A tissue-specific, Gata6-driven transcriptional program instructs remodeling of the mature arterial tree
Figures
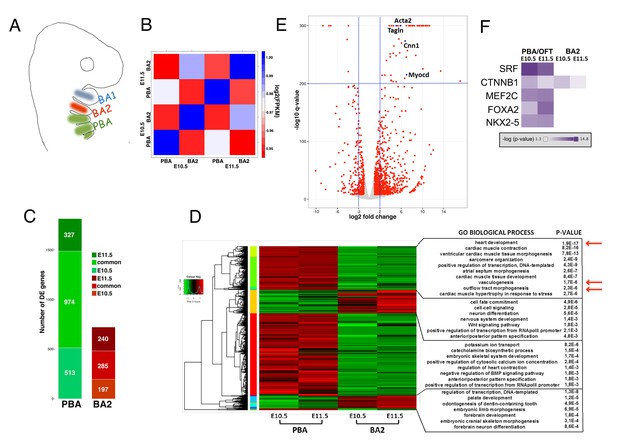
Global transcriptomes of developing BAs and OFT.
(A) In mammals, BA2 (red) hosts aortic arch (AA)2, which regresses, while posterior BAs (PBA, green) host AA3-6, which contribute to the main thoracic arteries. (B) Correlation plot of global expression profiling separates BA2 and PBA at E10.5 and E11.5. Heatmap shows the Spearman correlation coefficients for each pair of samples based on the normalized expression values (FPKM). (C) Count of DE genes between PBA and BA2 (fold change ≥±2; q-value <0.05) at E10.5–11.5. The bar plot shows PBA/OFT-enriched genes as the largest fraction of DE genes (color-coded as in A). (D) Hierarchical clustering of DE genes in any of the three pair-wise comparisons. DAVID analysis (Huang et al., 2009) of the clusters generated detects significant association with PBA/OFT-specific Biological Process GO terms (arrows). (E) Volcano plot. Top significant PBA/OFT-enriched genes are highly expressed in SMCs. (F) Ingenuity Pathway Analysis (IPA) identifies the TF SRF as the most likely upstream regulator of PBA/OFT-enriched genes (p-value<1e-20). SRF targets include Myocd and known Myocd targets (e.g. Acta2, Tagln).
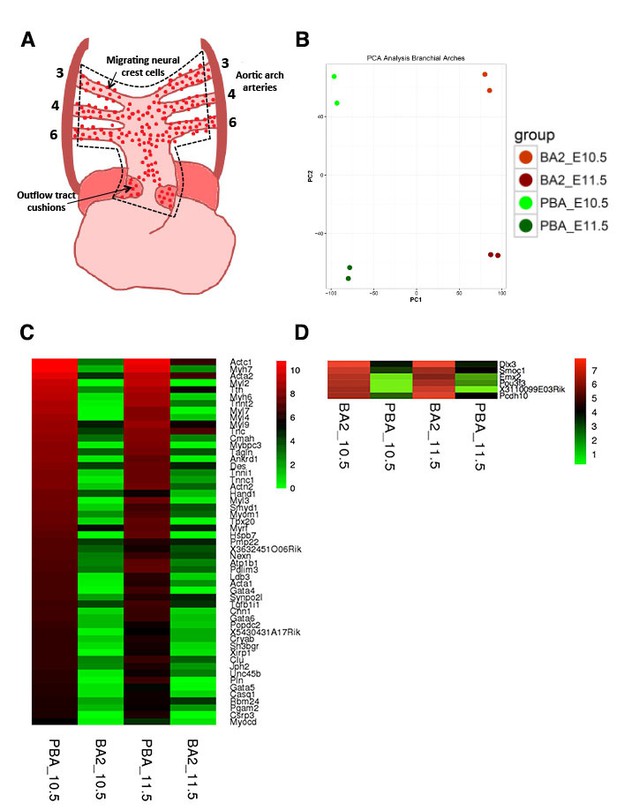
Differential expression across developing BAs.
