Dengue viruses cleave STING in humans but not in nonhuman primates, their presumed natural reservoir
Figures
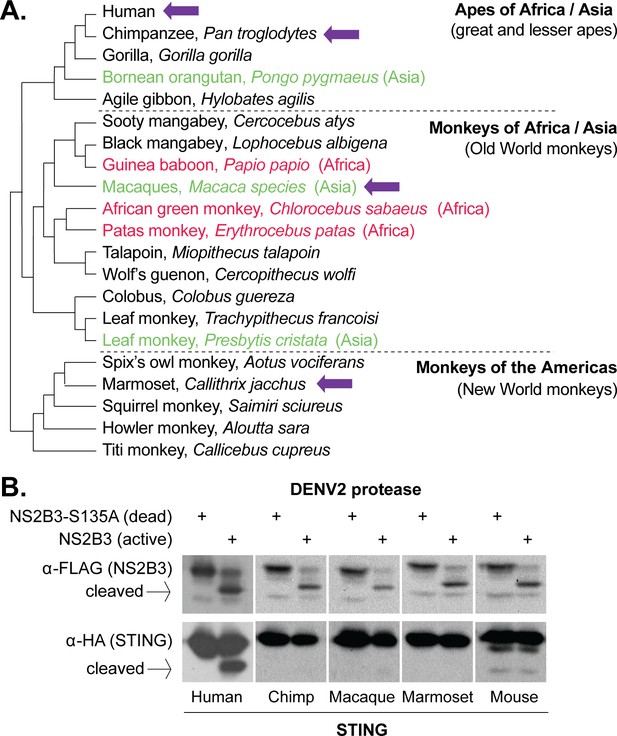
Dengue virus (DENV2) can cleave human but not nonhuman primate STING.
(A) A phylogeny of select primate species, showing the three main simian clades: apes, Old World monkeys, and New World monkeys (Perelman et al., 2011). The primate species from which STING is tested in this study are shown with purple arrows. Possible primate reservoir hosts for sylvatic dengue viruses, based on virus isolation from sentinel monkeys, or antibody detection, are shown in red (Africa) and green (Asia). The current evidence for these primate reservoir hosts is reviewed in the discussion section. (B) 293T cells were cotransfected with plasmids encoding STING-HA, and the NS2B3-Flag protease complex with or without the S135 inactivating mutation. Whole cell lysate isolated 24 hr post transfection was run on a protein gel and immunoblotted with anti-Flag or anti-HA antibodies. The encoded NS2B-NS3-Flag polyprotein auto-processes into the NS2B3 protease complex if the protease is active, as seen in the anti-Flag blot where in some samples the NS3-Flag protein has been liberated through cleavage. We sometimes see lower bands underneath the full-length mouse STING, but conclude that they are endogenous degradation products since they are equal in intensity in the presence of the active or dead protease.
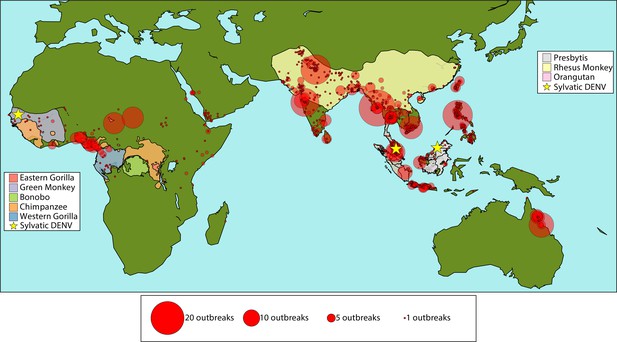
Many primate species reside in areas where dengue viruses are endemic in humans.
