∆N1∆N4 | <0.0001 | **** | |||||
| WT vs. ∆N1∆N2∆N4 | <0.0001 | **** | |||||
| WT vs. ∆cco1cco2 | <0.0001 | **** | |||||
| Figure 3A | Number of values (biological replicates) | mean | median | SD | SEM | Lower 95% confidence interval of mean | Upper 95% confidence interval of mean |
| WT-YFP | 12 | 54.95 | 54.92 | 4.387 | 1.266 | 52.16 | 57.74 |
| ∆N4-YFP | 3 | 29.92 | 30.83 | 2.234 | 1.29 | 24.37 | 35.46 |
| ∆N1∆N2-YFP | 3 | 30.49 | 31.91 | 3.527 | 2.036 | 21.73 | 39.25 |
| ∆N1∆N2∆N4-YFP | 3 | 4.408 | 4.296 | 3.23 | 1.865 | −3.617 | 12.43 |
| ∆cco1cco2-YFP | 3 | 7.097 | 5.306 | 4.093 | 2.363 | −3.072 | 17.27 |
| t-test | p value | p value summary | |||||
| WT-YFP vs. ∆N4- YFP | <0.0001 | **** | |||||
| WT-YFP vs. ∆N1∆N2-YFP | <0.0001 | **** | |||||
| ∆N1∆N2-YFP vs. ∆N1∆N2∆N4-YFP | 0.0007 | *** | |||||
| ∆N1∆N2-YFP vs. ∆cco1cco2-YFP | 0.0017 | ** | |||||
| Figure 3—figure supplement 1A | Number of values (biological replicates) | mean | median | SD | SEM | Lower 95% confidence interval of mean | Upper 95% confidence interval of mean |
| WT) | 12 | 45.05 | 45.08 | 4.387 | 1.266 | 42.26 | 47.84 |
| ∆N4 | 3 | 28.22 | 31.31 | 7.442 | 4.297 | 9.731 | 46.71 |
| ∆N1∆N2 | 3 | 27.81 | 28.57 | 2.514 | 1.451 | 21.56 | 34.05 |
| ∆N1∆N2∆N4 | 3 | 7.002 | 6.973 | 0.7508 | 0.4335 | 5.137 | 8.867 |
| ∆cco1cco2 | 3 | 5.38 | 4.183 | 2.146 | 1.239 | 0.05034 | 10.71 |
| t-test | p value | p value summary | |||||
| WT vs. ∆N4 | 0.0002 | *** | |||||
| WT vs. ∆N1∆N2 | <0.0001 | **** | |||||
| ∆N1∆N2 vs. ∆N1∆N2∆N4 | 0.0002 | *** | |||||
| ∆N1∆N2 vs. ∆cco1cco2 | 0.0003 | *** | |||||
| Figure 5 | Number of values (biological replicates) | mean | median | SD | SEM | Lower 95% confidence interval of mean | Upper 95% confidence interval of mean |
| WT | 8 | 150.3 | 151.2 | 10.31 | 3.644 | 141.7 | 158.9 |
| ∆N1∆N2 | 4 | 139.3 | 137.6 | 12.33 | 6.166 | 119.6 | 158.9 |
| ∆N4 | 7 | 131.9 | 127.8 | 8.915 | 3.369 | 123.7 | 140.2 |
| ∆N1∆N4 | 4 | 99.96 | 99.34 | 2.726 | 1.363 | 95.62 | 104.3 |
| ∆cco1cco2 | 4 | 95.19 | 95.56 | 1.559 | 0.7793 | 92.71 | 97.67 |
| ∆N1∆N2∆N4 | 4 | 102.8 | 99.79 | 8.664 | 4.332 | 88.98 | 116.6 |
| ∆phz | 7 | 84.98 | 84.23 | 10.93 | 4.131 | 74.87 | 95.09 |
| t-test | p value | p value summary | |||||
| WT vs. ∆N1∆N2 | 0.1302 | ns | |||||
| WT vs. ∆N4 | 0.0028 | ** | |||||
| WT vs. ∆N1∆N4 | <0.0001 | **** | |||||
| WT vs. ∆cco1cco2 | <0.0001 | **** | |||||
| WT vs. ∆N1∆N2∆N4 | <0.0001 | **** | |||||
| WT vs. ∆phz | <0.0001 | **** | |||||
| Figure 6 | Number of values (biological replicates) | mean | median | SD | SEM | Lower 95% confidence interval of mean | Upper 95% confidence interval of mean |
| WT | 9 | 27.44 | 39 | 18.48 | 6.16 | 13.24 | 41.65 |
| gacA::Tn | 9 | 92.56 | 93 | 8.546 | 2.849 | 85.99 | 99.12 |
| ∆N1∆N2 | 4 | 19 | 21.5 | 14.07 | 7.036 | −3.39 | 41.39 |
| ∆N1∆N2∆N4 | 6 | 64.17 | 68 | 18 | 7.35 | 45.27 | 83.06 |
| ∆cco1cco2 | 9 | 70.56 | 76 | 22.69 | 7.565 | 53.11 | 88 |
| t-test | p value | p value summary | |||||
| WT vs. ∆N1∆N2∆N4 | 0.0022 | ** | |||||
| ∆N1∆N2 vs. ∆N1∆N2∆N4 | 0.0030 | ** | |||||
| WT vs. ∆N1∆N2 | 0.4362 | ns |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30205.020