(A) Embryonic circulation, front view (adapted from High and Epstein, 2008). The broken line encloses the PBA/OFT, the embryonic area used as a source of RNA. The AA3, 4 and 6 are housed by BA3-6 (PBA in green in Figure 1A) (B) PCA shows reproducibility of the biological replicates and separates BA2 and PBA/OFT at both developmental stages. (C–D) Heatmaps of top significant DE genes (Volcano plot in Figure 1, upper right and left quadrants) in PBA/OFT (C) and BA2 (D).
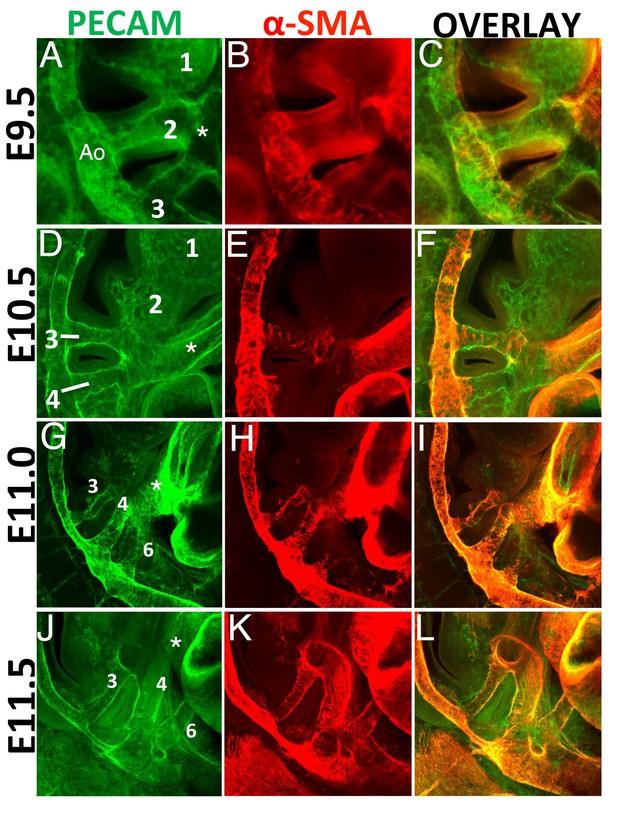
SMCs are generated exclusively in BA3-6, in a rostro-caudal fashion.
Confocal analysis of whole mount wild type embryos at E9.5 (A–C), E10.5 (D–F), E11.0 (G–I) and E11.5 (J–L) to visualize the endothelial network (PECAM, green) (A,D,G, J) and vascular SMCs (SMA, red) (B, E, H, K). C, F, I, L are overlays of PECAM and SMA staining. (A-C) At E9.5 (25 somites, (s), AAs 2 and 3 are connected to the dorsal aorta. AA1 has already started to regress into capillary beds. SMA-positive cells are visible in the dorsal aorta and OFT, but are not detected in the walls of AA2-3. (D–F) At E10.5 (37s), AA3 and AA4 are fully formed, but only AA3 is associated to vascular SMCs. Capillary beds replace AA2 in BA2. G-I. A clear AA6 is visible at E11.0 (43s). At this stage AA3-4 are fully covered with SMA-positive cells, while the newly formed AA6 is not associated with vascular SMCs. (J–L) At E11.5 (48s) formation of AA3, 4 and 6 is complete. All visible AAs (AA3-4-6) are covered by SMA-positive cells. Right view. Ao, aorta; asterisks label the OFT.
