Live cell-lineage tracing and machine learning reveal patterns of organ regeneration
Figures
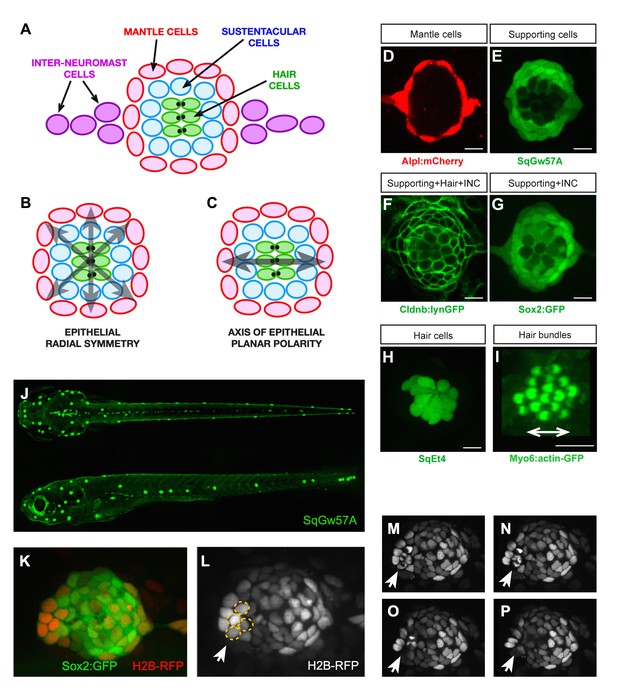
Geometric organization of the neuromast.
(A–C) Schematic representation of a neuromast depicting (A) cell classes identifiable by expression of transgenic markers. Grey arrows indicate, respectively, (B) radial symmetry and (C) epithelial planar polarity. (D–I) Confocal images of cell-specific transgenic markers. (D) Alpl:mCherry marks mantle and interneuromast cells, (E) SqGw57A shows all supporting cells, (F) Cldnb:lynGFP marks all neuromast cells, (G) Sox2-GFP marks supporting and interneuromast cells, (H) SqET4 labels hair cells, and (I) Myo6b:actin-GFP highlights the planar polarization of the hair cells by decorating their apical stereocilia. Scale bars: 10 µm. (J) Images of dorsal (top) and lateral (bottom) views of a SqGw57A transgenic zebrafish larva, revealing the full complement of superficial neuromasts and their stereotypic position. (K) A single confocal section of the lateral view of a neuromast expressing GFP in supporting cells (Sox2-GFP) and a RFP in all nuclei (H2B-RFP). (L) Same neuromast in K showing RFP-marked nuclei. The white arrow indicates 4 cells (circled), which are target of the laser beam for ablation. (M–P) Four still images of the neuromast in L over a period of five minutes, in which the laser-targeted cells are eliminated from the epithelium (white arrow).
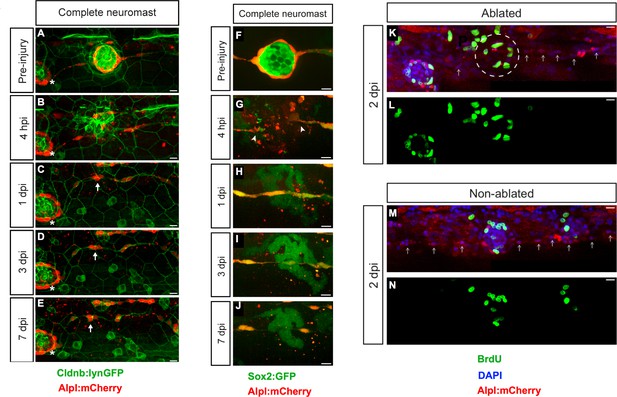
Zebrafish larvæ do not regenerate completely-ablated neuromasts.
(A–E) Confocal images of a 7 day follow-up of the complete ablation of a neuromast in the double transgenic line Tg[Cldnb:lynGFP; Alpl:mCherry]. (A) The site of damage was identified over subsequent days by the position of an intact reference neuromast (white asterisk). (B) Laser-mediated cell ablation produced a wound 4 hours-post-injury (hpi). (C–E) This wound was replaced by a thread of mCherry(+) cells (white arrow) 1 day-post-injury (dpi), which did not change over the subsequent 6 days. (F–J) Confocal images over a 7 day time course after the ablation of a neuromast in the double transgenic line Tg[Sox2:GFP; Alpl:mCherry]. Identically to A-E, the complete ablation of the target neuromast results in a thin trail of interneuromast cells (white arrowheads) covering the damaged area (K–N). Scale bars: 10 µm.
