Loss of functional BAP1 augments sensitivity to TRAIL in cancer cells
Figures
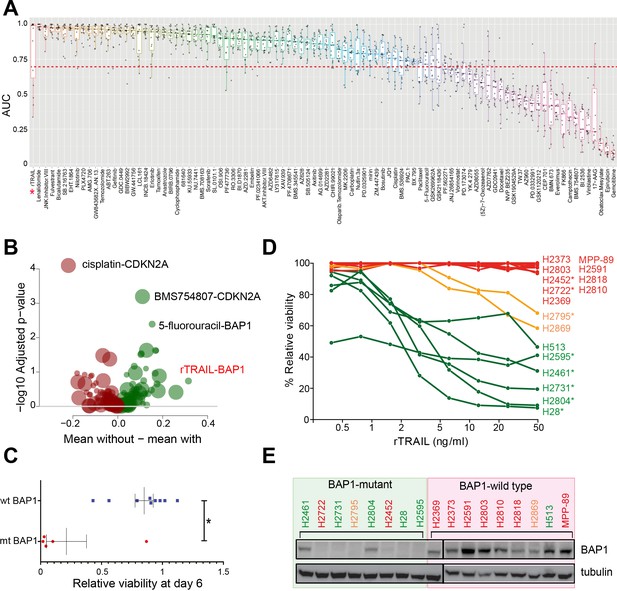
A chemical screen in mesothelioma cell lines identifies a BAP1-mutant population sensitised to the death receptor ligand rTRAIL.
(A) Area under the curve (AUC) values for 15 malignant mesothelioma (MM) cells treated for 6 days with 94 compounds. Each dot indicates the AUC value for an individual cell line treated. AUC <0.7 is indicated by the red dotted line — only those compounds with ≥2 cell lines below this value were analysed for statistically significant associations with gene mutations. The AUC values for rTRAIL are indicated by the red asterisk. (B) A Welch t-test was used to test for significant pharmacogenomics interactions between the 94 single agents in the screen and the presence of driver mutations in any of 5 MM cancer genes. Each volcano plot circle corresponds to a significant gene–drug interaction whose position on the x-axis indicates the corresponding effect size. Both half-axes are positive; the right side (green circles) indicates the effect sizes of sensitivity associations, whereas the left side (red circles) corresponds with the effect sizes of resistance associations. The position on the y-axis indicates the statistical significance of the identified interaction. The size of a given circle is proportional to the number of samples in which the selected functional event involved in the corresponding interaction occurs. Specific examples of associations are indicated where the effect size is large (rTRAIL and BAP1 mutations) or highly significant (cisplatin and CDKN2A mutations). (C) Cell viability between wild-type BAP1 (wt BAP1) (n = 10) and mutant BAP1 (mt BAP1) (n = 5) MM lines following 6 days of treatment with rTRAIL (t-test; *p=0.015). (D) Cell viability data for 17 MM lines treated for 6 days with a concentration range of rTRAIL (0.4–50 ng/ml). MM lines are coloured according to their sensitivity pattern (green = sensitive (S); orange = partially sensitive (PS); red = resistant (R)). *Indicates cell lines harbouring BAP1 mutations. (E) Immunoblot of BAP1 protein expression in BAP1-mutant versus BAP1-wild-type MM cell lines. Sensitivity to rTRAIL treatment is indicated as font colour: green (S); orange (PS); red (R).
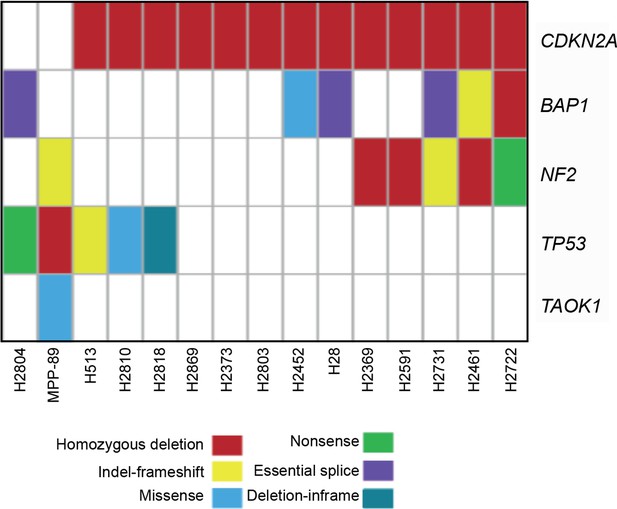
Mutation status of 5 candidate tumour driver genes in the 15 MM lines used in the combinatorial chemical inhibitor screen.
https://doi.org/10.7554/eLife.30224.004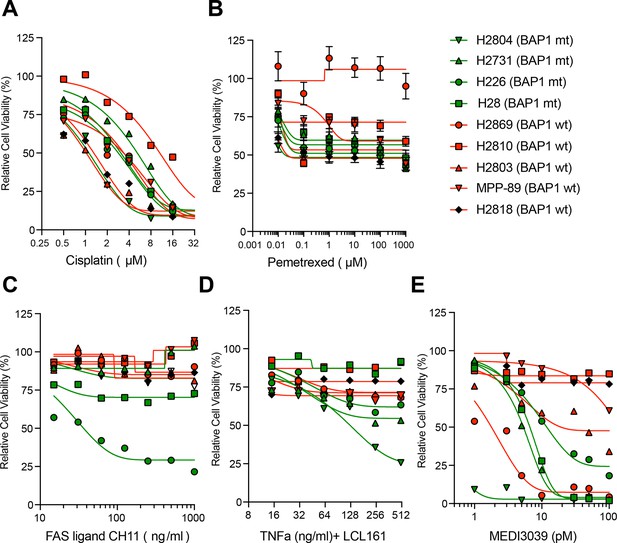
BAP1 and the response to alternative apoptotic stimuli in MM cells.