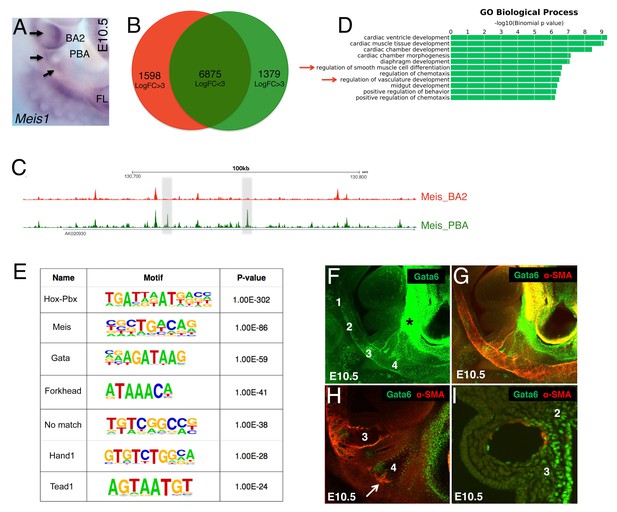
Unbiased identification of main TFs instructing PBA-specific transcription.
(A) Whole mount in situ hybridizations on E10.5 mouse embryo. Meis1 is expressed in the BA2 (arrow) and in the PBA (BA3 and BA4, arrows). (B) Diffrep analysis of Meis binding in PBA and BA2. Venn diagram shows ‘shared’ Meis binding sites (FE ≥ 10) with logFC < 3 signal (n = 6875) and with higher (logFC ≥ 3) signal in PBA (green) and BA2 (red). (C) UCSC browser tracks upstream of Nrp1 illustrate the largely overlapping binding of Meis in PBA and BA2, with instances of increased Meis binding signal (gray regions) in PBA relative to BA2. (D) Top over-represented biological processes associated to Meis peaks (FE ≥ 10) with higher signal in PBA (logFC ≥ 3) include ‘smooth muscle cell differentiation’ and ‘vasculature development’. (E) De novo motif discovery on Meis peaks (FE ≥ 10) with higher signal in PBA identifies high enrichment of the recognition sequence for GATA TFs, together with motifs recognized by Meis and Meis partners Hox/Pbx. (F–I) Confocal analysis of E10.5 embryos (36–37 s) whole mount (F–H) and sagittal sections (I) stained with Gata6 (F) and Gata6/SMA antibodies (G–I). Gata6 is detected in the BA3, BA4 (F–H) and the heart region (F–G). Individual z-stack (H) shows Gata6 co-localization with SMA-positive cells in the BA3 and the appearance of SMCs in Gata6-positive area in the BA4 (arrow). Gata6/SMA-positive cells surround AA3 (I). Numbers indicate corresponding BAs; asterisks label the OFT.
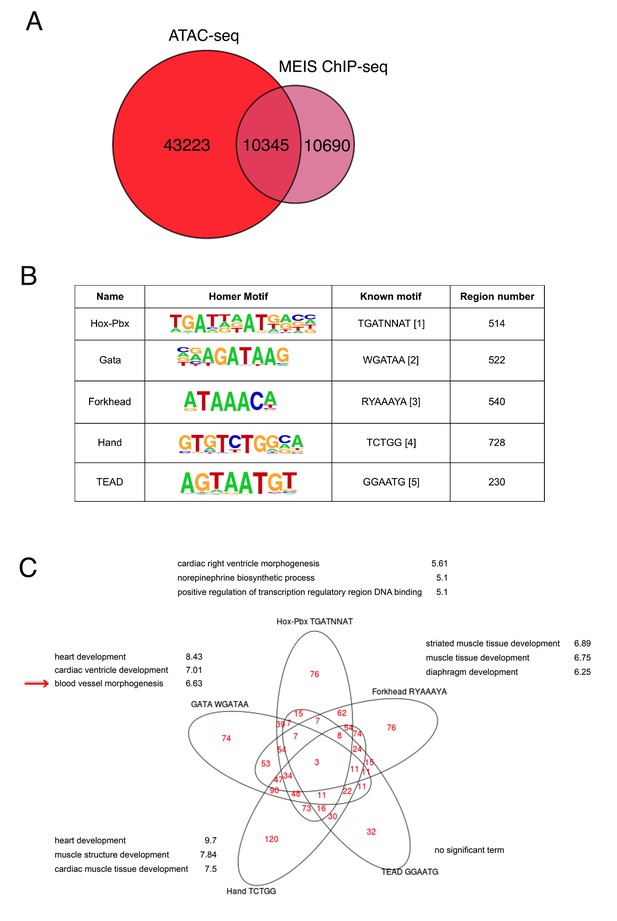
Analysis of Meis-bound regions containing different TF recognition motifs.