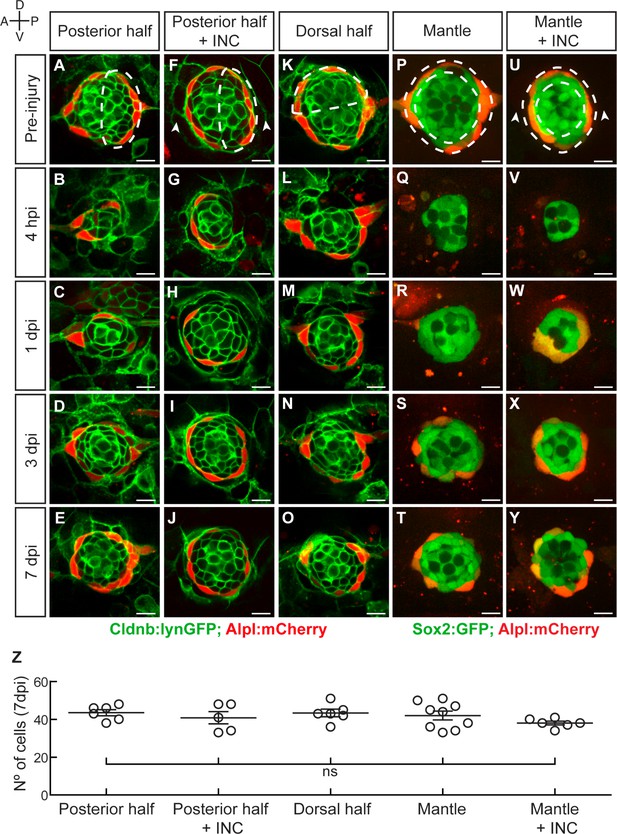
Neuromasts have isotropic regenerative capacity.
(A) Ablation of the posterior half of a neuromast. (B–C) The damage is resolved by cellular movement from the undamaged site 1dpi. (D) Neuromasts recover geometric order after 3 days and (J) return to homeostasis by 7dpi. Dashed lines in A,F,K,P,U delineate the ablated area. (F–J) Simultaneous ablation of the posterior half of a neuromast and the interneuromast cells flanking its anterior and posterior sides (n = 5) led to a regeneration outcome identical to that of the experiment in (A–E). Arrowheads in (F) point the location normally occupied by the interneuromast cells. (K–O) Neuromasts depleted from their dorsal half (n = 6) also recover epithelial size, proportions and geometry in a manner indistinguishable from equatorial-side ablation after 7 days. (P–T) 7 days after their complete laser-mediated ablation, mantle cells regenerated for neuromasts to recover the mantle. (U–Y) The ablation of interneuromast cells flanking both sides of neuromasts that were depleted of mantle cells resulted in the same outcome (n = 6). (Z) Quantification of the number of cells in regenerated neuromasts at 7 dpi. Number of neuromast cells was no statistically significant between groups of different damage regimes as determined by one-way ANOVA (F(4,27)=1.013, p=0.4183). Scatter plot shows mean ±s.e.m.ns: non-significant. Scale bars: 10 µm.
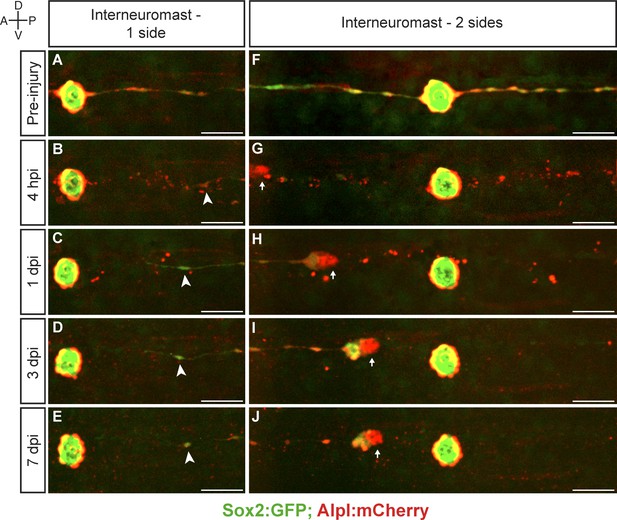
Interneuromast cells do not regenerate.