72 hour cell viability results for 9 MM cell lines (4 BAP1-mutant - green and five wild-type - red) treated with (A) cisplatin (B) pemetrexed (C) FAS receptor agonistic antibody CH11, (D) TNF-α and 5 μM LCL161 or (E) DR5 agonist MEDI3039 assessed by MTT assay.
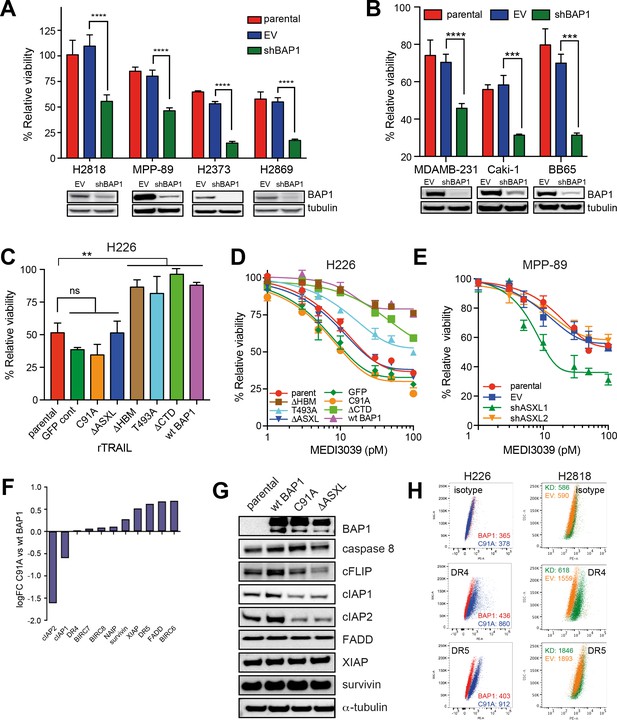
BAP1-induced TRAIL resistance extends to other cancer subtypes and is dependent upon functional deubiquitinase and ASXL-binding sites.
(A) BAP1-wild-type H2818, MPP-89, H2373 and H2869 MM lines were transduced with BAP1 (shBAP1) or empty vector (EV) shRNA. Immunoblot confirmed BAP1 knockdown in the BAP1 shRNA-transduced cells. Parental and transduced cells were treated with rTRAIL (1000 ng/ml) and cell viability assessed after 72 hr by MTT assay (t-test; ****p<0.0001). (B) The BAP1-wild-type breast cancer line MDAMB-231 and the renal cell carcinoma (RCC) lines Caki-1 and BB65 were transduced with BAP1 (shBAP1) or empty vector (EV) shRNA. Immunoblot confirmed BAP1 knockdown in the BAP1 shRNA transduced cells. Parental and transduced cells were treated with rTRAIL (1000 ng/ml) and cell viability assessed after 72 hr by MTT assay (t-test; ****p<0.0001). (C) The rTRAIL-sensitive H226 MM line, which harbours a homozygous deletion of BAP1, was transduced with either a GFP control, wild-type BAP1 or a mutant BAP1 containing an inactive functional domain: C91A — inactivating mutation of deubiquitinase catalytic site; ΔHBM — deletion of HCF-1-binding motif; T493A — inactivating mutation of FOXK2-binding site; ΔASXL — deletion of ASXL1/2 protein-binding site; ΔCTD — deletion of C-terminal domain containing nuclear localisation signal. These transduced lines were treated with 50 ng/ml rTRAIL and cell death assessed with XTT assay (one-way ANOVA; **p<0.01). (D) The parental and transduced H226 MM lines were treated with a concentration range (1–100 pM) of the small molecule death receptor agonist MEDI3039 and cell viability assessed with XTT assay. (E) The BAP1-wild-type MPP-89 MM line was transduced with ASXL1 (shASXL1), ASXL2 (shASXL2) or empty vector (EV) shRNA. qPCR confirmed a decrease in ASXL1 and ASXL2 mRNA expression in the ASXL1 shRNA and ASXL2 shRNA-transduced cells, respectively (Figure 2—figure supplement 6). Parental and transduced cells were treated with a concentration range (1–100 pM) of MEDI3039 and cell viability assessed with XTT assay. (F) Differential gene expression of apoptosis regulator genes in the catalytically inactive BAP1-mutant (C91A) relative to the wild-type BAP1-transduced (wt BAP1) H226 cells. (G) Immunoblot of apoptosis regulator proteins in the catalytically inactive BAP1-mutant (C91A), inactive ASXL1/2-binding site BAP1-mutant (ΔASXL) or wild-type BAP1-transduced (wt BAP1) H226 cells. (H) Flow cytometry analysis of death receptor 4 (DR4) and 5 (DR5) cell surface expression in H226 cells transduced with the catalytically inactive BAP1-mutant (C91A) or wild-type BAP1 (wt BAP1) and of BAP1-wild-type H2818 MM cells transduced with BAP1 (KD) or empty vector (EV) shRNA. The values represent the median fluorescence intensity (MFI).