https://doi.org/10.7554/eLife.31919.004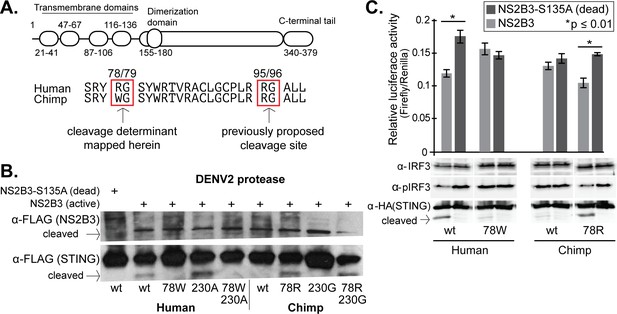
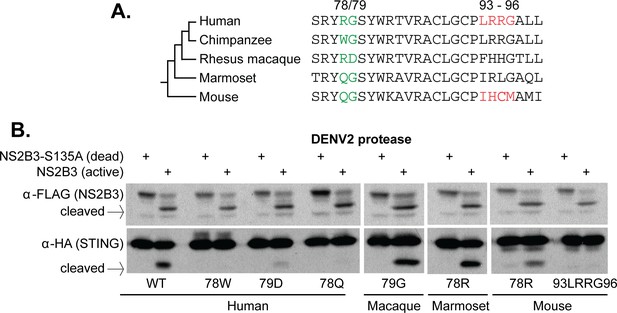
STING residue 78 determines susceptibility to NS2B3 cleavage in human versus chimpanzee STING comparisons.
(A) A domain diagram of human STING is shown, as defined in (Wu et al., 2014). An alignment of human and chimpanzee STING in the region of the newly identified cleavage determinant (78/79) and the one previously determined (95/96) (Aguirre et al., 2012; Yu et al., 2012). (B) Site-directed mutagenesis was performed on either human or chimpanzee STING at position 78, substituting the residue at this position in human (R) with that in chimpanzee (W) and vice versa. Plasmids encoding the NSB3 protease complex and STING were cotransfected into 293T cells, and 48 hr later lysates were collected and analyzed by anti-FLAG western blot. In this experiment, both the protease and STING are tagged with FLAG. Data presented are representative of at least two experiments. (C) (bottom) 293T cells were transfected with plasmids expressing the DENV2 NS2B3 protease and wildtype (wt) or mutated (78W or 78R) STING. IRF3 and phosphorylated IRF3 (pIRF3) were detected by western blot in lysates harvested 48 hr later. (top) The identical experiment, but performed in biological triplicate and with the addition of plasmids encoding a firefly luciferase gene driven by the interferon beta (IFNb) promoter, and a renilla luciferase gene driven by a CMV promoter. The relative luciferase activity (Y-axis) was calculated by normalizing the luciferase signal to the renilla signal in each replicate. A Welch’s T-test was used to compare the levels of luciferase produced in the presence of active versus dead protease. Data is representative of at least two experiments.
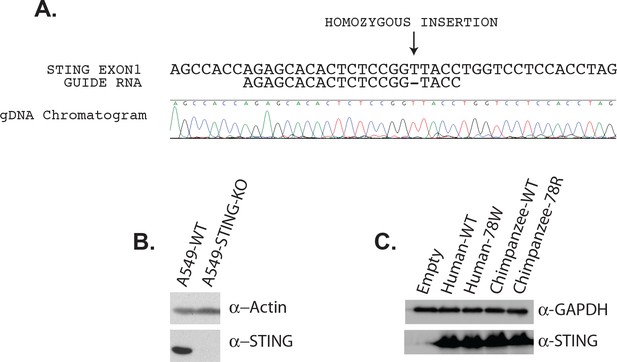
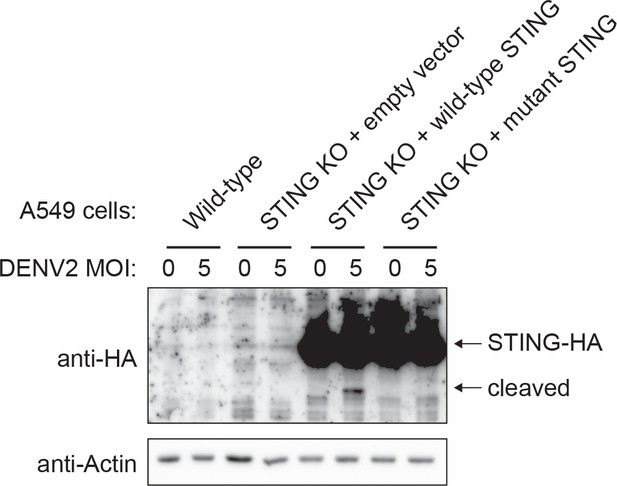
Generation of STING knockout cells using CRISPR-Cas9, and stable re-complementation of these lines.