(A–E) The ablation of interneuromast cells adjacent to one flank of a neuromast resulted in the stretching of the last undamaged interneuromast cell (arrowhead) but does not trigger interneuromast-cell proliferation or the reformation of interneuromast-cell strings (n = 14). (F–J) Likewise, the complete ablation of interneuromast cells in both flanks from one neuromast to the next, generates a corresponding gap of interneuromast cells that did not change over 7 days (n = 8). Scale bars: 50 µm.
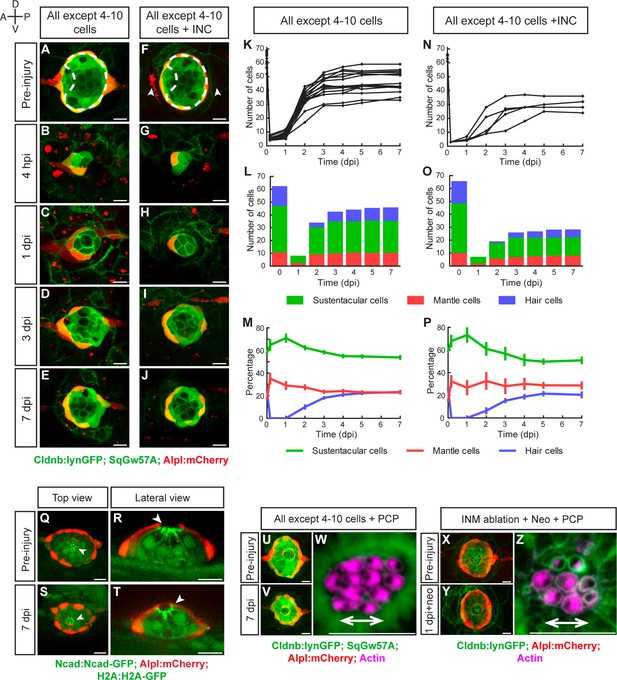
Recovery of organ architecture after loss of tissue integrity.
(A–E) Confocal images of a neuromast regenerating from 4 to 10 cells during a period of 7 days. Neuromasts recover radial symmetry 3 dpi (D), and original organ proportions at 7 dpi (E). (F–J) Neuromasts reduced to 4–10 cells that were previously deprived from adjacent interneuromast cells (INCs) (arrowheads in F), regenerated and reformed radial symmetry (H–I) and proportions 7 dpi, despite maintaining a reduced size (J). Dashed circles in (A,F) illustrate damaged areas. Scale bars: 10 µm. (K,N) Total cell numbers in regenerating neuromasts over 7 days in the two conditions depicted in (A–J). (L,O) In the first 2 dpi neuromast consist almost exclusively of supporting cells (green and red). Hair cells (blue) begin to appear between at 2dpi. (M,P) Percentages of cell classes during a 7 day regeneration period. Right after damage, neuromast experience an imbalance of cell proportions that is re-established over the course of 3 days. Afterwards the neuromasts continues to slowly increase total cell number at similar rates. The final proportion of cell classes recapitulates that of the starting condition. Time points show mean ± s.e.m. [All except 4–10 cells] n = 15, [All except 4–10 cells + INC] n = 6. (Q) Top and (R) lateral views of a triple-transgenic Tg[Ncad: Ncad-EGFP; Alp:mCherry; H2A:H2A-EGFP] neuromast before injury. (S) Top and (T) lateral views of a regenerated neuromast 7 days post injury (n = 4). Basal location of nuclei and apical N-cadherin enrichment evidence the apicobasal polarization of the organ. The accumulation of N-cadherin (white arrowheads) in the regenerated neuromast shows that apical constrictions are properly re-established during the process. (U–V) Maximal intensity projection of a neuromast in the triple transgenic line Tg[Cldnb:lynGFP; SqGw57A; Alpl:mCherry] prior to injury that eliminates all except 4 to 10 cells (U), and the same neuromast 7 days after damage (V). (W) Hair-bundle staining with rhodamine-phalloidin (colored in pink) reveals the coherent planar polarization of the hair cells in the regenerated neuromast shown in (V). (X) Confocal projection of a neuromast before the removal of flanking interneuromast cells. (Y) Maximal projection of a neuromast 48 hr after interneuromast-cell ablation and 24 hr after neomycin treatment. (Z) Phalloidin staining of hair bundles of hair cells regenerated in the absence of interneuromast cells, showing recovery of coherent epithelial planar polarity. Scale bars: 10 µm.