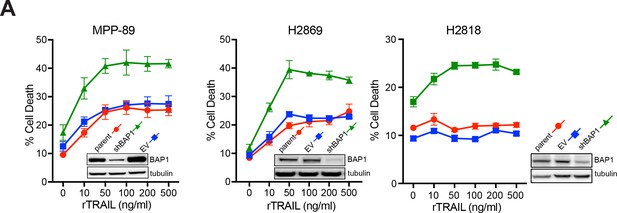
shRNA knockdown of BAP1 increases sensitivity to rTRAIL in MM cells.
Three BAP1-wild-type MM cell lines (A) MPP-89, (B) H2869 and (C) H2818 were transduced with empty vector (EV) or BAP1 shRNA (shBAP1). Immunoblot confirmed BAP1 knockdown. The parental, EV and shBAP1 cells were treated with rTRAIL for 24 hr and cell death measured by Annexin V/DAPI flow cytometry assay.
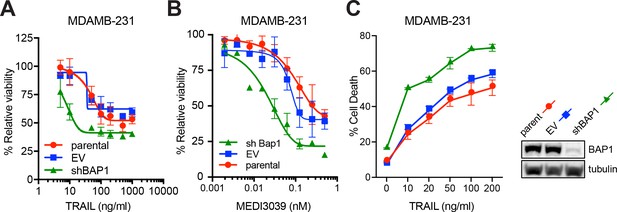
shRNA knockdown of BAP1 increases sensitivity to DR agonists in breast cancer cells.
The MDAMB-231 breast cancer cell line was transduced with empty vector (EV) or BAP1 shRNA (shBAP1). Immunoblot confirmed BAP1 knockdown. Cells were treated with (A) rTRAIL and (B) MEDI3039 and cell viability measured with MTT assay at 72 hr. (C) Cells were treated with rTRAIL for 24 hr and cell death measured with Annexin V/DAPI flow cytometry assay.
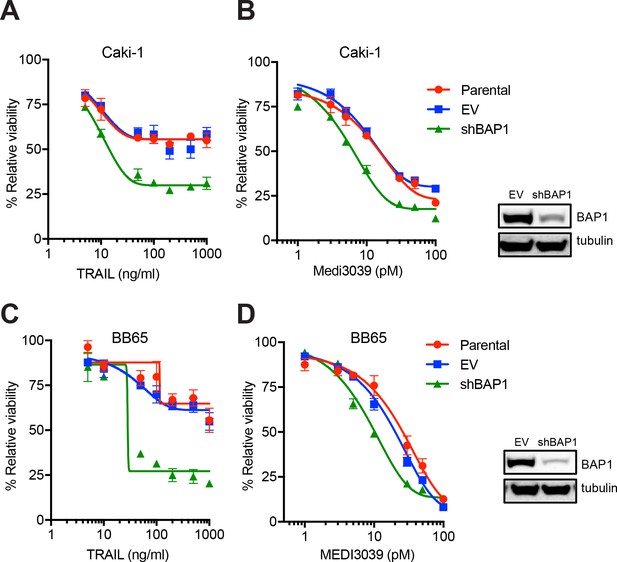
shRNA knockdown of BAP1 increases sensitivity to DR agonists in clear cell renal carcinoma cells.
Clear cell renal carcinoma cell lines, Caki-1 and BB65, were transduced either with either empty vector (EV) or BAP1 shRNA (shBAP1). Immunoblot confirmed BAP1 knockdown. Cells were treated with rTRAIL (A and C) or MEDI3039 (B and D) for 72 hr and cell viability measured by MTT assay.
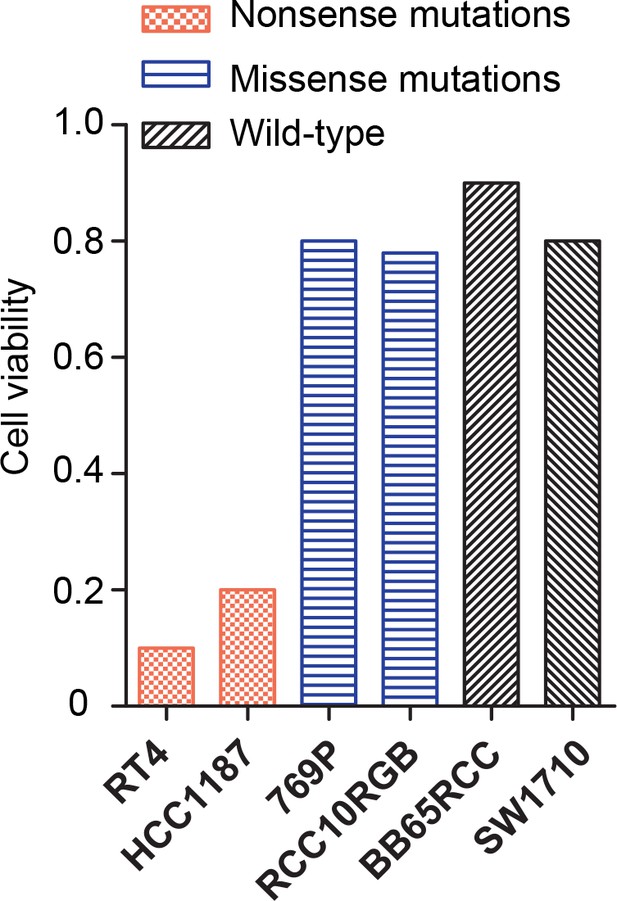
Cell viability of non-mesothelioma BAP1-mutant cell lines following rTRAIL treatment.
