eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast
Figures
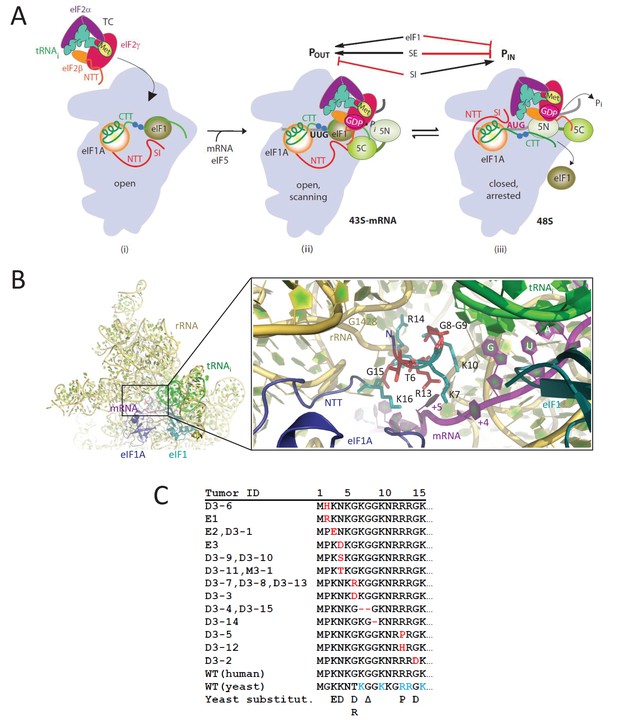
eIF1A-NTT residues associated with UM are predicted to participate in stabilizing the PIN state of the closed conformation of the 48S PIC.
(A) Model describing known conformational rearrangements of the PIC during scanning and start codon recognition. (i) eIF1 and the scanning enhancers (blue balls) in the C-terminal tail (CTT) of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly binds. (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAiMet bound in the POUT state. eIF2 can hydrolyze GTP to GDP•Pi, but release of Pi is blocked by eIF1. The N-terminal tail (NTT) of eIF1A interacts with the eIF5-CTD. (iii) On AUG recognition, Met-tRNAiMet moves from the POUT to PIN state, clashing with eIF1 and the CTT of eIF1A, provoking displacement of the eIF1A CTT from the P site, dissociation of eIF1 from the 40S subunit, and Pi release from eIF2. The NTT of eIF2β interacts with the eIF5-CTD, and the eIF1A-NTT, harboring scanning inhibitor (SI) elements, interacts with the codon:anticodon helix. (Above) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and impede transition to PIN state, whereas the eIF1A SI element in the NTT stabilizes the PIN state. (Adapted from (Hinnebusch, 2014)). Results presented below show that this function of the eIF1A-NTT is impaired by uveal melanoma (UM)-associated substitutions and others that disrupt direct contacts with the mRNA or codon:anticodon helix shown in (B). (B) Magnified portion of the py48S PIC structure (PDB 3J81) showing contacts made by the eIF1A-NTT (shades of blue and cyan) in the closed/PIN conformation. Side-chains of NTT residues substituted in UM (red) or directly contacting 18S rRNA (yellow), tRNAi (green) or mRNA (purple) are shown as sticks. (C) Sequence of human eIF1A NTT residues 1–15 showing the substitutions (red) or deletions (dash) found in the indicated UM tumors. Substitutions in yeast eIF1A corresponding to those found in UM tumors are listed on the last line. The five basic residues of the yeast NTT making direct contacts in the PIC and substituted here in addition to the UM-associated substitutions are shown in cyan.
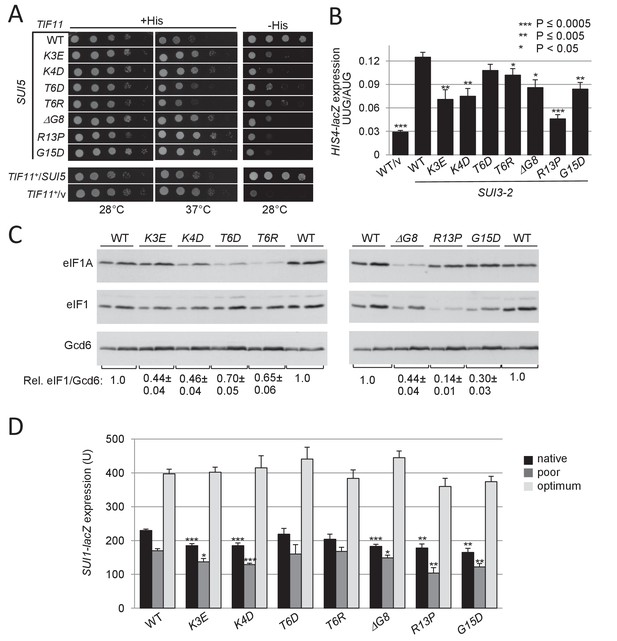
UM-associated substitutions in the yeast eIF1A NTT suppress Sui- phenotypes conferred by mutations SUI5 and SUI3-2 and increase discrimination against the poor, native start codon of SUI1 mRNA.
(A) Ten-fold serial dilutions of tif11Δ his4-301 strain H3582 containing the indicated TIF11 (eIF1A) alleles on single copy (sc) plasmids and either episomal SUI5 (p4281/YCpTIF5-G31R-W) or empty vector (/v) were analyzed for Slg- and His+/Sui- phenotypes on SC lacking leucine (Leu) and tryptophan (Trp) supplemented with 0.3 mM His and incubated at 28°C or 37°C for 2 days (+His), or on SC-Leu-Trp plus 0.003 mM His (-His) and grown at 28°C for 4 days. (B) Derivatives of strain H3582 containing the indicated TIF11 alleles and episomal SUI3-2 (p4280/YCpSUI3-S264Y-W) or empty vector (/v) and also harboring HIS4-lacZ reporters with AUG or UUG start codons (plasmids p367 and p391, respectively) were cultured in synthetic dextrose minimal medium (SD) supplemented with His at 28°C to A600 of ~1.0, and β-galactosidase activities (in units of nanomoles of ο-nitrophenyl-β-D-galactopyranoside cleaved per min per mg) were measured in whole cell extracts (WCEs). The ratio of expression of the UUG to AUG reporter was calculated from at least four different measurements, and the mean and S.E.M.s were plotted. (C) Derivatives of H3582 containing the indicated TIF11 alleles were cultured in SD supplemented with His, Trp and uracil (Ura) at 28°C to A600 of ~1.0, and WCEs were subjected to Western analysis using antibodies against eIF1A/Tif11, eIF1/Sui1 or eIF2Bε/Gcd6 (analyzed as loading control). Two different amounts of each extract differing by 2-fold were loaded in successive lanes. (D) Same strains as in (C) harboring the sc plasmids with SUI1-lacZ fusions containing the native suboptimal (-3CGU-1, pPMB24), poor (-3UUU-1, pPMB28) or optimum (-3AAA-1, pPMB25) AUG contexts were cultured in SD +His + Trp at 28°C to A600 of ~1.0, and assayed for β-galactosidase activities as in (B).
-
Figure 2—source data 1
Source data for Figure 2 and Figure 2—figure supplement 1.
Effects of UM-associated substitutions in the yeast eIF1A NTT on HIS4-lacZ UUG:AUG expression ratios in SUI3-2 cells, eIF1 levels, expression of SUI1-lacZ fusions containing the native, poor or optimum AUG contexts and GCN4-lacZ expression in SUI3-2 cells.