(A) A depiction of the insertion induced in the first exon of STING in A549 cells. Below, a Sanger sequencing chromatogram from a this genomic locus shows the homozygous insertion. (B) Both wildtype (wt) A549 cells and A549 cells that were knocked out for STING were probed by western blot for Actin or STING. An antibody recognizing the endogenous STING was used (anti-STING; Abcam 92605). (C) STING-KO A549 cells were re-complimented with HA-tagged versions of STING using lentiviral transduction. Cell lines were generated to express either wildtype (wt) or mutant STING from human or chimpanzee. A mutation was made in human STING to render it cleavage-resistant (78W). A mutation was made in chimpanzee STING to make it cleavage-susceptible (78R). Western blots where performed with antibodies against GAPDH or STING.
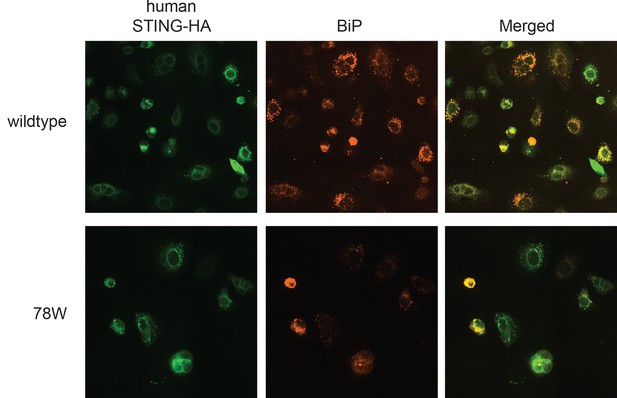
Wildtype and mutated human STING both colocalize with the ER-resident protein BiP.
Immunofluorescence imaging was performed on STING and the ER-resident protein BiP. A549 cells knocked out for STING were reconstituted with either wildtype human STING (top) or human STING 78W (cleavage resistant) by retroviral transduction (as shown in Figure 2—figure supplement 1). These cells were fixed in a 4% paraformaldehyde solution. Antibodies against the HA epitope tag (STING) or BiP were incubated with the fixed cells. Cells were imaged at 40x magnification on the Nikon A1R confocal microscope.
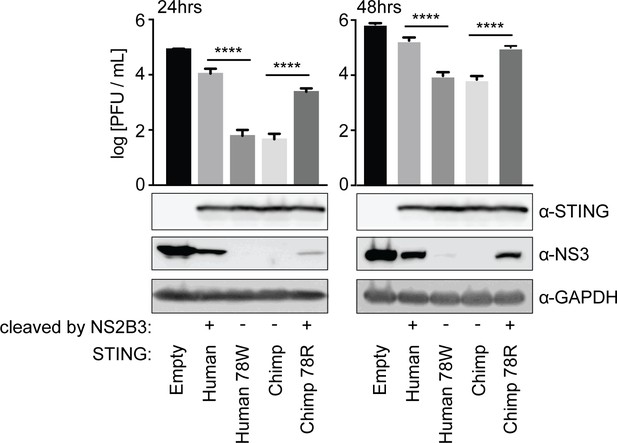
Cleavage of STING at position 78/79 promotes virus replication.
The endogenous copies of STING in A549 cells were knocked out using the Cas9 nuclease (see Figure 2—figure supplement 1). These cells were re-complemented by retroviral transduction with no gene (pLPCX-empty), wildtype human STING, cleavage-resistant human STING (human 78W), wildtype chimpanzee STING, or cleavage-susceptible chimpanzee STING (chimp 78R). These cell lines were infected at MOI of 0.3 with dengue virus 2 (DENV2 16681). After 24 and 48 hr the virus supernatant was removed and titrated on BHK21 cells. At the same time, cells were collected in RIPA buffer, lysed, and run on a gel for western blotting using antibodies against STING, dengue virus NS3, and GAPDH (loading control). A Tukey's multiple comparisons test indicated significant differences in infectious virus in the presence of each mutant STING compared to wildtype STING, as shown (****=p < 0.0001), after significant one-way ANOVA. Data are representative of at least two independent experiments.