Bladder (RT4) and breast (HCC1187) cancer cell lines harbouring nonsense mutations in BAP1 show increased sensitivity to rTRAIL compared with renal cell carcinoma or bladder cancer cell lines harbouring missense (769P and RCC10RGB) or wild-type BAP1 (BB65RCC and SW1710). Cell viability was measured after 6 days of treatment with 100 ng/ml rTRAIL.
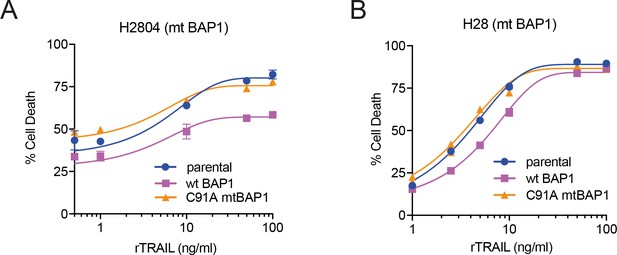
Overexpression of wild-type BAP1 induces resistance to rTRAIL in BAP1 mutant MM cells.
The rTRAIL-sensitive H2804(A) and H28(B) mesothelioma cell lines, which harbour mutations in BAP1, were transduced with wild-type BAP1 (wt BAP1) or BAP1 with an inactive deubiquitinase catalytic domain (C91A) and treated with a dose range of rTRAIL.
Cell death was assessed with Annexin V/DAPI apoptosis assay.
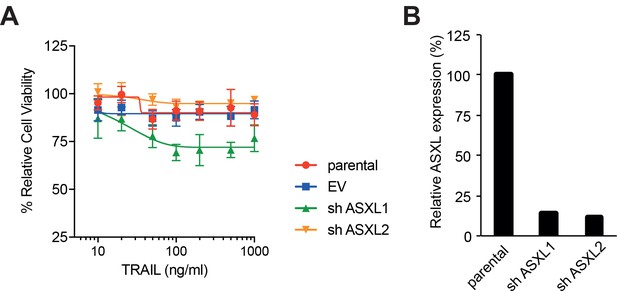
shRNA knockdown of ASXL1 increases sensitivity of MM cells to rTRAIL.
(A) Cell viability of parental, empty vector, ASXL1 and ASXL2 shRNA-transduced MPP-89 cells treated with rTRAIL (0–1000 ng/ml) for 3 days measured with XTT assay. (B) Efficacy of ASXL1 and ASXL2 shRNA knockdown assessed by qPCR.
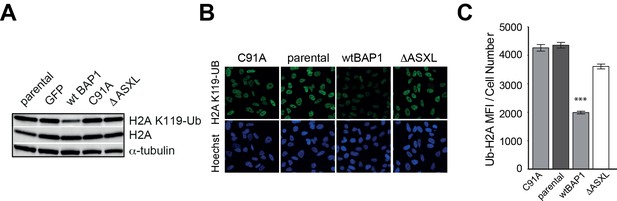
Ubiquitinated histone 2A at K119 (H2AK119Ub) expression and BAP1 function.
(A) Immunoblot analysis of H2AK119Ub levels in the parental, GFP-, wild-type BAP1 (wt BAP1)-, deubiquitinase mutant BAP1 (C91A)- and ASXL-binding mutant BAP1 (ΔASXL)-transduced H226 MM cell lines. (B) Immunofluorescence images of H2AK119Ub staining in the parental, deubiquitinase mutant-transduced (C91A), ASXL-binding mutant-transduced (ΔASXL) and wild-type BAP1-transduced H226 cell lines. (C) Quantification of immunofluorescence staining in 2B (normalised to cell number; one-way ANOVA; ***p<0.001).
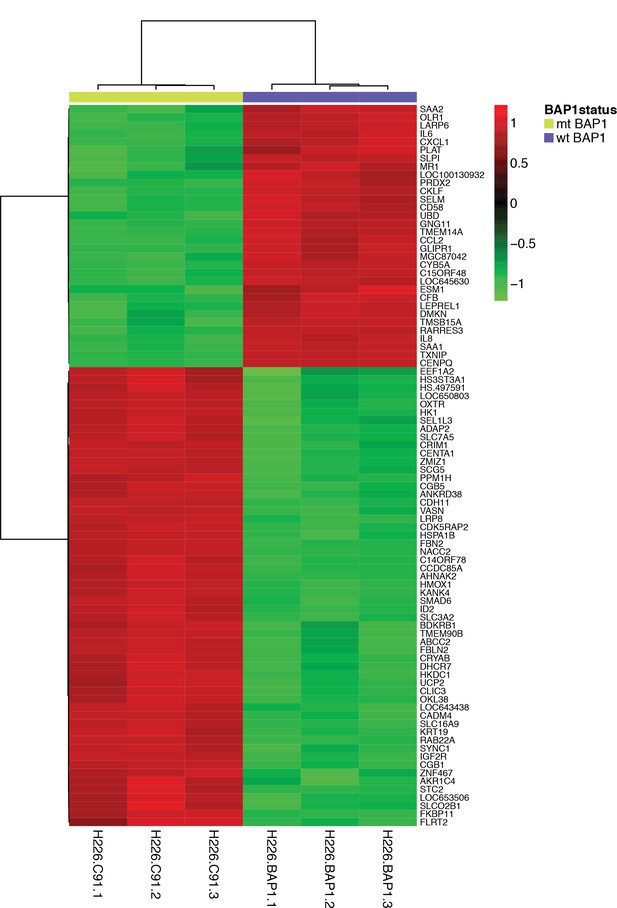
Differential gene expression data from H226 MM cells expressing C91A-mutant (mt BAP1) or wild-type BAP1 (wt BAP1).