- https://doi.org/10.7554/eLife.31250.005
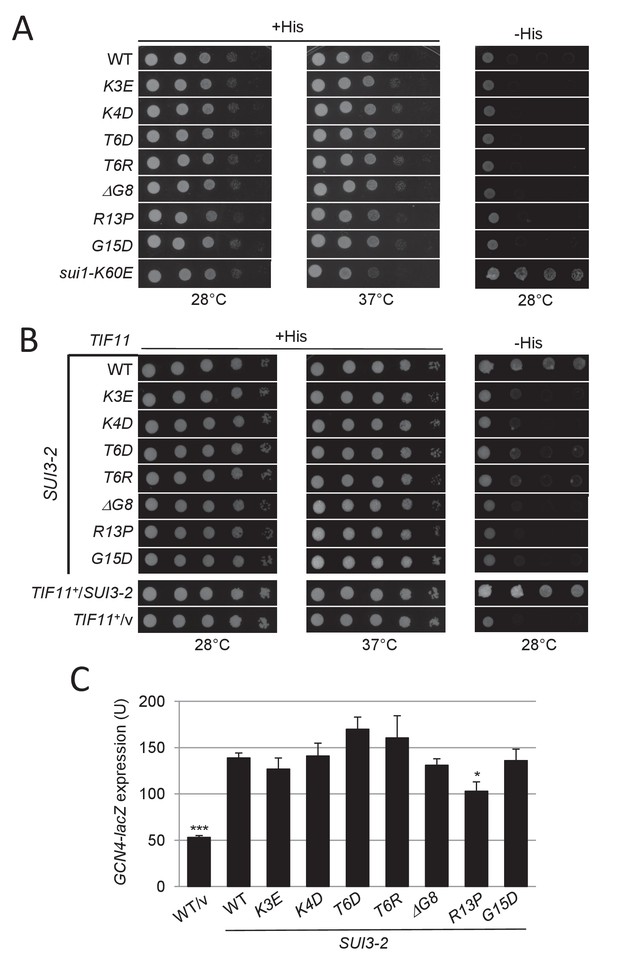
UM-associated eIF1A NTT substitutions reduce the His+/Sui- and Gcd- phenotypes of SUI3-2.
(A) Ten-fold serial dilutions of tif11Δ his4-301 strain H3582 containing the indicated TIF11 (eIF1A) alleles on single copy (sc) plasmids were grown at 28°C or 37°C for 1.5 days on synthetic complete (SC) medium lacking leucine (Leu) supplemented with 0.3 mM histidine (+His) and at 28°C for 7 days on SC-Leu plus 0.003 mM His (-His). Strain pPMY32 harboring the Sui- variant eIF1-K60E is shown for comparison. (B) Transformants of H3582 with the indicated plasmid-borne TIF11 alleles also containing episomal SUI3-2 (p4280/YCpSUI3-S264Y-W) or empty vector (/v) were analyzed for Slg- and His+/Sui- phenotypes by spotting 10-fold serial dilutions on SC lacking Leu and tryptophan (Trp) supplemented with 0.3 mM His and incubated at 28°C or 37°C for 2 days (+His), or on SC-Leu -Trp plus 0.003 mM His (-His) grown at 28°C for or 7 days. (C) Transformants of the strains from (A) harboring the GCN4-lacZ fusion on plasmid p180 were cultured and analyzed as in Figure 2B.
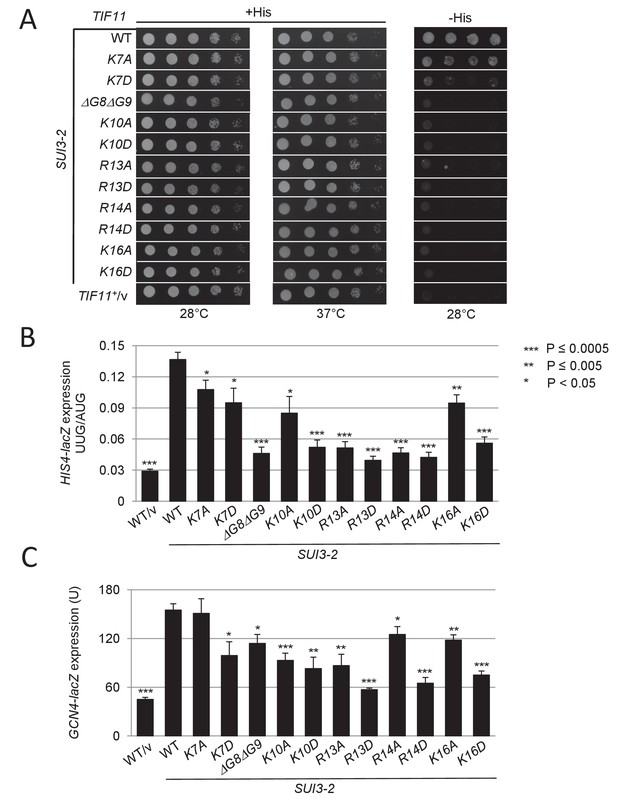
Substitutions in key eIF1A NTT basic residues reduce the elevated UUG initiation and derepressed GCN4-lacZ expression conferred by Sui- mutation SUI3-2.
(A) Derivatives of strain H3582 containing the indicated TIF11 alleles and episomal SUI3-2 (p4280/YCpSUI3-S264Y-W) or empty vector (/v) were analyzed for Slg- and His+/Sui- phenotypes by spotting 10-fold serial dilutions on SC-Leu-Trp plus 0.3 mM His and incubated at 28°C or 37°C for 2 days (+His), or on SC-Leu-Trp plus 0.003 mM His (-His) and grown at 28°C for 7 days, as in Figure 2—figure supplement 1B. (B–C) Transformants of the strains from (A) harboring HIS4-lacZ reporters with AUG or UUG start codons (B) or the GCN4-lacZ reporter (C) were cultured and assayed for β-galactosidase activities as in Figure 2B.
-
Figure 3—source data 1
Source data for Figure 3 and Figure 3—figure supplement 1.
Effects of substitutions in eIF1A NTT basic residues on HIS4-lacZ UUG:AUG expression ratios and GCN4-lacZ expression both in SUI3-2 cells. Effects of eIF1A-R13P and K16D substitutions on FLUC/RLUC UUG:AUG expression ratios in SUI3-2 cells.
- https://doi.org/10.7554/eLife.31250.008
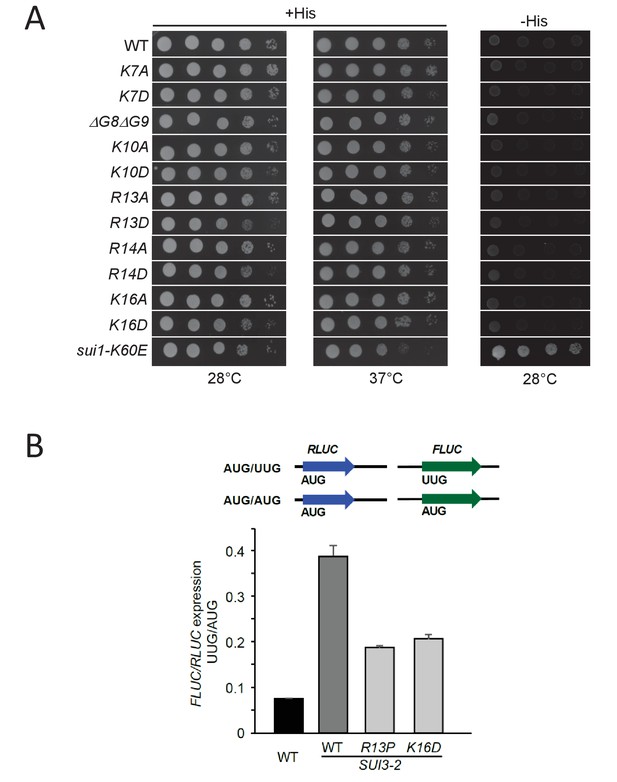
Certain eIF1A NTT substitutions affecting PIC interactions confer slow-growth phenotypes andConfirmation of Ssu-phenotypes of R13P and K16D substitutions using UUG- or AUG-initiated luciferase reporters.