STING is cleaved during dengue virus infection.
The indicated A549 cells lines (wild-type, STING knockout, or re-complemented) were plated in F-12K media with 10% FBS. Dengue virus 2 (16681) was diluted in F-12K media with 2% FBS, added to cells at an MOI of 5, and allowed to attach to cells for 1 hr at room temperature. Uninfected wells were incubated with F-12K media with 2% FBS not containing virus. F-12K media with 10% FBS was added to cells and they were maintained at 37°C with 5% CO2. After 20 hr the media in all wells was replaced with F-12K media with 2% FBS containing 20 μM proteasome inhibitor MG-132 (Sigma-Aldrich M7449). 4 hr following MG-132 treatment (24 hr following infection) the cells were collected and lysed in RIPA buffer supplemented with protease inhibitor (Roche, 4693159001). Protein concentration was calculated using the Bradford method. 15% 37.5:1 Acrylamide/Bisacrylamide gels were used to separate 30 μg of whole cell lysate for each sample. Protein was transferred overnight at 30 volts onto a nitrocellulose membrane. Blocking was performed with a 10% milk solution in tris-buffered saline supplemented with 0.1% TWEEN20. Primary antibodies were used against HA (3F10 clone Sigma 11867423001) and β-actin (Santa Cruz Biotechnology Sc47778).
Residues 78 and 79 of STING define a dengue virus cleavage determinant in both primate and mouse STING.
(A) A phylogeny and multiple sequence alignment of STING from various primate species and mouse. Shown in green is the 78/79 motif in STING that is mutated in panel B. Shown in red is the motif changed in mouse STING, only, in panel B. (B) Site directed mutagenesis was performed on human, rhesus macaque, marmoset, or mouse STING at sites 78/79 or 93–96 (mouse only). 293T cells were cotransfected with mammalian expression plasmids encoding STING along with wildtype or mutant NS2B3. 24 hr after transfection, whole-cell lysate was harvested and probed for FLAG or HA by western blot. Data presented are representative of at least two experiments.
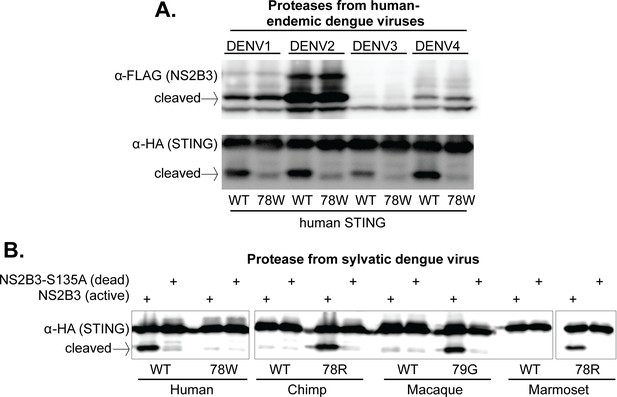
The 78/79 cleavage determinant in STING is targeted by proteases encoded by all endemic human dengue viruses, and by at least one sylvatic dengue virus.
(A) 293T cells were cotransfected with plasmids encoding NS2B3 from DENV1-4 along with human STING with or without a mutation at site 78. Western blotting was performed on lysate harvested 24 hr post transfection to detect NS2B3 (anti-FLAG) or STING (anti-HA). Data presented are representative of at least two experiments. (B) 293T cells were cotransfected with plasmids encoding the indicated STING and the NS2B3 from a sylvatic isolate of dengue virus (DakAr-141069). 24 hr post transfection, lysates where harvested, run on a gel, and western blotting was performed with an anti-HA antibody to detect STING. All data presented are representative of at least two experiments.