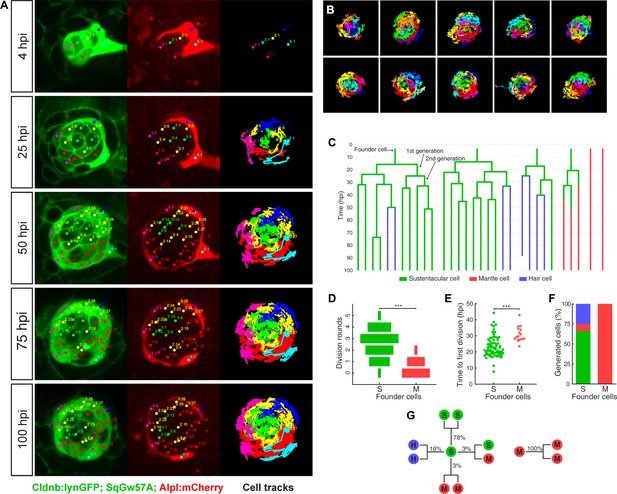
Long-term whole-organ single-cell tracking reveals cell-clone formation during neuromast regeneration.
(A) Still images showing a representative 100 hr time-lapse recording of a regenerating neuromast in Tg[Clndb:lynGFP; SqGw57A; Alpl:mCherry] larva (left and middle panels). Cellular clones that share a common founder cell are clustered and color-coded. Cell trajectories reveal a concentric growth pattern (right panel). (B) Cell trackings at the last recorded timepoints for 10 out of the total of 15 regenerated neuromasts. (C) Cell-lineage tracing from time-lapse movie shown in (A). Branching points symbolize cell divisions. The division of a founder cell generates two cells of the 1 st generation. Subsequent divisions produce cells of the 2nd, 3rd and 4th generation. Cell classes are indicated with green (sustentacular), blue (hair) and red (mantle) colors. (D) Sustentacular founder cells undergo significantly more (p=3.59e-06, Mann-Whitney test) division rounds than mantle founder cells during 100 hr of neuromast regeneration. (E) The first division of sustentacular founder cells (n = 76) occurs significantly earlier (p=1.13e-5, Mann-Whitney test) than that of mantle founder cells (n = 16). (F) Sustentacular founder cells (n = 76) generate all three neuromast cell classes whereas mantle founder cells (n = 30) produce only mantle cells. (G) Out of 307 sustentacular cell divisions, 78% were self-renewing, 16% produced a pair of hair cells, 3% produced sustentacular cells that both became mantle cells within the next generation and 3% generated two sustentacular cells of which only one transited to mantle cell fate within the next generation. All 20 observed mantle cell divisions were self-renewing.
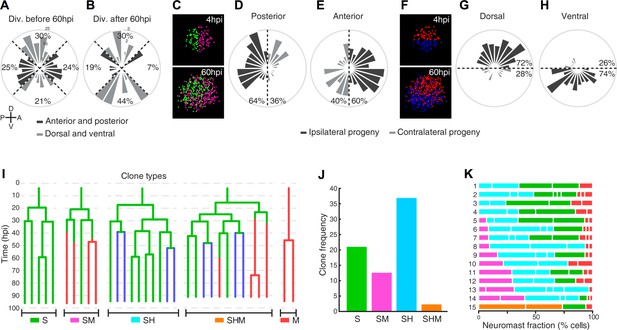
Neuromast regeneration is not stereotypic and reveals different clone type compositions.