(A) H3582 derivatives harboring the indicated TIF11 alleles in sc plasmids were grown at 28°C and 37°C for 2 days on +His, and at 28°C for 7 days on –His as in Figure 2—figure supplement 1A. Strain pPMY32 harboring the Sui- variant eIF1-K60E is shown for comparison. (B) Derivatives of strain H3582 containing the indicated TIF11 alleles and episomal SUI3-2 and also harboring pairs of dual luciferase reporters LUCRenilla(AUG)/LUCfirefly(UUG) or LUCRenilla(AUG)/LUCfirefly(AUG) on sc URA3 plasmids pRaugFFuug and pRaugFFaug, respectively, were cultured as in Figure 2B (except at 30°C) and the luminescence was measured in WCEs. The LUCfirefly (UUG):LUCRenilla (AUG) ratio of luminescence, in relative light units, was normalized to the LUCfirefly (AUG):LUCRenilla (AUG) ratio to calculate the FLUC(UUG)/RLUC(AUG) from six independent transformants, and the mean and S.E.M.s were plotted.
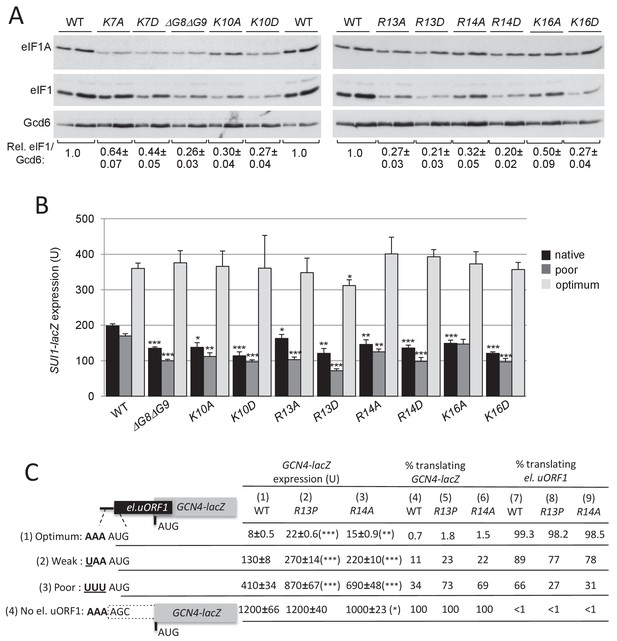
Substitutions in key eIF1A NTT basic residues increase discrimination against poor AUG contexts.
(A) Western blot analysis of eIF1 expression in derivatives of H3582 with the indicated TIF11 alleles, as in Figure 2C. (B) Transformants of strains in (A) with SUI1-lacZ fusions containing the native suboptimal, poor or optimum AUG contexts were assayed for β-galactosidase activities as in Figure 2D. (C) H3582 derivatives, harboring WT, R13P or R14A TIF11 alleles and el.uORF1 GCN4-lacZ reporters containing the depicted optimum (pC3502, row1), weak (pC4466, row2) or poor (pC3503, row3) context of uAUG-1, or uORF-less GCN4-lacZ reporter with a mutated uAUG-1 (pC3505, row4), were assayed for β-galactosidase activities as in Figure 2D. Mean expression values with S.E.M.s were determined from four transformants (columns 1, 2 and 3). The percentages of scanning ribosomes that translate el.uORF1 (columns 7, 8 and 9) or leaky-scan uAUG-1 and translate GCN4-lacZ instead (columns 4, 5 and 6) were calculated from results in columns 1, 2 and 3 by comparing the amount of expression observed for each uORF-containing reporter to the uORF-less construct. Statistically significant differences between mutant and WT are marked with asterisks (*p<0.05; **p<0.005; ***p<0.0005).
-
Figure 4—source data 1
Source data for Figure 4 and Figure 4—figure supplements 1, 2 and 3.
Effects of substitutions in eIF1A NTT basic residues on eIF1 levels, expression of SUI1-lacZ fusions with the native, poor or optimum AUG contexts and eIF1-R13P or R14A substitutions on leaky scanning of el.uORF1 in GCN4-lacZ reporters. Effects of selected eIF1A NTT variants overexpression on eIF1 levels, SUI1-lacZ fusions expression and HIS4-lacZ UUG:AUG expression ratios or GCN4-lacZ expression in SUI3-2 cells. Effects of eIF1-K16A, K16D and overexpression of eIF1A-ΔG8ΔG9 and K10D substitutions on leaky scanning of el.uORF1 in GCN4-lacZ reporters.
- https://doi.org/10.7554/eLife.31250.013
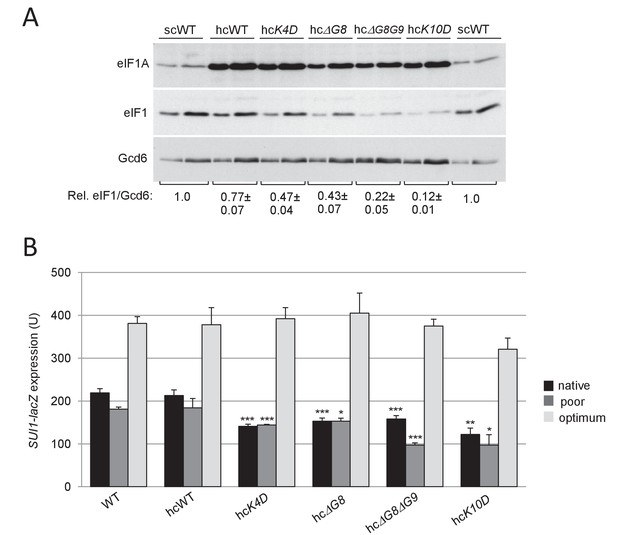
Overexpression of selected eIF1A NTT variants confers reduced expression of eIF1 and SUI1-lacZ fusions with native and poor AUG contexts.
(A) Western analysis of derivatives of H3582 harboring the indicated TIF11 alleles on high copy (hc) was conducted as in Figure 2C. (B) Same strains as in (A) containing the indicated SUI1-lacZ fusions were assayed for β-galactosidase activities as in Figure 2D.