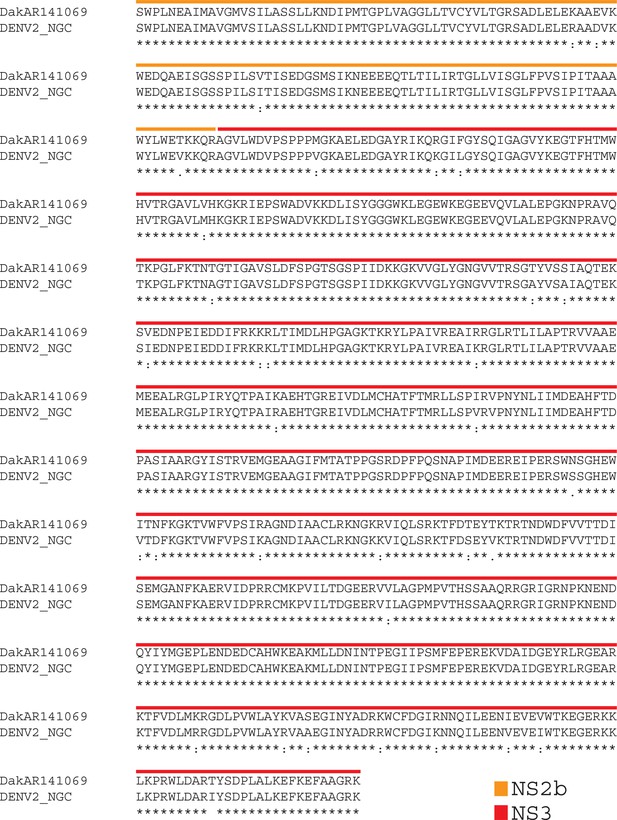
Alignment of the NS2B3 protease from sylvatic (top) versus human (bottom) dengue viruses.
https://doi.org/10.7554/eLife.31919.012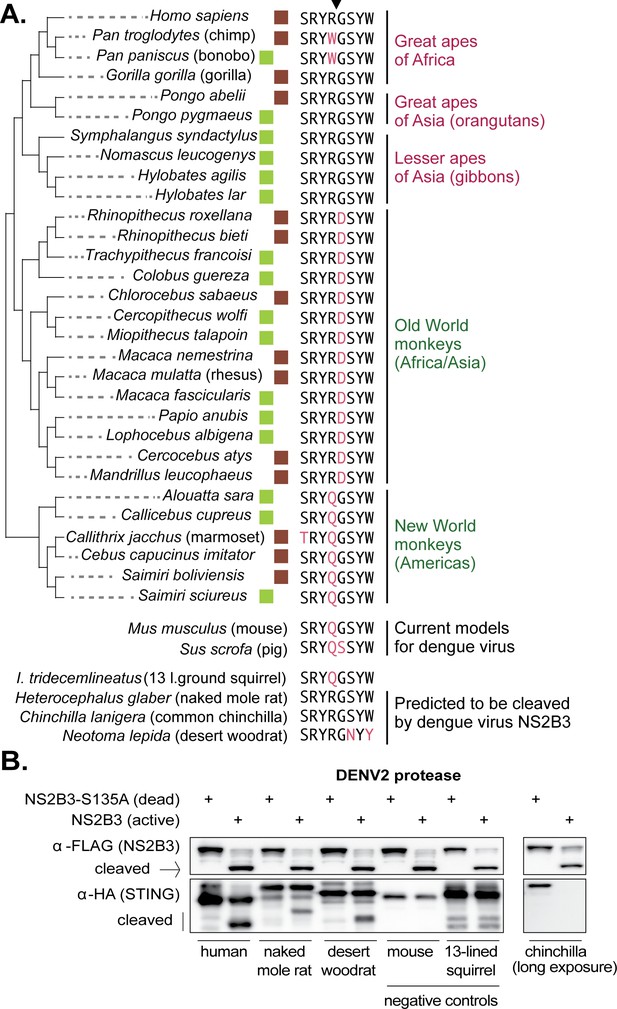
The dengue virus cleavage determinant in STING of various species.