Only genes with logFC ≥2 and adj.p <0.05 are displayed.
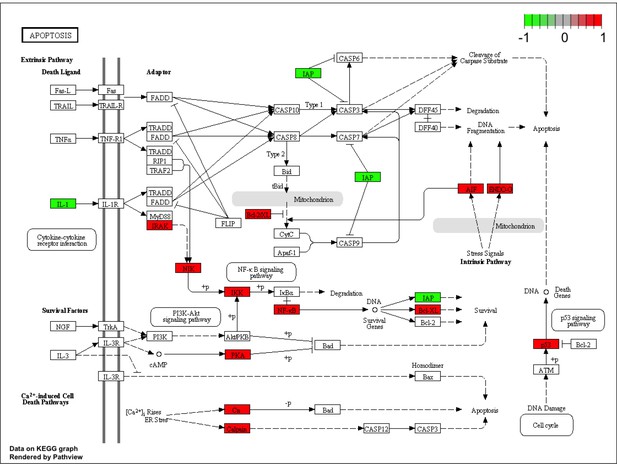
Signalling pathway impact analysis of gene expression data from H226 MM cells expressing C91A-mutant (mt BAP1) or wild-type BAP1 (wt BAP1).
The proteins in the pathway are highlighted in green if the expression in mt BAP1 is significantly less than wt BAP1 and red if the expression in mt BAP1 is significantly more than wt BAP1.
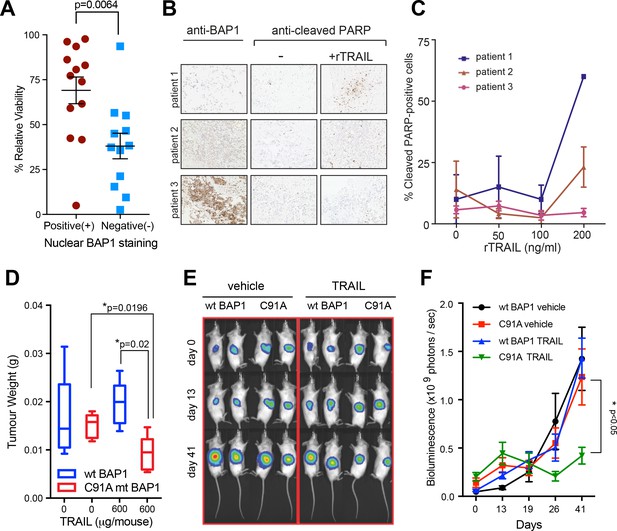
Loss of functional BAP1 leads to TRAIL sensitivity in early passage mesothelioma cell lines, human tumour explants and mouse xenograft models.
(A) Mean cell viability effect between human early passage MM cell lines (positive nuclear BAP1 staining; n = 13 and negative nuclear BAP1 staining; n = 12) as assessed by immunohistochemistry following 3 days of treatment with rTRAIL (50 ng/ml) (t-test, p=0.0067). (B) Immunohistochemical images of tumour explants derived from three MM patients treated with either vehicle or rTRAIL for 24 hr. Explants were stained with anti-BAP1 and anti-cleaved PARP (marker for apoptosis) antibodies. (C) The percentage of cleaved PARP-positive cells in tumour explants derived from three patients and treated with either vehicle or 0, 50, 100 and 200 ng/ml of rTRAIL for 24 hr was scored based on the percentage of cells with nuclear cleaved PARP-positive staining. (D) Weights of tumour xenografts derived from BAP1-wild-type (wt BAP1) versus catalytically inactive BAP1-mutant (C91A mt BAP1) transduced MM cells following treatment with either vehicle or TRAIL (600 μg per mouse) at the time of sacrifice (day 42) (t-test). (E) Serial bioluminescence imaging of BAP1-wild-type (wt BAP1) and catalytically inactive BAP1-mutant (C91A) MM xenografts in mice treated with either vehicle or TRAIL. Mice were imaged on day 0 (after tumour inoculation), day 13 (before TRAIL administration) and day 41 (time of sacrifice). The intensity of luminescence is denoted by colour: red - high luciferase signal (high tumour burden) and blue - low luciferase signal (low tumour burden). (F) A time-course of bioluminescence scores in BAP1-wild-type (wt BAP1) versus catalytically inactive BAP1-mutant (C91A) MM tumour xenografts. Bioluminescence was measured on days 0, 13, 19, 26 and 41, 15 min after injecting the mice with 0.2 ml luciferin intraperitoneally. The number of photons emitted per second indicates the tumour burden (two way ANOVA).
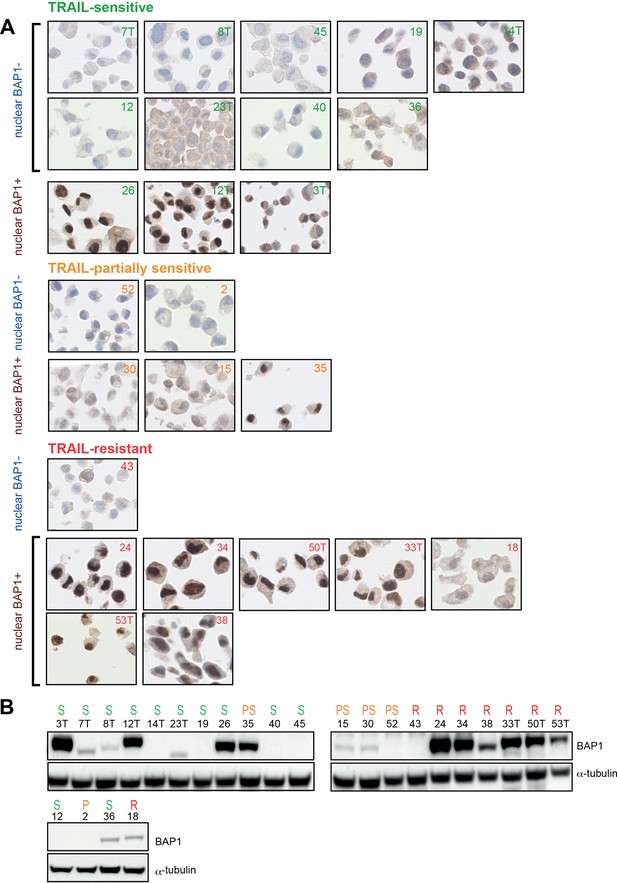
BAP1 expression in early passage MM cultures.