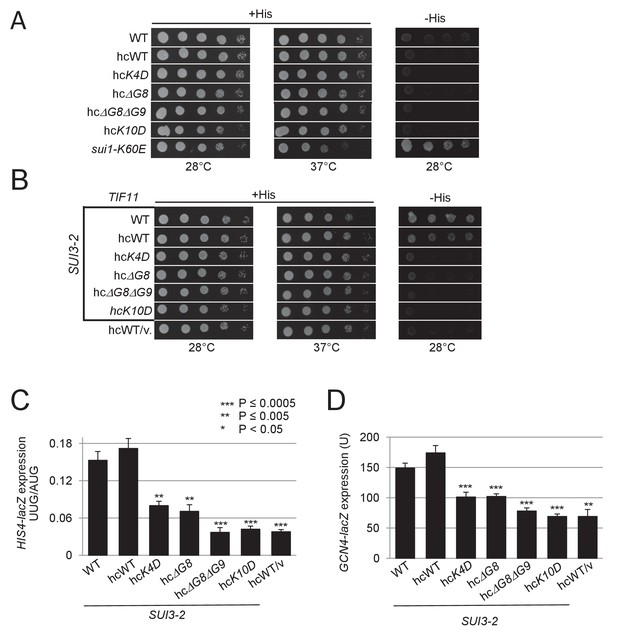
Selected eIF1A NTT variants when overexpressed still suppress the His+/Sui- phenotype and elevated UUG initiation and GCN4-lacZ expression conferred by SUI3-2.
(A) H3582 derivatives harboring the indicated TIF11 alleles in hc plasmids were grown at 28°C and 37°C for 2 days on +His and at 28°C for 7 days on –His as in Figure 2—figure supplement 1A. (B) Same strains as in (A) also harboring episomal SUI3-2 (p4280/YCpSUI3-S264Y-W) or empty vector were analyzed for Slg- and His+/Sui- phenotypes as in Figure 2—figure supplement 1B. (C) Same strains as in (B) also harboring HIS4-lacZ reporters with AUG or UUG start codons were cultured and assayed for β-galactosidase activities as in Figure 2B. (D) Transformants of the strains from (B) harboring the GCN4-lacZ fusion on plasmid p180 were cultured and analyzed as in Figure 2B.
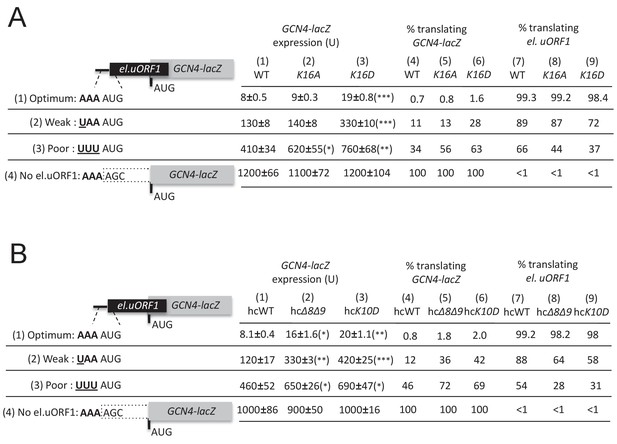
Additional eIF1A targeted and UM-associated NTT mutations increase leaky scanning of GCN4 uAUG-1 in vivo.
(A–B) H3582 derivatives harboring the indicated TIF11 alleles and el.uORF1 GCN4-lacZ reporters were analyzed as in Figure 4C.
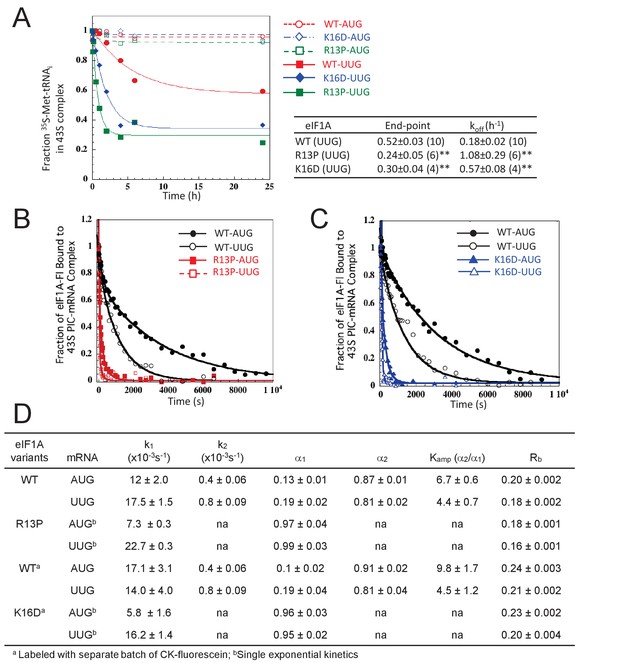
UM-associated mutant eIF1A-R13P and targeted mutant eIF1A-K16D destabilize the closed/PIN conformation of the 48S PIC at UUG codons in vitro.
(A) Effects of R13P and K16D on TC dissociation kinetics from reconstituted partial 43S•mRNA(AUG) or mRNA(UUG) complexes formed with TC containing [35S]-Met-tRNAiMet and WT eIF1A, eIF1A-R13P or eIF1A-K16D, as indicated. Representative curves are shown for each measurement. Tabulated rate constants (koff) and reaction end-points with S.E.M.s are averages of between 4–10 replicate experiments (number in parenthesis); asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student’s t-test (*p<0.05; **p<0.01). (B–D) Effects of R13P and K16D on the dissociation kinetics of fluorescein-labeled eIF1A from reconstituted partial 43S•mRNA complexes, monitored by following changes in fluorescence anisotropy over time after addition of a large excess of unlabeled WT eIF1A. The data for WT eIF1A were fit with a double exponential decay equation, where the fast phase (rate constant k1) corresponds to dissociation of eIF1A from the ‘open’ conformation of the PIC and the second phase (rate constant k2) corresponds to dissociation from the ‘closed’ state (Maag et al., 2006). The ratio of amplitudes of the second phase (α2, closed state) to the first phase (α1, open state) is defined as Kamp. Data for both R13P/K16D were fit to a single exponential equation with rate constant k1. Rb is the anisotropy of PIC-bound eIF1A. (B–C) Representative eIF1A dissociation kinetics from PICs assembled with WT (circles), R13P (squares, panel B), or K16D (triangles, panel C) with mRNA(AUG) (filled symbols) or mRNA(UUG) (open symbols). (D) Summary of kinetic parameters from experiments in (B–C). Different preparations of labeled WT eIF1A were employed for the experiments examining R13P and K16D, as indicated. All experiments were performed at least two times and errors are average deviations.
-
Figure 5—source data 1
Source data for Figure 5 and Figure 5—figure supplement 1.
Effects of eIF1A-R13P and K16D on TC dissociation kinetics from partial 43S•mRNA complexes: rate constants (koff) and reaction end-points, and on the dissociation kinetics of fluorescein-labeled eIF1A from partial 43S•mRNA complexes reconstituted with WT eIF2 or the eIF2β-S264Y variant of eIF2: rate constants (k1 and k2), amplitudes (α1 and α2,), ratio of amplitudes (Kamp) and anisotropy of PIC-bound eIF1A (Rb).