(A) Proliferation is markedly isotropic during the first 60 hr of neuromast regeneration (n = 348). (B) Homeostatic, dorso-ventral (DV) proliferative territories are restored after 60hpi (n = 27). (C) 40% and 36% of the progeny from anterior (n = 202) and posterior (n = 173) founder cells crossed to the contralateral side (light grey) after 60 hr of regeneration. (D) Only 28% and 26% of the progeny from dorsal (n = 199) and ventral (n = 176) founder cells crossed to the contralateral side (light grey) during the same period of time. (E) Representative examples of different clone types extracted from time-lapse data. Sustentacular cells give rise to S, SM, SH, and SHM clones (color coded respectively with green, pink, cyan and orange) whereas mantle cells produce only pure mantle cell clones. (F) The clone composition of the 15 regenerated neuromasts is not stereotypic. The length of each bar represents the proportion of neuromast cells that belong to each clone. Neuromast eight has been shown in Figure 5A,B. (G) The most frequent clones contain sustentacular and hair cells (SH, n = 37 clones), followed by those with only sustentacular cells (S, n = 21 clones). The third most frequent are composed by sustentacular and mantle cells (SM, n = 12 clones). Clones containing all three cell classes were rare (SHM, n = 2 clones).
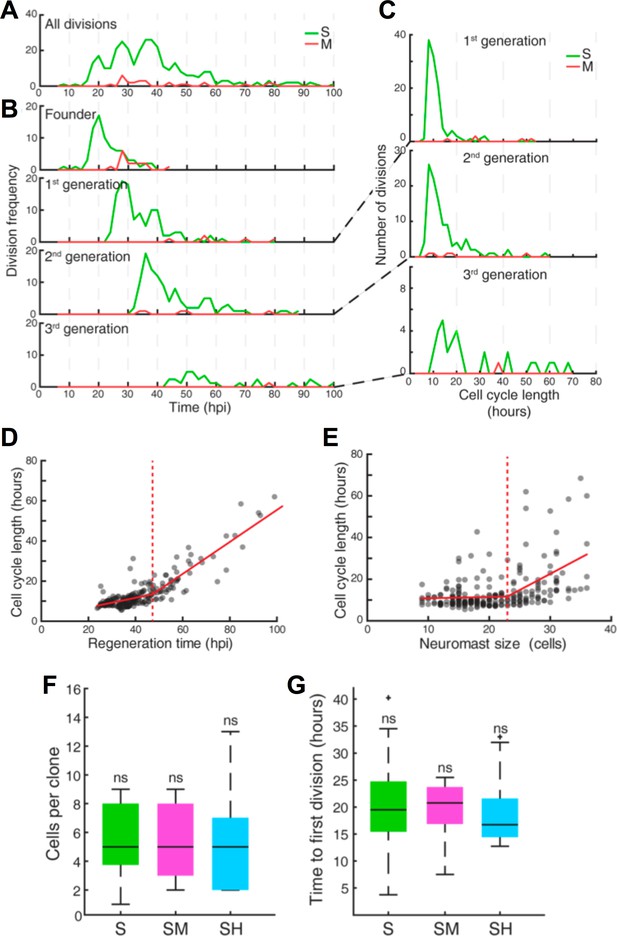
Quantification of cell divisions during neuromast regeneration.
(A–B) Equally spaced waves of coordinated sustentacular cell divisions (green) underlie the recovery of neuromast cell size. Mantle cell divisions (red) occur occasionally and do not follow the pattern of sustentacular cells. Proliferative waves correspond to the coordinated divisions of cells from independent generations. (C) Cells from the 1 st and 2nd generation divide on average after cell cycles of 11 ± 5 and 14 ± 9 hr respectively (mean ±s.d.). Coordination is lost at the 3rd generation when cell cycles start to lengthen (26 ± 18 hr, mean ±s.d.). (D) Cell cycle length (11 ± 3 hr, mean ±s.d.) is marginally influenced by regeneration time until 47 hr after injury, when cycle length starts increasing proportionally with regeneration time. (E) Cell cycle lengths (12 ± 6 hr, mean ±s.d.) do not correlate directly with neuromast size until 24 neuromast cells. (F) S, SM and SH clones produce similar number of cells (p=0.68, Kruskal Wallis test). In the box plots, the boundary of the box indicates the 25th and 75th percentile, respectively the black line within the box marks the median. Whiskers above and below the box include points that are not outliers. (G) Sustentacular founder cells of S, SM, and SH clones divide similarly early (p=0.42, Kruskal Wallis test) after approximately 18 hr after neuromast injury. (H) Sustentacular founder cells that produce SH (cyan) and S clones (green) are distributed similarly around the center of the organ (at x = y = 0). Those that generate SM clones (pink) are localized further away from the center and are biased towards the posterior side.