(A) Comparison of Meis ChIP-seq (FE ≥ 10 peaks) in E11.5 BA2 and ATAC-seq in E10.5 BA2. Half of Meis peaks overlap with accessible chromatin in the BA2. (B) Experimentally identified motifs (known motifs), which correspond to Homer identified motifs, were used to count the instances of highly enriched (logFC <3)Meis binding (200nt summit regions, FE ≥ 10) containing at least one motif. (C) Functional enrichment of motifs regions (-log10 p-value) and their intersections. Venn diagram shows the intersection of Meis-bound regions containing each motif and their corresponding top three GO biological processes. In Meis regions containing GATA motifs, the term ‘muscle development’ did not appear in the full list. ‘Blood vessel morphogenesis’ was also associated to Forkhead-containing regions. TEAD- containing regions did not return any significant term. 1. (Donaldson et al., 2012); 2. (Han et al., 2016); 3. (Nakagawa et al., 2013); 4. (Hollenberg et al., 1995); 5. (Diepenbruck et al., 2014).
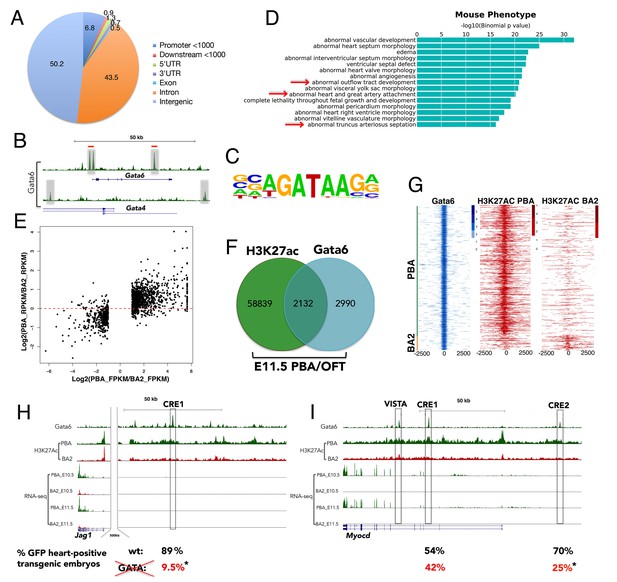
Gata6 occupies active enhancers in PBA/OFT.
(A) CEAS analysis of the distribution of Gata6 peaks relative to Reference Sequence (RefSeq) gene structures. The pie chart and corresponding percentage values indicate the proportion of reads. (B) UCSC browser tracks shows Gata6 binding (gray regions) at Gata4 and Gata6 loci. Red lines indicate Gata4 binding in E12.5 ventricles. (C) Sequence logo of the most significant motifs identified using de novo motif discovery. (D) Top over-represented mouse phenotypes associated to Gata6 peaks. The x axes values correspond to the binomial raw (uncorrected) p-values. (E) Scatterplot of the Log2(ratio of FPKM) values for DE genes between PBA and BA2 versus the Log2(ratio of RPKM) values of their H3K27ac signals at promoters and distal regions (Correlation: 0.694; p<2.2e-16). (F) Venn diagram (not proportional) of Gata6 peaks (200nt summits) and H3K27Ac-positive regions in the PBA/OFT. Almost half of Gata6 peaks overlap regions acetylated in the PBA/OFT. (G) Heatmap of Gata6 peaks and corresponding H3K27ac peaks (within 5000 nt of the summit) shows most Gata6 peaks overlap PBA/OFT-specific enhancers (n = 820), and only a minority of BA2-specific enhancers (n = 98). Regions detected as H3K27Ac-positive in both PBA/OFT and BA2 were excluded from this analysis. (H, I) UCSC browser tracks. RNA-seq and ChIP-seq profiles for Gata6 binding and H3K27Ac in the PBA/OFT and H3K27Ac in the BA2 at Jag1 (H) and Myocd (I) loci. Gata6 binds regions highly acetylated in PBA/OFT, but not BA2 (boxed; VISTA highlights a heart-positive enhancer). Numbers correspond to the % of embryos injected with wild-type (upper row, in black) and mutant (lower row, in red) enhancers, displaying reporter activity in the heart in addition to the midbrain (positive control); asterisks denote p-value<0.05 (Fisher's Exact Test).