- https://doi.org/10.7554/eLife.31250.016
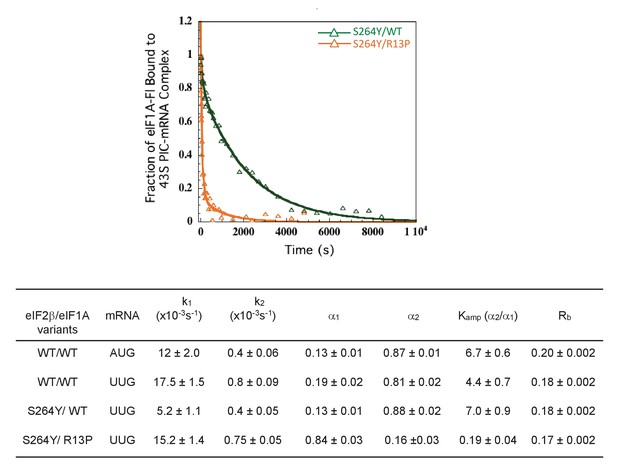
UM mutant eIF1A-R13P suppresses the stabilizing effect of eIF2 Sui- variant containing eIF2β-S264Y on the closed conformation of the 48S PIC at UUG codons in vitro.
Effect of R13P on the dissociation kinetics of fluorescein-labeled eIF1A from partial 43S•mRNA complexes reconstituted with eIF2 containing the eIF2β-S264Y subunit (encoded by SUI3-2), conducted as in Figure 5B–D.
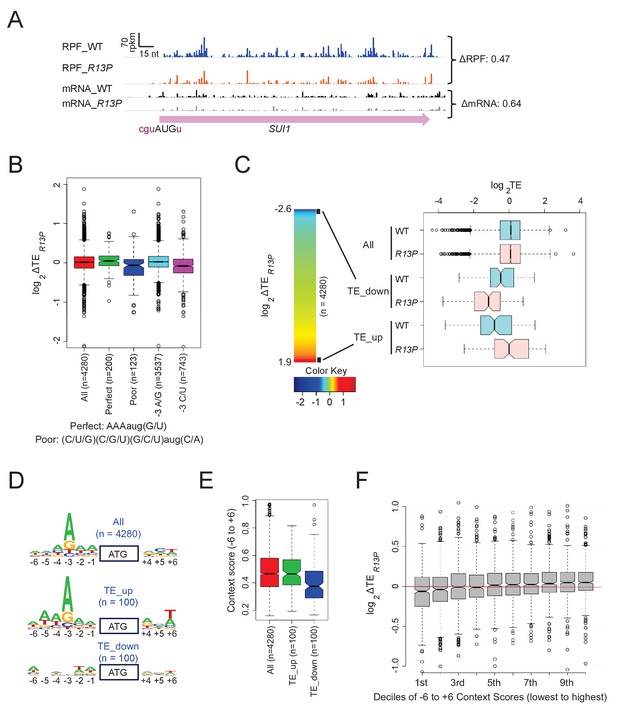
UM mutant eIF1A-R13P increases discrimination against poor Kozak context of main CDS AUG codons genome-wide.
(A) Ribosome-protected fragments (RPFs) and mRNA reads on the SUI1 gene in WT and R13P cells in units of rpkm (reads per 1000 million mapped reads), showing schematically the position of the CDS (pink) and the −3 to −1 and +4 context nucleotides of the AUG codon (in brick). ΔRPF and ΔmRNA give the ratios of RPFs and total mRNA fragments, respectively, in R13P versus WT cells for SUI1. The Integrated Genomics Viewer (Broad Institute) was employed to display ribosome/mRNA reads. (B) Notched box-plot of the ratios of log2TE values in R13P vs. WT cells (ΔTER13P) for groups of genes (number, n, indicated) with perfect or poor AUG context (as defined in figure), preferred (A/G) or non-preferred (C/U) bases at −3, and all 4280 genes with >10 RPF reads and >32 mRNA reads (average of 4 samples, two replicates of WT and two replicates of tif11-R13P) in the main CDS, and 5’UTR length >5 nt. (C) left: Heat-map of TE changes in R13P versus WT cells for 4280 genes. Black boxes at the top and bottom of the map demarcate the groups of 100 genes designated as TE_down and TE_up, respectively. right: Box-plots of log2TE values in R13P versus WT cells for the ‘TE_down’ and ‘TE_up’ groups of genes. (D) Logos of AUG context sequences for the 4280 genes in (B), and the ‘TE_up’ and ‘TE_down’ groups of genes defined in (C). (E) Box-plots of AUG context scores calculated for positions −6 to −1 and +4- + 6 for the ‘TE_up’ and ‘TE_down’ groups of genes. (F) Box-plot analysis of ΔTER13P values for the same 4280 genes analyzed in (B–E) for deciles of equal size binned according to the AUG context scores calculated as in (E).
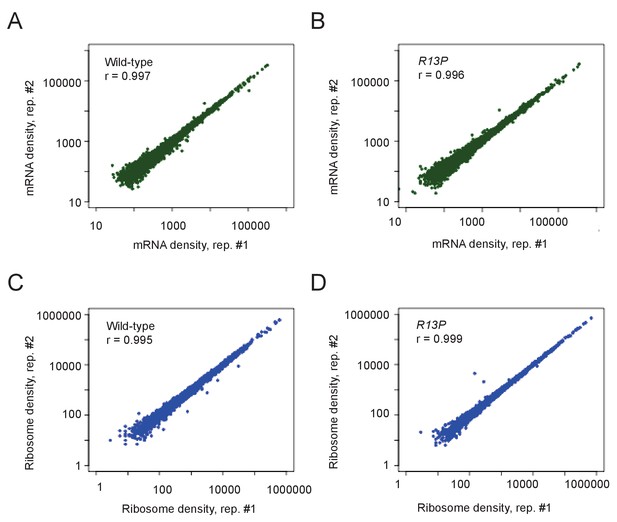
Genome-wide ribosome footprint and mRNA reads for WT and tif11-R13P strains.
(A–D) Scatterplots of total mRNA reads (A–B) and ribosome protected fragments (RPFs) (C–D) from WT and tif11-R13P strains for genes with >128 total mRNA reads or >40 RPF reads in the four samples combined (two strains and their two replicates). Pearson correlation coefficients (R) were calculated for plotted genes.
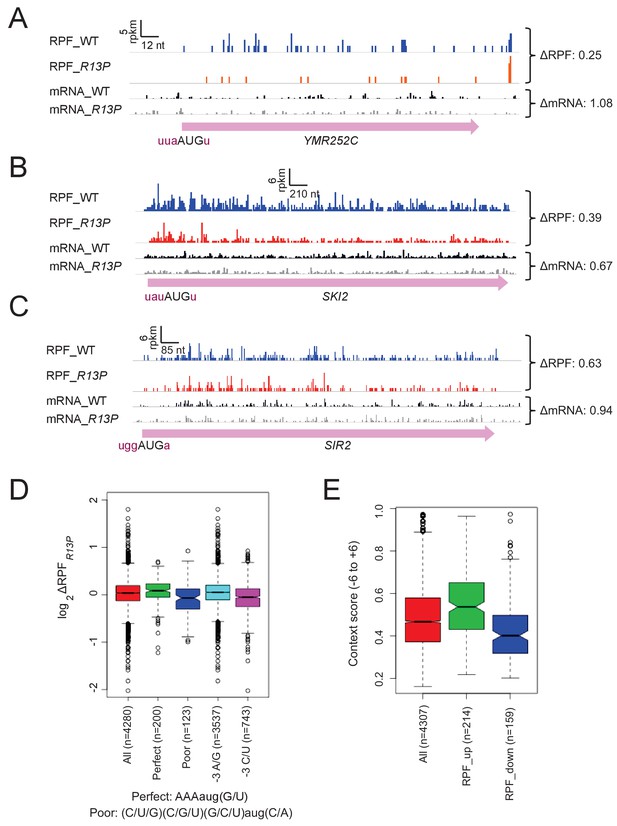
Supporting evidence that eIF1A-R13P increases discrimination against poor Kozak context of main CDS AUG codons genome-wide.