(A) An alignment of the eight amino acid region in STING surrounding residues 78R/79G, the newly identified dengue virus cleavage determinant (downward arrow at top). Deviations from the human motif are highlighted. The green boxes indicate STING orthologs sequenced as part of this study. The brown boxes indicate STING sequences obtained from Genbank. Apes are shown at the top of the tree (pink type), monkeys at the bottom (green type). Depicted below are sequences from the same region of STING from two animal models for dengue virus (mouse, pig [Cassetti et al., 2010]), several small rodent species which encode the correct cleavage motif at 78/79 (naked mole rat, common chinchilla, desert woodrat), and one that does not (13 lined ground squirrel). Genbank accession numbers of sequences shown: mouse (XP_017173483), pig (XP_005661761), 13-lined ground squirrel (XM_005327275), naked mole rat (JAO02071), chinchilla (XP_005382124), and desert woodrat (OBS58238). (B) STING-HA genes were synthesized for the rodent species discussed in panel A. Cleavage assays were performed by co-transfecting plasmids encoding the dengue protease (dead or active) as well as each STING, and then performing immunoblotting as described in the methods. The data presented are representative of at least two independent experiments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (Homo sapiens) | STING; TMEM173 | NA | GENBANK:NM_198282 | GENBANK:MF622062 |
| gene (Homo sapiens) | STING; TMEM173 | this study | GENBANK:MF622062 | |
| gene (Pan troglodytes) | STING; TMEM173 | NA | GENBANK:XM_016953921 | |
| gene (Pan paniscus) | STING; TMEM173 | this study | GENBANK:MF616339 | |
| gene (Gorilla gorilla) | STING; TMEM173 | NA | GENBANK:XM_0040426 | |
| gene (Pongo abelii) | STING; TMEM173 | NA | GENBANK:XM_002815952 | |
| gene (Hylobates agilis) | STING; TMEM173 | this study | GENBANK:MF616342 | |
| gene (Symphalangus syndactylus) | STING; TMEM173 | this study | GENBANK:MF616343 | |
| gene (Nomascus leucogenys) | STING; TMEM173 | this study | GENBANK:MF616344 | |
| gene (Hylobates lar) | STING; TMEM173 | this study | GENBANK:MF616341 | |
| gene (Rhinopithecus roxellana) | STING; TMEM173 | NA | GENBANK:XM_010388119 | |
| gene (Rhinopithecus bieti) | STING; TMEM173 | NA | GENBANK:XM_017895026 | |
| gene (Trachypithecus francoisi) | STING; TMEM173 | this study | GENBANK:MF616352 | |
| gene (Colobus guereza) | STING; TMEM173 | this study | GENBANK:MF616351 | |
| gene (Chlorocebus sabaeus) | STING; TMEM173 | NA | GENBANK:XM_008014636 | |
| gene (Cercopithecus wolfi) | STING; TMEM173 | this study | GENBANK:MF616350 | |
| gene (Miopithecus talapoin) | STING; TMEM173 | this study | GENBANK:MF616349 | |
| gene (Macaca nemestrina) | STING; TMEM173 | NA | GENBANK:XM_011716377 | |
| gene (Macaca mulatta) | STING; TMEM173 | NA | GENBANK:XM_015141010 | |
| gene (Macaca mulatta) | STING; TMEM173 | this study | GENBANK:MF622060 | |
| gene (Macaca fascicularis) | STING; TMEM173 | this study | GENBANK:MF616346 | |
| gene (Papio papio) | STING; TMEM173 | this study | GENBANK:MF616348 | |
| gene (Lophocebus albigena) | STING; TMEM173 | this study | GENBANK:MF616347 | |
| gene (Cercocebus atys) | STING; TMEM173 | NA | GENBANK:XM_012090448 | |
| gene (Mandrillus leucophaeus) | STING; TMEM173 | NA | GENBANK:XM_011997224 | |
| gene (Aloutta sara) | STING; TMEM173 | this study | GENBANK:MF616355 | |
| gene (Callicebus cupreus) | STING; TMEM173 | this study | GENBANK:MF616354 | |
| gene (Callithrix jacchus) | STING; TMEM173 | NA | GENBANK:XM_00898588 | |
| gene (Callithrix jacchus) | STING; TMEM173 | this study | GENBANK:MF622061 | |