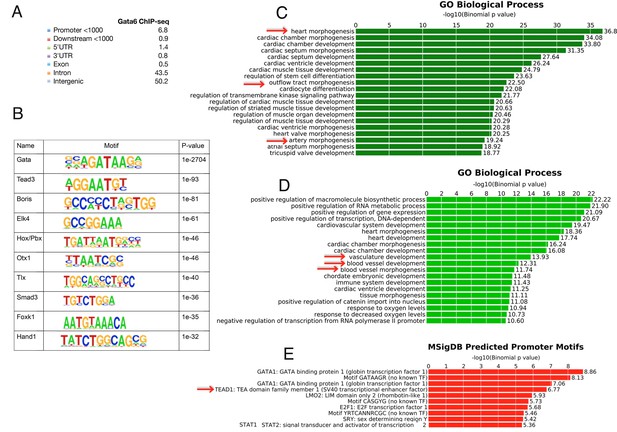
Extended analysis of Gata6 peaks.
(A) CEAS analysis of the distribution of Gata6 peaks relative to Reference Sequence (RefSeq) gene structures. Numbers correspond to percentage values; the proportion of reads is shown in the pie chart In Figure 4A. (B) Sequence logo of motifs identified by Homer. (C) Top over-represented biological processes associated to Gata6 peaks. The x axes values correspond to the binomial raw (uncorrected) p-values. (D) Top over-represented biological processes associated to PBA/OFT-specific enhancers bound by Gata6 (n = 820). (E) Predicted motifs in promoters associated to PBA/OFT-specific enhancers bound by Gata6 (n = 820).
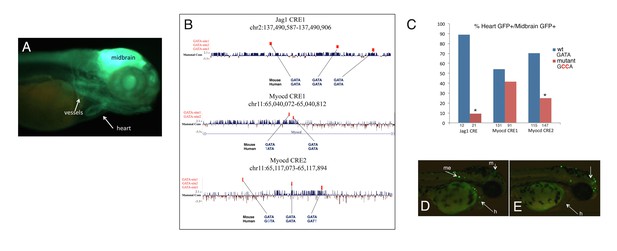
Disrupting GATA motifs affects the activity of GATA6-bound enhancers.
(A) GFP reporter activity in Myocd CRE1 lines. Lines were derived from embryos injected with Myocd CRE1. Myocd CRE1 drives GFP in the heart and in vessels-like structures. The construct also contains a control enhancer, which drives GFP activity in the midbrain. (B) Genomic coordinates (mm9) and nucleotide sequence of the regions injected in zebrafish. The overall mammal conservation is shown, together with the sequence conservation and position of the GATA motifs. In mutant enhancers, each GATA was replaced by GCCA. (C) Activity of the wild-type (blue) and corresponding mutant (red) enhancers expressed as percentage of midbrain GFP-positive embryos (positive control) also showing GFP expression in the heart. For each wild-type and mutant enhancer, numbers on the x axis correspond to the total number of midbrain-positive embryos; asterisks denote p-value<0.05 (Fisher's Exact Test: Myocd CRE1 p-value=0.07686; Myocd CRE2 p-value=1.838e-13; Jag1 CRE: p-value=3.978e-05). (D, E) GFP reporter activity in embryos injected with wild-type (H) and mutant (I) Jag1 CRE1. M, midbrain; me, melanocytes; h, heart.