(A–C) RPFs and mRNA reads for genes resembling SUI1 in containing poor context for their AUG codons and decreased RPFs in R13P versus WT cells, presented as in Figure 6A. (D) Same analysis as conducted in Figure 6B except using RPFs versus TE values. (E) Similar analysis as in Figure 6E except conducted with groups of genes showing significant changes in RPFs in R13P versus WT cells using DESEQ2 instead of DESEQ. (at FDR < 0.1 as judged by DESEQ2 analysis).
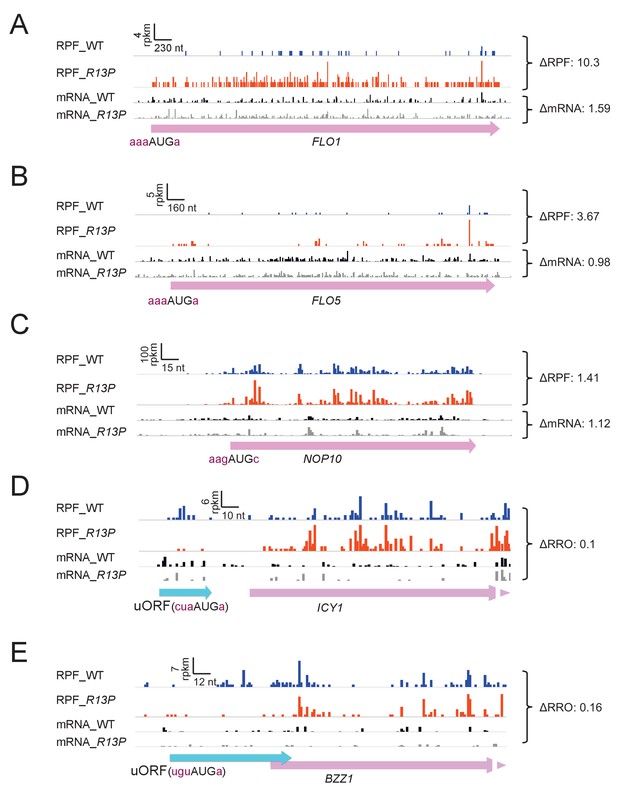
Supporting evidence that eIF1A-R13P increases discrimination against poor Kozak context of both main CDS and uORF AUG codons.
(A–C) RPFs and mRNA reads on genes containing preferred context for their AUG codons and behaving oppositely of SUI1 in showing increased RPFs in R13P versus WT cells. (D–E) RPFs and mRNA reads on genes resembling CPA1 in containing an uORF with an AUG in poor context and displaying a decreased ratio of RPFs in the uORF vs. CDS (RRO) in R13P vs. WT cells (ΔRRO values of 0.10 or 0.16).
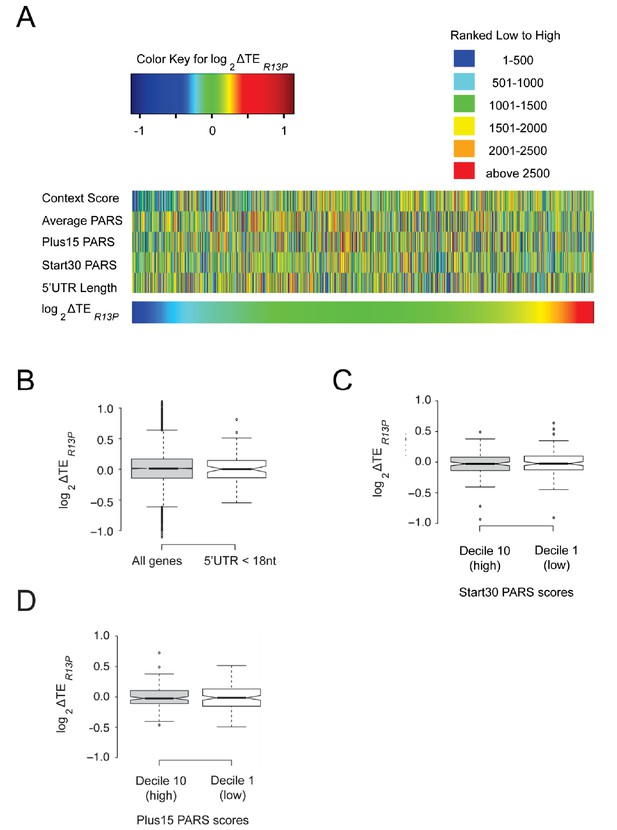
Changes in TE conferred by R13P are not correlated with 5’UTR length or propensity for structure.
(A) Heat-map comparison of Z-scores calculated from TE values in WT and R13P mutant cells, PARS scores for selected intervals, and 5’UTR lengths for 2355 genes taken from the group of 4280 described in Figure 6 for which PARS data are also available (Kertesz et al., 2010). See main text for definition of PARS categories. (B) Boxplot comparison of TE changes conferred by R13P for 79 genes with 5’UTR lengths ≤ 17 nt in at least three out of four genome-wide measurements of 5’UTR lengths (Nagalakshmi et al., 2008; Lawless et al., 2009; Xu et al., 2009; Pelechano et al., 2013). (C–D) Boxplot comparison of TE changes conferred by R13P for two groups of 249 genes with the highest or lowest Start30 PARS scores (C) or Plus15 PARS scores (D).
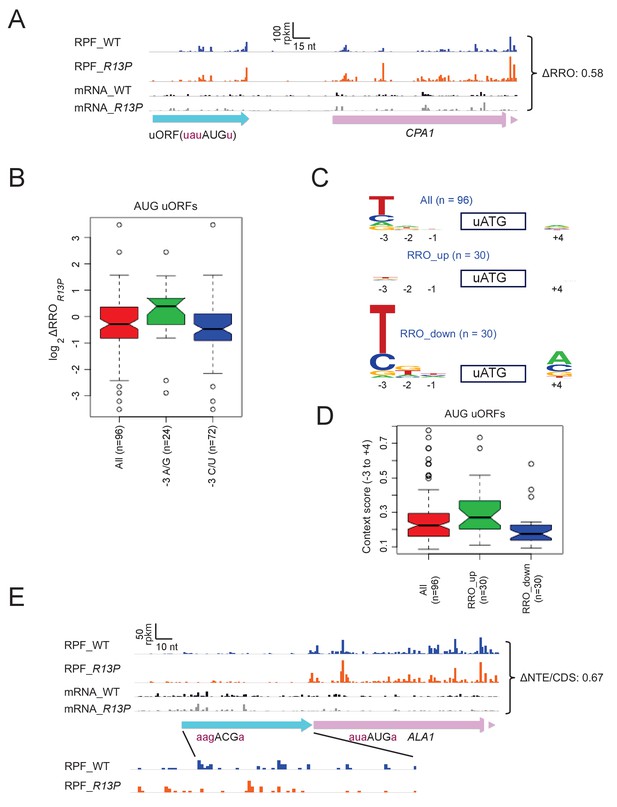
eIF1A-R13P increases discrimination against poor Kozak context of uORF AUG codons genome-wide.