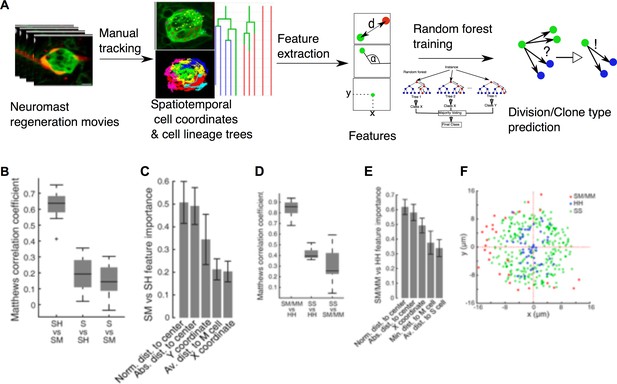
Implementation of predictive machine-learning analysis.
(A) Overview from experiments to prediction. Movies of neuromast regeneration allow us to track every single cell over 100hpi and to generate a cell lineage from these track points. Information covered in all tracks and lineages can be extracted as features with which we train our random forest machine-learning classifier to predict division or cell lineage fate. (B) Sustentacular founder cell choices between SH vs. SM clones can be predicted with high accuracy (MCC = 0.63 ± 0.09, mean ± s.d., n = 15 bootstrapped samples) whilst choices between S and SH or SM clones are highly inaccurate (MCC = 0.19 ± 0.11 and 0.15 ± 0.10, mean ± s.d., respectively, n = 15 bootstrapped samples), based on 32 calculated features. (C) Features relative to the position of the founder cells and their nearest cellular environment can discriminate between SM and SH clone types. (D) Choices between SM/MM and HH divisions can be predicted with high accuracy (MCC = 0.91 ± 0.07, mean ± s.d., n = 15 bootstrapped samples) while those between SS and HH or SM/MM have low accuracy (MCC = 0.50 ± 0.05 and 0.38 ± 0.15, respectively, mean ± s.d., n = 15 bootstrapped samples) (E) Features describing the cell’s position in relation to the neuromast center and their proximity to other mantle cells have the highest influence on the cell fate choices of a sustentacular cell. (F) SM/MM divisions (red) appear predominantly at the periphery of the organ whereas HH divisions (blue) appear proximal to the center. Sustentacular cell self-renewing divisions (SS, green) occur mostly around the neuromast center, generating a ring-like pattern.
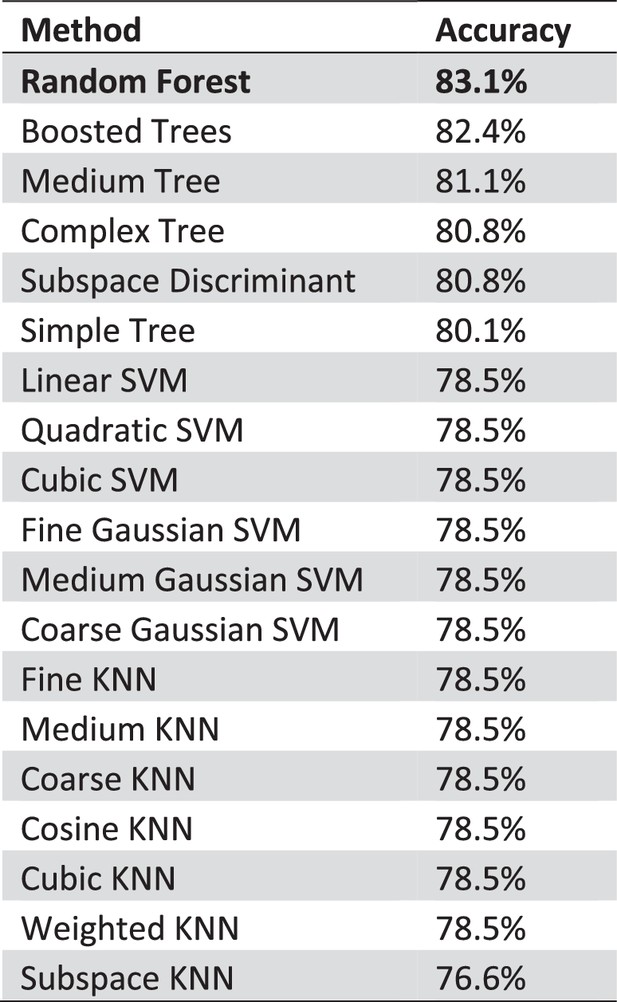
Comparison of different classification methods.