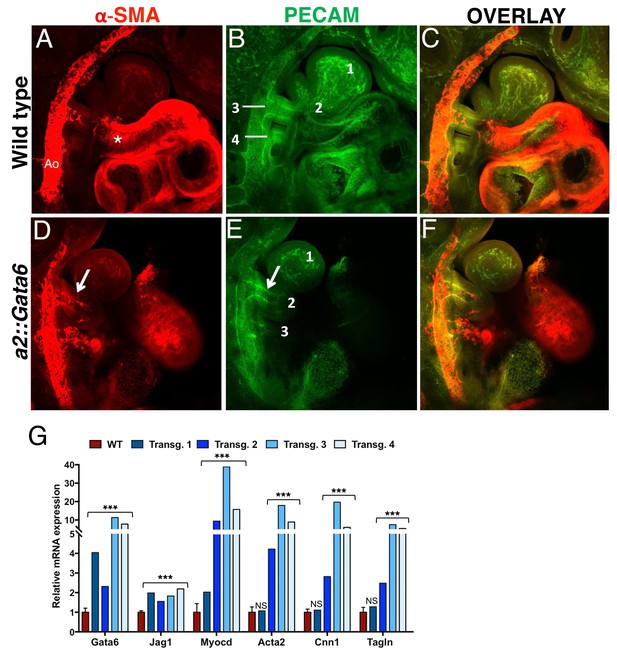
Gata6 is sufficient to induce SMCs in BA2 and to preserve AA2.
Confocal analysis of whole mount wild type (A–C) and a2::Gata6 transgenic mouse embryos (D–F) at E10.5 (s ≥ 32) to visualize vascular SMCs (a-SMA, red) (A, D), the endothelial network (PECAM, green) (B, E) and SMA/PECAM overlay (C, F); see also Figure 5—videos 1–4. (A-C) In wild-type embryos, SMA-positive cells are visible in the AA3, the dorsal aorta and the heart, but are not detected in BA2, where AA2 has regressed to a capillary bed (see also Figure 2). D-F, In a2::Gata6 embryos, vascular SMCs are detected in the BA2 (arrows in D). In the majority of transgenic embryos analyzed, the BA2 hosts a persistent artery connected to the heart (arrow in E). For best visualization of the ectopic AA2, an individual z-stack is shown in (D–F); the complete z-stack series is shown in Figure 5—videos 3–4. (G) Quantitative RT-PCR analysis of changes in gene expression in a::Gata6 embryos (s ≥ 37) and littermate controls. The wild-type values are presented as a mean ± SEM of two technical replicates of four independent embryos. For each transgenic embryo, the average of two technical replicates ± SEM is shown. Transgenic embryos 2–4 showed evidence of a functional AA2. Conversely, transgenic embryo one did not show significant upregulation of SMC-markers relative to wild-type; this embryo had no evidence of ectopic AA2. Asterisks (***) correspond to pvalues < 0.005; NS = not significant. All the results of transgenic embryos analyses are summarized in Figure 5—source data 1. Ao, dorsal aorta; numbers indicate BAs and corresponding AAs. Asterisks label the OFT.
-
Figure 5—source data 1
Summary of transgenic embryos results.
- https://doi.org/10.7554/eLife.31362.012
-
Figure 5—source data 2
Data file for Figure 5G.
- https://doi.org/10.7554/eLife.31362.013
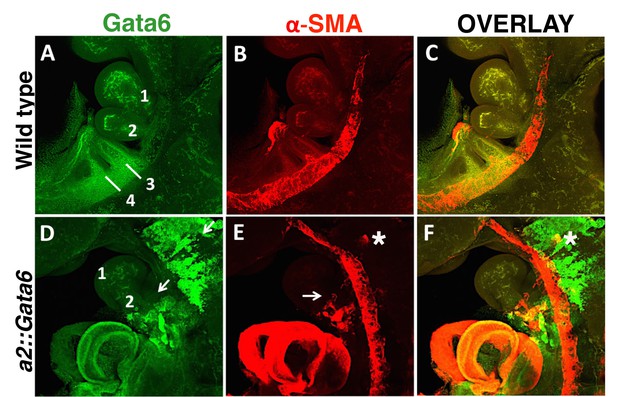
Gata6 ectopic expression in a2::Gata6 embryos.