(A) Immunohistochemical analysis and (B) immunoblot. Cultures are grouped by sensitivity to rTRAIL.
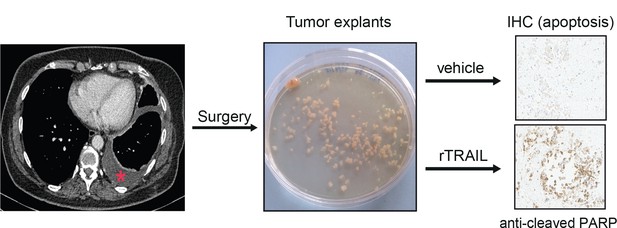
Ex vivo experimental protocol.
Tumour explants were obtained by cutting primary pleural tissue from patients with MM who underwent pleurectomy into fragments of approximately 2 mm3.
The explants were treated with vehicle or rTRAIL (50 ng/ml, 100 ng/ml or 200 ng/ml) for 24 hr, following which time explants were fixed and stained for cleaved-PARP (which is a marker of apoptosis).
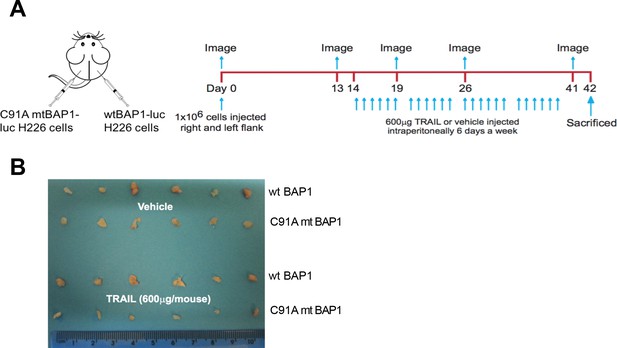
In vivo experimental protocol.
(A) Schematic of in vivo experimental protocol. Mice were injected with H226 cells transduced with wild-type BAP1 and luciferase or catalytically inactive BAP1-mutant (C91A) and luciferase on the right and left flanks, respectively. Mice were divided into two groups, each of which received 600 μg TRAIL or vehicle 6 days a week (day 14–40). Tumour size was assessed longitudinally with bioluminescence imaging on days 0, 13, 19, 26 and 41. (B) Size of tumours derived from BAP1-wild-type (wt) versus catalytically inactive (C91A) BAP1-mutant (mt) MM cells following treatment with either vehicle or TRAIL (600 μg per mouse) at time of sacrifice (day 42). A centimetre scale is included in the photograph for comparison.
Tables
BAP1 immunoblot status, nuclear BAP1 staining and rTRAIL sensitivity (50 ng/ml) of the 25 human early passage MM cultures.
https://doi.org/10.7554/eLife.30224.016| Sample name | Western blot | Nuclear BAP1-IHC | Sensitivity |
|---|---|---|---|
| 7T | − | − | Sensitive |
| 8T | − | − | Sensitive |
| 45 | − | − | Sensitive |
| 19 | − | − | Sensitive |
| 14T | − | − | Sensitive |
| 12 | − | − | Sensitive |
| 23T | − | − | Sensitive |
| 40 | − | − | Sensitive |
| 36 | Low Expression | − | Sensitive |
| 26 | + | + | Sensitive |
| 12T | + | + | Sensitive |
| 3T | + | + | Sensitive |
| 52 | − | − | Partially Sensitive |
| 2 | − | − | Partially Sensitive |
| 30 | Low Expression | + | Partially Sensitive |
| 15 | Low Expression | + | Partially Sensitive |
| 35 | + | + | Partially Sensitive |
| 24 | + | + | Partially Sensitive |
| 43 | − | − | Resistant |
| 34 | + | + | Resistant |
| 50T | + | + | Resistant |
| 33T | + | + | Resistant |
| 18 | + | + | Resistant |
| 53T | + | + | Resistant |
| 38 | + | + | Resistant |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene | ||||
| BRCA associated protein-1 (human) | BAP1 | Entrez Gene NCBI | Gene ID: 8314 | |
| Additional sex combs like 1 (human) | ASXL1 | Entrez Gene NCBI | Gene ID: 171023 | |
| strain, strain background | ||||
| NOD.CB17-Prkdcscid/NcrCrl | NOD SCID mice | Charles River Laboratories, UK | RRID:IMSR_CRL:394 | |
| cell line | ||||
| Early passage mesotheliomacell cultures | 7T, 8T, 45, 19, 14T, 23T, 40, 36, 26, 12T, 3T, 52, 2, 30, 15, 35, 24, 43, 34, 50T, 33T, 18, 53T, 38 | MesobanK, Mesothelioma UK | www.mesobank.com Mesothelioma Tissue Bank, Papworth Hospital NHS Trust, UK | |
| NCI-H2373 | H2373 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A533 | |
| NCI-H2803 | H2803 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U997 | |
| NCI-H2452 | H2452 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_1553 | |
| NCI-H2722 | H2722 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U994 | |
| NCI-H2369 | H2369 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A532 | |
| NCI-H2795 | H2795 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U996 | |
| NCI-H2869 | H2869 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_V001 | |
| NCI-H2591 | H2591 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A543 | |
| MPP 89 | MPP-89 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_1427 | |
| NCI-H2810 | H2810 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U999 | |
| NCI-H2818 | H2818 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_V000 | |
| NCI-H513 | H513 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A570 | |