| gene (Cebus capucinus imitator) | STING; TMEM173 | NA | GENBANK:XM_017536735 | |
| gene (Samiri boliviensis) | STING; TMEM173 | NA | GENBANK:XM_003933913 | |
| gene (Saimiri sciureus) | STING; TMEM173 | this study | GENBANK:MF616353 | |
| gene (Mus musculus) | STING; TMEM173 | NA | GENBANK:NM_001289591 | |
| gene (Sus scrofa) | STING; TMEM173 | NA | GENBANK:XP_005661761 | |
| gene (Heterocephalus glaber) | NA | GENBANK:JAO02071 | ||
| gene (Chinchilla lanigera) | NA | GENBANK:XP_005382124 | ||
| gene (Neotoma lepida) | NA | GENBANK:OBS58238 | ||
| gene (Dengue viurs 2) | NS2B3 | NA | GENBANK:M29095 | |
| cell line (Homo sapiens) | 293T cells | ATCC | CRL-3216 | |
| cell line (Homo sapiens) | A549 cells | ATCC | CCL-185 | |
| antibody | Rat anti-HA-HRP (3F10) | Sigma | 11867423001 | |
| antibody | Mouse anti-Flag (M2) | Sigma | F3165 | |
| antibody | Rabbit anti-pIRF3 | abcam | ab76493 | |
| antibody | Rabbit anti-IRF3 | Santa Cruz Biotech | sc-9082 | |
| antibody | Rabbit anti-GAPDH | Cell Signaling | 14C10 | |
| antibody | Rabbit anti-STING | abcam | ab92605 | |
| antibody | Mouse anti-Actin (C4) | Santa Cruz Biotech | Sc47778 | |
| recombinant DNA reagent | DENV2 NS2B3 WT (plasmid) | PMID: 1642612 | Progenitors: DENV2 NGC (GENBANK:M29095), pCR3.1 | |
| recombinant DNA reagent | DENV2 NS2B3 S135A (plasmid) | PMID: 1642612 | Progenitors: DENV2 NS2B3 WT pCR3.1 plasmid, SDM | |
| recombinant DNA reagent | DENV1 (Hawaii) cDNA | this paper | Progenitors: World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) Catalog number NR-4287 | |
| recombinant DNA reagent | DENV2 (New Guinea C) cDNA | this paper | Progenitors: World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) Catalog number NR-4288 | |
| recombinant DNA reagent | DENV3 (Philippines/ H87/1956) cDNA | this paper | Progenitors: World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) Catalog number NR-2771 | |
| recombinant DNA reagent | DENV4 (H241) cDNA | this paper | Progenitors: World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) Catalog number NR-4289 | |
| recombinant DNA reagent | DENV1 (Hawaii) NS2B3 WT (plasmid) | this paper | Progenitors: DENV1 (Hawaii) cDNA, pCR3.1 | |
| recombinant DNA reagent | DENV2 (New Guinea C) NS2B3 WT (plasmid) | this paper | Progenitors: DENV2 (New Guinea C) cDNA, pCR3.1 | |
| recombinant DNA reagent | DENV3 (Philippines/ H87/1956) NS2B3 WT (plasmid) | this paper | Progenitors: DENV3 (Philippines/H87/1956) cDNA, pCR3.1 | |
| recombinant DNA reagent | DENV4 (H241) NS2B3 WT (plasmid) | this paper | Progenitors: DENV4 (H241) cDNA, pCR3.1 | |
| recombinant DNA reagent | Sylvatic (DakAr-141069) Dengue NS2B3 Protease (WT) | this paper | Progenitors: DakAr-141069 NS2B3 sequence (GenBank EF105389) | |
| recombinant DNA reagent | Sylvatic (DakAr-141069) Dengue NS2B3 Protease (S135A) | this paper | Progenitors: Sylvatic (DakAr-141069) Dengue NS2B3 Protease (WT) SDM product | |
| recombinant DNA reagent | human cDNA | this paper | Progenitors: A549 cell line (ATCC CCL-185) | |
| recombinant DNA reagent | chimpanzee cDNA | this paper | Progenitors: Coriell PR00748 | |
| recombinant DNA reagent | rhesus macaque cDNA | this paper | Progenitors: Mm265-95 | |
| recombinant DNA reagent | marmoset cDNA | this paper | Progenitors: Coriell PR07404 | |
| recombinant DNA reagent | mouse cDNA | this paper | Progenitors: RNA extracted from mouse liver | |
| recombinant DNA reagent | human STING-HA (plasmid) | this paper | Progenitors: human cDNA, pcDNA3.1 plasmid | |
| recombinant DNA reagent | human STING-HA (plasmid) | this paper | Progenitors: human cDNA, pLPCX plasmid | |