(A) RPFs and mRNA reads on the CPA1 gene and its uORF with AUG in poor context, displaying a decreased ratio of RPFs in the uORF vs. CDS (RRO) in R13P vs. WT cells (ΔRRO = 0.58). The pink arrow missing a portion of the arrowhead designates the beginning of the CPA1 main CDS. (B) Notched box-plot of the ratios of log2TE values in R13P vs. WT cells (ΔTER13P) for a group of 96 genes containing an AUG-initiated uORF and exhibiting >32 RPFs in the main CDS and >2 RPFs in the uORF and a 5’UTR for the uORF of >2 nt in length; and of the subsets of 24 genes from this group with preferred A/G at −3, or the 72 genes with non-preferred C/U at −3, relative to the uORF AUG codon. (C) Logos of upstream AUG context sequences for the 96 genes in (B), and the subsets of 30 genes with the greatest increase (RRO_up) or decrease (RRO_down) in uORF relative to CDS RPFs (RRO values) in R13P versus WT cells. (D) Box-plots of upstream AUG context scores calculated for positions −3 to −1 and +4 for the same genes analyzed in (C). (E) RPFs and mRNA reads on the beginning of the ALA1 main CDS (pink) and N-terminal extension (NTE, cyan schematic), displaying a decreased ratio of NTE/CDS RPFs in R13P vs. WT cells (ΔNTE/CDS = 0.67). Note that the ΔNTE/CDS ratio reflects the ratio of initiation at the upstream AUG to the combined initiation events at upstream AUG and main CDS AUG.
Tables
Plasmids used in this study
https://doi.org/10.7554/eLife.31250.023| Plasmid | Description | Source or reference |
|---|---|---|
| YCplac111 | sc LEU2 cloning vector | (Gietz and Sugino, 1988) |
| YEplac181 | hc LEU2 cloning vector | (Gietz and Sugino, 1988) |
| YCplac22 | sc TRP1 cloning vector | (Gietz and Sugino, 1988) |
| p3390/pDSO9 | sc LEU2 TIF11 in YCplac111 | (Choi et al., 2000) |
| p5633 | sc LEU2 tif11-K3E in YCplac111 | This study |
| p5635 | sc LEU2 tif11-K4D in YCplac111 | This study |
| p5638 | sc LEU2 tif11-T6D in YCplac111 | This study |
| p5637 | sc LEU2 tif11-T6R in YCplac111 | This study |
| p5640 | sc LEU2 tif11-ΔG8 in YCplac111 | This study |
| p5642 | sc LEU2 tif11-R13P in YCplac111 | This study |
| p5644 | sc LEU2 tif11-G15D in YCplac111 | This study |
| pDH469 | sc LEU2 tif11-K7A in YCplac111 | This study |
| pDH468 | sc LEU2 tif11- K7D in YCplac111 | This study |
| pDH481 | sc LEU2 tif11-ΔG8ΔG9 in YCplac111 | This study |
| pDH471 | sc LEU2 tif11-K10A in YCplac111 | This study |
| pDH470 | sc LEU2 tif11-K10D in YCplac111 | This study |
| pDH473 | sc LEU2 tif11-R13A in YCplac111 | This study |
| pDH472 | sc LEU2 tif11-R13D in YCplac111 | This study |
| pDH475 | sc LEU2 tif11-R14A in YCplac111 | This study |
| pDH474 | sc LEU2 tif11-R14D in YCplac111 | This study |
| pDH478 | sc LEU2 tif11-K16A in YCplac111 | This study |
| pDH476 | sc LEU2 tif11-K16D in YCplac111 | This study |
| p3400/pDSO23 | hc LEU2 TIF11 in YEplac181 | (Choi et al., 2000) |
| pPMB167 | hc LEU2 tif11-K4D in YEplac181 | This study |
| pPMB168 | hc LEU2 tif11-ΔG8 in YEplac181 | This study |
| pPMB169 | hc LEU2 tif11-ΔG8ΔG9 in YEplac181 | This study |
| pPMB170 | hc LEU2 tif11- K10D in YEplac181 | This study |
| p4281/YCpTIF5-G31R-W | sc TRP1 TIF5-G31R in YCplac22 | (Valásek et al., 2004) |
| p4280/YCpSUI3-S264Y-W | sc TRP1 SUI3-S264Y in YCplac22 | (Valásek et al., 2004) |
| p367 | sc URA3 HIS4(ATG)-lacZ | (Donahue and Cigan, 1988) |
| p391 | sc URA3 HIS4(TTG)-lacZ | (Donahue and Cigan, 1988) |
| p180 | sc URA3 GCN4-lacZ | (Hinnebusch, 1985) |
| pPMB24 | sc URA3 SUI1-lacZ | (Martin-Marcos et al., 2011) |
| pPMB25 | sc URA3 SUI1-opt-lacZ | (Martin-Marcos et al., 2011) |
| pPMB28 | sc URA3 SUI1UUU-lacZ | (Martin-Marcos et al., 2011) |
| pC3502 | sc URA3 -3AAA−1 el.uORF1 GCN4-lacZ in YCp50 | (Visweswaraiah et al., 2015) |
| pC4466 | sc URA3 -3UAA−1 el.uORF1 GCN4-lacZ in YCp50 | (Visweswaraiah et al., 2015) |
| pC3503 | sc URA3 -3UUU−1 el.uORF1 GCN4-lacZ in YCp50 | (Visweswaraiah et al., 2015) |
| pC3505 | sc URA3 el.uORF1-less GCN4-lacZ in YCp50 | (Visweswaraiah et al., 2015) |
| pTYB2-TIF11 | TIF11 in pTYB2 | (Acker et al., 2007) |
| p6013 | tif11-R13P in pTYB2 | This study |
| p6015 | tif11-K16D in pTYB2 | This study |
| pRaugFFuug | Dual luciferase reporter LUCren(aug)-LUCfirefly (uug) in URA3 vector | (Kolitz et al., 2009) |
| pRaugFFuug | Dual luciferase reporter LUCren(aug)-LUCfirefly (uug) in URA3 vector | (Kolitz et al., 2009) |
Yeast strains used in this study
https://doi.org/10.7554/eLife.31250.024| Strain | Genotype | Source |
|---|---|---|
| H3582 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3392 (sc URA3 TIF11) | (Fekete et al., 2005) |
| PMY318 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3390 (sc LEU2 TIF11) | This study |
| PMY284 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5633 (sc LEU2 tif11-K3E) | This study |
| PMY285 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5635 (sc LEU2 tif11-K4D) | This study |
| PMY286 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5638 (sc LEU2 tif11-T6D) | This study |
| PMY287 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5637 (sc LEU2 tif11-T6R) | This study |
| PMY289 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5640 (sc LEU2 tif11-ΔG8) | This study |
| PMY290 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5642 (sc LEU2 tif11-R13P) | This study |
| PMY291 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5644 (sc LEU2 tif11-G15D) | This study |
| PMY320 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH469 (sc LEU2 tif11-K7A) | This study |
| PMY321 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH468 (sc LEU2 tif11-K7D) | This study |
| PMY322 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH481 (sc LEU2 tif11-ΔG8ΔG9) | This study |
| PMY323 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH471 (sc LEU2 tif11-K10A) | This study |
| PMY324 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH470 (sc LEU2 tif11-K10D) | This study |
| PMY325 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH473 (sc LEU2 tif11-R13A) | This study |
| PMY326 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH472 (sc LEU2 tif11-R13D) | This study |
| PMY327 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH475 (sc LEU2 tif11-R14A) | This study |
| PMY329 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH474 (sc LEU2 tif11-R14D) | This study |
| PMY330 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH478 (sc LEU2 tif11-K16A) | This study |
| PMY332 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH476 (sc LEU2 tif11-K16D) | This study |
| PMY354 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3400 (hc LEU2 TIF11) | This study |
| PMY355 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB167 (hc LEU2 tif11-K4D) | This study |
| PMY357 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB168 (hc LEU2 tif11-ΔG8) | This study |
| PMY358 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB169 (hc LEU2 tif11-ΔG8ΔG9) | This study |
| PMY359 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB170 (hc LEU2 tif11- K10D) | This study |
| PMY32 | MATa ura3-52 leu2-3 leu2-112 trp1Δ−63 his4-301(ACG) sui1Δ::hisG pPMB02 (sc LEU2 sui1-K60E) | (Martin-Marcos et al., 2011) |