| NCI-H2595 | H2595 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A545 | |
| NCI-H2461 | H2461 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_A536 | |
| NCI-H2731 | H2731 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U995 | |
| NCI-H2804 | H2804 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_U998 | |
| NCI-H28 | H28 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_1555 | |
| NCI-H226 | H226 | Szlosarek lab, Barts Cancer Institute, UK | RRID:CVCL_1544 | |
| MDA-MB-231 | MDAMB-231 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_0062 | |
| Caki-1 | Caki-1 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_0234 | |
| BB65 | BB65 | Wellcome Trust Sanger Institute, UK | RRID:CVCL_1078 | |
| antibody | ||||
| BAP1 (C-4) mouse mAb | anti-BAP1 | Santa Cruz Biotechnology, Santa Cruz, CA | Cat# sc-28383 RRID:AB_626723 | 1:500 in milk; 1:50 for flow cytometry |
| Caspase-8 (1C12) mouse mAb | anti-caspase 8 | Cell Signaling Technology, Danvers, MA | Cat# 9746 RRID:AB_2275120 | 1:1000 in BSA |
| FLIP (7F10) mouse mAb | anti c-FLIP | Enzo Life Sciences, Farmingdale, NY | Cat# ALX-804-961-0100 RRID:AB_2713915 | 1:1000 in milk |
| c-IAP1 (D5G9) rabbit mAb | anti-cIAP1 | Cell Signaling Technology,Danvers, MA | Cat# 7065S RRID:AB_10890862 | 1:1000 in BSA |
| c-IAP2 (58C7) rabbit mAb | anti-cIAP2 | Cell Signaling Technology, Danvers, MA | Cat# 3130S RRID:AB_10693298 | 1:1000 in BSA |
| FADD rabbit pAb | anti-FADD | Cell Signaling Technology, Danvers MA | Cat# 2782 RRID:AB_2100484 | 1:1000 in BSA |
| XIAP (3B6) rabbit mAb | anti-XIAP | Cell Signaling Technology, Danvers, MA | Cat# 2045 RRID:AB_2214866 | 1:1000 in milk |
| survivin rabbit pAb | anti-survivin | Cell Signaling Technology, Danvers, MA | Cat# 2803 RRID:AB_490807 | 1:1000 in BSA |
| α-Tubulin (11H10) Rabbit mAb | anti-α-tubulin | Cell Signaling Technology, Danvers, MA | #2125 | 1:2000 in milk |
| Ubiquityl-Histone H2A (Lys119) (D27C4) XPRabbit mAb | anti-H2AK119Ub | Cell Signaling Technology, Danvers, MA | Cat# 8240P RRID:AB_10891618 | 1:2000 in BSA |
| Histone H2A (D6O3A) Rabbit mAb | anti-H2A | Cell Signaling Technology, Danvers, MA | Cat# 12349 RRID:AB_2687875 | 1:1000 in BSA |
| Anti-mouse IgG, HRP-linked antibody | anti-mouse HRP | Cell Signaling Technology, Danvers, MA | Cat# 7076 RRID:AB_330924 | 1:2000 in milk |
| Anti-rabbit IgG, HRP-linked antibody | anti-rabbit HRP | Cell Signaling Technology, Danvers, MA | Cat# 7074 RRID:AB_2099233 | 1:2000 in milk |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, AlexaFluor 488 | AlexaFluor 488-conjugated anti-mouse antibody | Thermo Fisher Scientific, UK | Cat# A-21202 RRID:AB_141607 | 1:200 for flow cytometry |
| Annexin V, AlexaFluor 647 conjugate | Annexin V AlexaFluor 647-conjugated antibody | Thermo Fisher Scientific, UK | Cat# A23204 RRID:AB_2341149 | 1:100 for flow cytometry |
| PE anti-human CD261 (DR4, TRAIL-R1) antibody | PE-conjugated antibody to DR4 | Biolegend, UK | Cat# 307205 RRID:AB_314669 | 1:100 for flow cytometry |
| PE anti-human CD262 (DR5, TRAIL-R2) antibody | PE-conjugated antibody to DR5 | Biolegend, UK | Cat# 307405 RRID:AB_314677 | 1:100 for flow cytometry |
| PE Mouse IgG1, κ Isotype Ctrl Antibody | PE isotype control antibody | Biolegend, UK | Cat# 400112 | 1:100 for flow cytometry |
| Goat anti-Rabbit IgG (H + L) Secondary Antibody, AlexaFluor 488 conjugate | AlexaFluor 488-conjugated anti-rabbit secondary antibody | Thermo Fisher Scientific, UK | Cat# R37116 RRID:AB_2556544 | 1:200 for flow cytometry |
| Anti-Cleaved PARP1 (E51) mAb | cleaved PARP primary antibody; anti-cleaved PARP | Abcam, UK | Cat# ab32064 RRID:AB_777102 | (1:6000) for immunohistochemistry |
| recombinant DNA reagent | ||||
| BAP1 (NM_004656) Human cDNA Clone | pCMV6-AC BAP1 plasmid | Origene, Rockville, MD | Cat# SC117256 | |
| pHIV-Luc-ZsGreen | ZS-green luciferase plasmid, pHIV-Luc-ZsGreen | Bryan Welm Lab, University of Utah, Addgene, Logan, UT | Cat# 39196 | |
| pCMVR8.74 | pCMV-dR8.74 | Thrasher lab, UCL, Addgene, UK | Cat# 22036 | |