| recombinant DNA reagent | human STING(R78W)-HA (plasmid) | this paper | Progenitors: human STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | human STING(R78W)-HA (plasmid) | this paper | Progenitors: human STING-HA pLPCX SDM product | |
| recombinant DNA reagent | human STING(R79D)-HA (plasmid) | this paper | Progenitors: human STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | human STING(R78Q)-HA (plasmid) | this paper | Progenitors: human STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | chimpanzee STING-HA (plasmid) | this paper | Progenitors: chimpanzee cDNA, pcDNA3.1 plasmid | |
| recombinant DNA reagent | chimpanzee STING-HA (plasmid) | this paper | Progenitors: chimpanzee cDNA, pLPCX plasmid | |
| recombinant DNA reagent | chimpanzee STING(W78R)-HA (plasmid) | this paper | Progenitors: chimpanzee STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | chimpanzee STING(W78R)-HA (plasmid) | this paper | Progenitors: chimpanzee STING-HA pLPCX SDM product | |
| recombinant DNA reagent | rhesus macaque STING-HA (plasmid) | this paper | Progenitors: rhesus macaque cDNA, pcDNA3.1 plasmid | |
| recombinant DNA reagent | rhesus macaque STING(D79G)-HA (plasmid) | this paper | Progenitors: rhesus macaque STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | marmoset STING-HA (plasmid) | this paper | Progenitors: marmoset cDNA, pcDNA3.1 plasmid | |
| recombinant DNA reagent | marmoset STING(Q78R)-HA (plasmid) | this paper | Progenitors: marmoset STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | mouse STING-HA (plasmid) | this paper | Progenitors: mouse cDNA, pcDNA3.1 plasmid | |
| recombinant DNA reagent | mouse STING(Q78R)-HA (plasmid) | this paper | Progenitors: mouse STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | mouse STING(93LRRG96)-HA (plasmid) | this paper | Progenitors: mouse STING-HA pcDNA3.1 SDM product | |
| recombinant DNA reagent | human STING-3xFLAG (plasmid) | this paper | Progenitors: human cDNA, pLPCX plasmid | |
| recombinant DNA reagent | human STING(R78W)-3xFLAG (plasmid) | this paper | Progenitors: human STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | human STING(G230A)-3xFLAG (plasmid) | this paper | Progenitors: human STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | human STING (R78W, G230A)-3xFLAG (plasmid) | this paper | Progenitors: human STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | chimpanzee STING-3xFLAG (plasmid) | this paper | Progenitors: chimpanzee cDNA, pLPCX plasmid | |
| recombinant DNA reagent | chimpanzee STING (W78R)-3xFLAG (plasmid) | this paper | Progenitors: chimpanzee STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | chimpanzee STING(A230G)-3xFLAG (plasmid) | this paper | Progenitors: chimpanzee STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | chimpanzee STING(W78R, A230G)-3xFLAG (plasmid) | this paper | Progenitors: chimpanzee STING-3xFLAG pLPCX SDM product | |
| recombinant DNA reagent | IFN-ß1-luc (plasmid) | PMID: 21512573 | ||
| recombinant DNA reagent | pRL-CMV (plasmid) | Promega: AF025843 | Progenitors: pRL-null | |
| commercial assay or kit | Dual-Glo Luciferase Assay System | Promega | Cat#E2920 | |
| commercial assay or kit | Superscript III First-Strand Synthesis System | Thermo Scientific | Cat#18080051 | |
| software, algorithm | MEGA7 | http://www.megasoftware.net/ | ||
| software, algorithm | ImageJ version 1.43u | http://rsb.info.nih.gov/ij | ||
| software, algorithm | Python 2.7.11 | https://www.python.org | ||
| software, algorithm | Sequencher | https://www.genecodes.com |
Additional files
-
Supplementary file 1
Alignment of primate STING proteins.
- https://doi.org/10.7554/eLife.31919.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31919.015