| PMY293 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3390 (sc LEU2 TIF11) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY295 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5633 (sc LEU2 tif11-K3E) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY296 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5635 (sc LEU2 tif11-K4D) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY297 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5638 (sc LEU2 tif11-T6D) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY298 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5637 (sc LEU2 tif11-T6R) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY300 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5640 (sc LEU2 tif11-ΔG8) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY301 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5642 (sc LEU2 tif11-R13P) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY302 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5644 (sc LEU2 tif11-G15D) p4281 (sc TRP1 TIF5-G31R) | This study |
| PMY335 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3390 (sc LEU2 TIF11) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY310 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5633 (sc LEU2 tif11-K3E) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY311 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5635 (sc LEU2 tif11-K4D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY312 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5638 (sc LEU2 tif11-T6D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY313 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5637 (sc LEU2 tif11-T6R) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY315 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5640 (sc LEU2 tif11-ΔG8) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY316 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5642 (sc LEU2 tif11-R13P) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY317 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5644 (sc LEU2 tif11-G15D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY339 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH469 (sc LEU2 tif11-K7A) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY340 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH468 (sc LEU2 tif11-K7D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY341 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH481 (sc LEU2 tif11-ΔG8ΔG9) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY342 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH471 (sc LEU2 tif11-K10A) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY343 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH470 (sc LEU2 tif11-K10D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY344 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH473 (sc LEU2 tif11-R13A) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY345 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH472 (sc LEU2 tif11-R13D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY346 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH475 (sc LEU2 tif11-R14A) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY348 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH474 (sc LEU2 tif11-R14D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY349 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH478 (sc LEU2 tif11-K16A) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY351 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pDH476 (sc LEU2 tif11-K16D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY337 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3390 (sc LEU2 TIF11) YCplac22 (sc TRP1) | This study |
| PMY338 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3390 (sc LEU2 TIF11) YCplac22 (sc TRP1) | This study |
| PMY360 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3400 (hc LEU2 TIF11) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY362 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB167 (hc LEU2 tif11-K4D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY364 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB168 (hc LEU2 tif11-ΔG8) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY365 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB169 (hc LEU2 tif11-ΔG8ΔG9) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY366 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ pPMB170 (hc LEU2 tif11- K10D) p4280 (sc TRP1 SUI3-S264Y) | This study |
| PMY361 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p3400 (hc LEU2 TIF11) YCplac22 (sc TRP1) | This study |
| GP3511 | MATα ura3-52 leu2-3 leu2-112 ino1 sui2∆ gcn2∆ pep4::LEU2 < HIS4 lacZ,ura3−52 > pAV1089 (SUI2,SUI3,GCD11-HIS,URA3) | (Pavitt et al., 1998) |
| H4560 | MATα ura3-52 leu2-3 leu2-112 ino1 sui2Δ gcn2Δ pep4::leu2::natMX sui3Δ::kanMX < HIS4 lacZ,ura3−52 > p5321 (SUI2,SUI3-2,GCD11-HIS,LEU2) | (Martin-Marcos et al., 2014) |
| YAS2488 | MATa leu2-3,−112 his4-53a trp1 ura3-52 cup1::LEU2/PGK1 pG/MFA2 pG | (Algire et al., 2002) |
| FZY010 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5642 (sc LEU2 tif11-R13P) YCplac22 (sc TRP1) | This study |
| FZY011 | MATa ura3-52 trp1Δ63 leu2-3, leu2-112 his4-301(ACG) tif11Δ p5642 (sc LEU2 tif11-R13P) YCplac22 (sc TRP1) | This study |
Additional files
-
Supplementary file 1
Supplementary Tables.
Table S1: Oligonucleotide primers employed for TIF11 mutagenesis in this study Table S2: Ribosome profiling datasets used for uORF identification
- https://doi.org/10.7554/eLife.31250.025
-
Supplementary file 2
Excel file containing results and analyses from ribosome footprint profiling of WT and tif11-R13P cells.
Spreadsheet 1, ‘CDS_all Expr’, tabulates log2 values of the following parameters for the 5037 expressed genes listed in columns A-B for WT and tif11-R13P cells: Ribosome footprint sequencing reads (RPF_WT and RPF_R13P); mRNA sequencing reads (mRNA_WT and mRNA_R13P); the ratios RPF_R13P/RPF_WT (ΔRPF_ R13P) and mRNA_R13P/mRNA_WT (ΔmRNA_R13P); and the ratio ΔRPF_R13P/ΔmRNA_R13P (ΔTE_R13P). Spreadsheet 2, ‘Context_score’, contains a subset of genes in Spreadsheet 1 (4280 genes) with 5’UTR length >5 nt examined for additional parameters: 5’UTR length, sequences between positions −6 and +6, and the context scores. Spreadsheet 3, ‘AUG_uORFs_identified’, contains all 564 AUG uORFs identified using the yassour-uorf program from multiple datasets listed in Table S2, listing uORF chromosome coordinates, distances of the uORF AUG from the 5’ end of the mRNA and the main CDS start codon, uORF sequence contexts between the −3 and +4 positions and the context scores (NA, for uORF 5’ UTR <3 nt). Spreadsheet 4, ‘uORF_Expr’, tabulates log2 values of the following parameters for the 97 expressed uAUG uORFs listed in column A for WT and tif11-R13P cells: Ribosome footprint sequencing reads on CDS (RPF_CDS_WT and RPF_CDS_R13P); Ribosome footprint sequencing reads on uORFs (RPF_uORF_WT and RPF_uORF_R13P); the ratios RPF_CDS_R13P/RPF_CDS_WT (ΔRPF_CDS_ R13P) and RPF_uORF_R13P/RPF_uORF_WT (ΔRPF_uORF_R13P); Relative Ribosome Occupancy (RRO) for WT (the ratio of RPF_uORF_WT/RPF_CDS_WT, RRO_WT) and R13P (the ratio of RPF_uORF_R13P/RPF_CDS_R13P, RRO_R13P) and the ratio RRO_R13P/RRO_WT (RRO_R13P); Spreadsheet 5, PARS score analysis of 5’ UTRs of 2642 genes, conducted as described in (Sen et al., 2016) and their context scores as listed in Spreadsheet 2. Spreadsheet 6 ‘CDS_RPF_Change’, tabulates log2 values of the ratio RPF_R13P/RPF_WT (ΔRPF_R13P (log2)) and adjusted p-value (padj) for the 5083 expressed genes detected by the DESEQ2 package listed in columns A-B
- https://doi.org/10.7554/eLife.31250.026
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31250.027