| pMD2.G | pMD2.G | Thrasher lab, UCL, Addgene, UK | Cat# 12259 | |
| sequence based reagent | ||||
| BAP1 GIPZ Lentiviral shRNA | BAP1 shRNA | UCL RNAi Library (Dharmacon, Lafayett, CO) | V2LHS 41473 | |
| ASXL1 GIPZ Lentiviral shRNA | ASXL1 shRNA | UCL RNAi Library (Dharmacon, Lafayett, CO) | V2LHS 78829 | |
| ASXL2 GIPZ Lentiviral shRNA | ASXL2 shRNA | UCL RNAi Library (Dharmacon, Lafayette, CO) | V3LHS_313940 | |
| peptide, recombinant protein | ||||
| Recombinant Human sTRAIL | rTRAIL | Peprotech, UK | Cat# 310–04 | |
| commercial assay or kit | ||||
| Cell Proliferation Kit XTT | XTT reagent | Applichem, UK | A8088 | |
| Q5 Site-Directed Mutagenesis Kit | Site directed mutagenesis | New England Biolabs, Ipswich, MA | Cat# E0554 | |
| Rabbit specific HRP/DAB (ABC) Detection IHC Kit | rabbit-specific HRP/DAB (ABC) detection IHC kit | Abcam, UK | Cat# ab64261 | |
| chemical compound, drug | ||||
| MEDI3039 | MEDI3039 | MedImmune, UK | ||
| software, algorithm | ||||
| GraphPad Prism software | Graphpad Prism | GraphPad Software, CA, USA | ||
| CaVEMan algorithm | CaVEMan | https://github.com/cancerit/CaVEMan | ||
| Pindel algorithm | Pindel | https://github.com/genome/pindel | ||
| Predicting Integral Copy Numbers In Cancer algorithm | PICNIC | http://www.sanger.ac.uk/science/tools/picnic | ||
| FlowJo software | Flowjo | FlowJo LLC | ||
| Other | ||||
| RIPA buffer | RIPA | Sigma-Aldrich, St. Louis, MO | Cat# R0278 | |
| Syto™ 60 red fluorescent nucleic acid stain | Syto 60 | Thermo Fisher Scientific, UK | Cat# S11342 | |
| Thiazolyl Blue Tetrazolium Bromide (MTT) | MTT reagent | Sigma-Aldrich, St. Louis, MO | Cat# M2128 | |
| jetPEI DNA transfection reagent | jetPEI | Source Bioscience, UK | Cat# 101–10 | |
| Polybrene | Polybrene | Sigma-Aldrich, St Louis, MO | Cat# 107689 | 8 μg/ml |
| Hoechst 33342 Solution (20 mM) | Hoechst 33342 | Thermo Fisher Scientific, UK | Cat# 62249 | |
| 4’, 6-diamidino-2-phenylindole | DAPI | Sigma-Aldrich, St Louis, MO | Cat# D9542 | 200 μg/ml |
Additional files
-
Supplementary file 1
List of 94 compounds used either as single agents or in combination with rTRAIL.
Listed are the unique ID number, the compound name and target, the cellular process targeted and the minimum and maximum concentration (micromolar) of the 5-point concentration range used for each compound.
- https://doi.org/10.7554/eLife.30224.021
-
Supplementary file 2
Name and histological subtype (where known) of the 15 mesothelioma cell lines.
- https://doi.org/10.7554/eLife.30224.022
-
Supplementary file 3
1425 area under the curve (AUC) viability scores for 94 experimental agents tested against 15 mesothelioma cell lines after 6 days of treatment.
- https://doi.org/10.7554/eLife.30224.023
-
Supplementary file 4
Results of Welch’s two sample t-test from analysis of 45 single compounds that ≥2 cell lines demonstrated sensitivity to (AUC <0.7) and using the mutation status of eight genes implicated as drivers in mesothelioma in each cell line.
A 6 day viability assay was used to determine cell line sensitivity. False discovery associations < 0.2 are highlighted as red font. Whether a mutation is associated with resistance or sensitivity to that compound is indicated by red or green in the ‘effect’ column, respectively.
- https://doi.org/10.7554/eLife.30224.024
-
Supplementary file 5
Description of BAP1 mutations detected in 15 mesothelioma cell lines and the sensitivity of the cell lines to rTRAIL (as measured by a 6 day viability assay).
The sensitivity of each cell line is indicated in the last column as sensitive (green), partially sensitive (orange) or resistant (red).
- https://doi.org/10.7554/eLife.30224.025
-
Supplementary file 6
Differential gene expression values of apoptotic genes in H226 mesothelioma cells transduced with either the catalytically inactive C91A BAP1 mutant (C91A) or wild-type BAP1 (WT).
- https://doi.org/10.7554/eLife.30224.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30224.027