With 83.1% accuracy random forests perform best comparing features based ML algorithms on our data. We used the standard classification learners in MATLAB to obtain a first impression of the performance of possible ML approaches. We a 5-fold cross-validation we tested and compared the described methods. With 83.1% accuracy random forests perform best comparing feature based ML algorithms on our data. We used the standard classification learner in MATLAB to get a first impression of the performance of possible ML approaches. With a 5-fold cross-validation we tested and compared the provided methods.
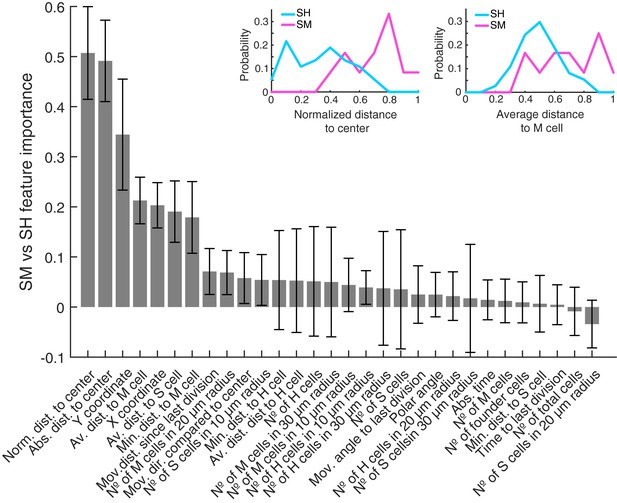
Features used to predict SM vs SH clones sorted by predictive importance.
The bar plot shows all features used to predict SM vs. SH clones. They are sorted by their predictive importance and their error bars are generated by the used leave-one-out approach. The plots above exemplary show feature distributions of normalized distance to neuromast center (left) and average distance to mantle cells (right) for SM and SH clones.
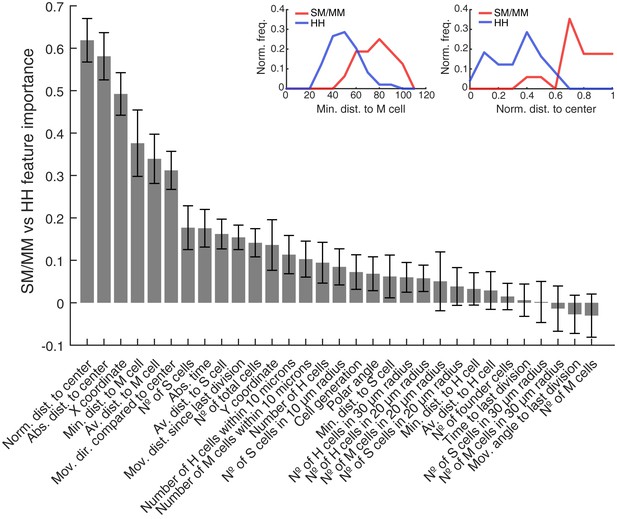
All features used to predict SM/MM vs HH divisions sorted by predictive importance.
The bar plot shows all features used to predict SM/MM vs HH divisions. They are sorted by their predictive importance and their error bars are generated by the used leave-one-out approach. The plots above exemplary show feature distributions of minimal distance to mantle cells (left) and normalized distance to neuromast center (right) for SM/MM and HH divisions.