Confocal analysis of whole mount wild type (A–C) and a2::Gata6 embryos (D–F) at E10.5 to visualize Gata6 (A, D, green) and VSMCs (C, E a-SMA, red) and Gata6/SMA overlay (C, F). (A–C) High levels of Gata6 are detected in the PBAs (see also Figure 3G). SMA-positive cells are visible in the AA3, the dorsal aorta and the heart, but are not detected in BA2. (D–F) Expression of Gata6 in the NC migrating to the BAs (arrow), in BA2 (2, arrow) and in PBAs in a2::Gata6 transgenic embryos. Ectopic SMCs were exclusively detected in transgenic embryos displaying Gata6 overexpression. Gata6 expression largely overlaps with SMA staining in the aortic arches of transgenic embryos. A cluster of Gata6-positive, SMA-positive cells is observed along NC migration route (asterisk).
Reconstruction of wild-type embryo, stained by PECAM.
https://doi.org/10.7554/eLife.31362.014Reconstruction of wild-type embryo, stained by a-SMA.
https://doi.org/10.7554/eLife.31362.015Reconstruction of a transgenic a2::Gata6 embryo, stained by PECAM to reveal the persistent AA2.
https://doi.org/10.7554/eLife.31362.016Reconstruction of a transgenic a2::Gata6 embryo, stained by a-SMA to reveal the ectopic SMCs in BA2.
https://doi.org/10.7554/eLife.31362.017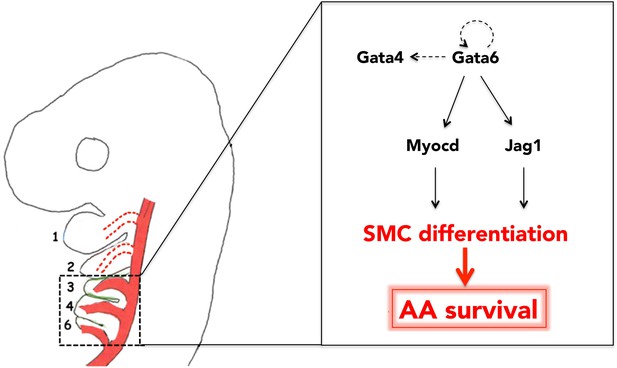
Gata6-driven SMC differentiation promotes survival of the posterior AAs.
Gata6 activates Myocd and Jag1 to initiate SMC differentiation in the cardiac NC, migrating to BA3-6. Generation of SMCs in BA3-6, and their recruitment by AA3-6, results in the stabilization of these vessels and their subsequent incorporation into the mature aortic tree. Conversely, because Gata6 is not expressed in BA1-2, AA1-2 do not ‘see’ any SMC and eventually regress. Auto-regulatory loops involving GATA TFs are likely features of Gata6 network. Arrows and double arrow indicate regulation and interaction, respectively; broken arrows indicate links that are not experimentally validated.
Additional files
-
Supplementary file 1
DE genes in PBA and BA2
- https://doi.org/10.7554/eLife.31362.020
-
Supplementary file 2
Meis differential binding in PBA and BA2
- https://doi.org/10.7554/eLife.31362.021
-
Supplementary file 3
Gata6 peaks and overlap with H3K27Ac peaks
- https://doi.org/10.7554/eLife.31362.022
-
Supplementary file 4
RNA-seq and ChIP-seq statistics
- https://doi.org/10.7554/eLife.31362.023
-
Supplementary file 5
Flowchart of the RNA-seq and ChIP-seq downstream analyses
- https://doi.org/10.7554/eLife.31362.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31362.025