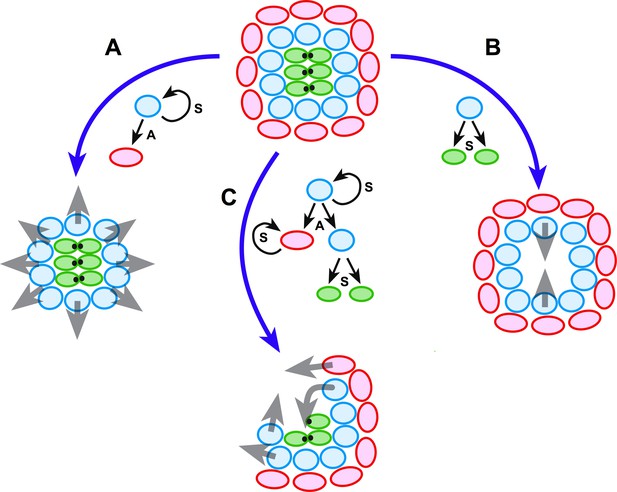
Schematic model of neuromast regeneration.
The top diagram exemplifies the architecture of an intact neuromast. A, B and C indicate three types of injury: A when mantle cells are lost, B when hair cells are ablated, and C when a localized combination of all three cell classes is lost. Under the model that we present, radial symmetry serves to localize damage and canalize regeneration spatially. If central hair cells are lost (A), radial symmetry is maintained for sustentacular progenitors to regenerate hair cells centripetally (grey arrows in A). If outer cells are lost (B), radial symmetry is also maintained for the generation of progeny that will acquire mantle fate and propagate centrifugally to reform the outer rim of the neuromast (grey arrows in B). Upon asymmetric damage, however, the radial symmetry is partially broken (C). The neuroepithelium repolarizes along an injured-intact axis, which canalizes regeneration towards the damaged areas (grey arrows in C). Individual cells are color-coded (mantle cells in red, sustentacular cells in light blue, and hair cells in green), and in each case we indicate the type of division that the intact cells undergo: symmetric (S) when they produce two equivalent cells or self-renew, and asymmetric (A) when their division generates sibling cells that differentiate into different classes.
Videos
A 20 min videomicroscopic recording of a neuromast after laser-mediated ablation of supporting cells.
Four laser-targeted cells (showing a dark spot in the nuclei from focal fluorescent-protein bleaching) are eliminated from the epithelium, which closes the wound. There is no noticeable collateral damage. Time resolution is one image per 30 s.
100 hr time-lapse recording of a regenerating neuromast after severe ablation.
A neuromast regenerates its original architecture from as few as six founder cells. Founder cells are identified by 1–6 (n) and their daughter cells receive 2 n and 2n + 1 identities. Recording starts 4 hr post injury (hpi) and shows single focal planes. Time is in hours post injury.
Tables
List of prediction features with description.
We used 32 mainly spatial and neighborhood specific features for the classification. Features are explained in the description column.
| Feature name | Description |
|---|---|
| Absolut time | Hours post induction (hpi) |
| Absolute distance to center | Euclidean distance to the neuromast center |
| Average distance to H cell | - |
| Average distance to M cell | - |
| Average distance to S cell | - |
| Cell generation | Number of divisions that the cell has undergone |
| Founder Cell Type | - |
| Minimum distance to H cell | - |
| Minimum distance to M cell | -- |
| Minimum distance to S cell | - |
| Movement angle to last division | Angle between current cell location, neuromast center and location of last cell division (or start of the movie in case of founder cell division) |
| Movement direction compared to center | Radial distance between current cell location and location of last cell division (or start of the movie in case of founder cell division). If the current location is nearer to the center the value is (+) in case it is further away the value is (-) |
| Movement distance since last division | Euclidean distance between current cell location and last cell division (or start of the movie in case of founder cell division) |
| Normalized distance to center | Radial distance of current cell location divided by the radial distance of the current furthest cell (to approximate the neuromast size) |
| Number of founder cells | - |
| Number of H cells | - |
| Number of H cells in 10 µm radius | - |
| Number of H cells in 20 µm radius | - |
| Number of H cells in 30 µm radius | - |
| Number of M cells | - |
| Number of M cells in 10 µm radius | - |
| Number of M cells in 20 µm radius | - |
| Number of M cells in 30 µm radius | - |
| Number of S cells | - |
| Number of S cells in 10 µm radius | - |
| Number of S cells in 20 µm radius | - |
| Number of S cells in 30 µm radius | - |
| Number of total cells | - |
| Polar angle | Polar angle is the counterclockwise angle between the x-axis, the neuromast center and the current cell location |
| Time to last division | Time to last division (or start of the movie in case of founder cell division) |
| X coordinate | - |
| Y coordinate | - |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30823.018





