Daam2 driven degradation of VHL promotes gliomagenesis
Figures
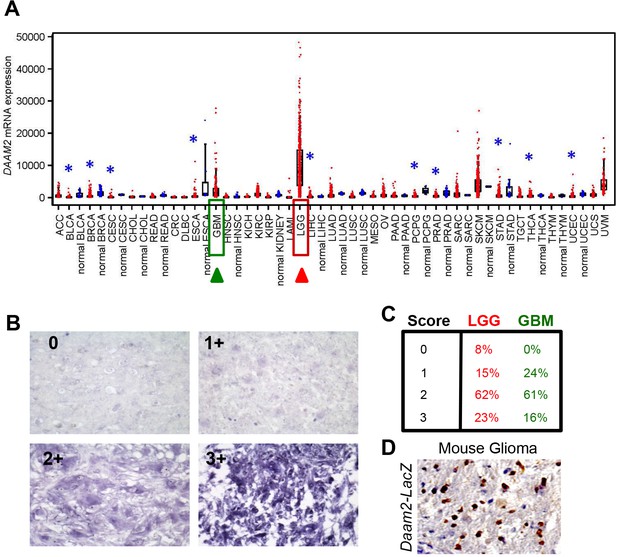
Daam2 is expressed in human glioma.
(A) Analysis of Daam2 RNA expression across a spectrum of cancers. Data was generated by TCGA Research Network and is publicly available (see methods). Box plots represent 5%, 25%, 50%, 75%, and 95%. Asterisk denotes tumor groups significantly higher (red) or lower (blue) as compared to corresponding non-tumor (‘normal’) group. P-values were generated by t-test on log-transformed data. (B) Examples of pathological grading of in situ hybridization analysis of Daam2 expression in human glioma tissue microarrays. Note that a score of 0 demonstrates no expression, while a score of 3 denotes very high expression. (C) Graded pathological scoring (0-low intensity, 3-high intensity) of Daam2 expression in the samples in B; values are the percentage of LGG and GBM tumors from the tissue microarray that were assigned respective score. (D) Immunohistochemistry analysis of Daam2-LacZ expression in mouse glioma generated in Daam2LacZ/+ mice.
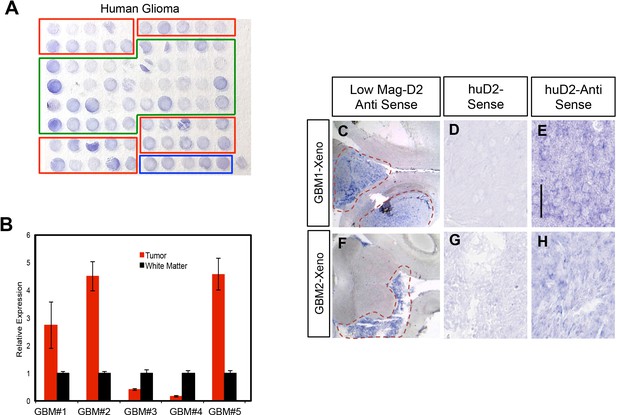
Daam2 is expressed in experimental models of human glioma.
(A) Tissue microarray showing in situ hybridization expression analysis of human DAAM2 across 35 LGG (red boxes) and 40 GBM (green boxes). Blue box denotes normal human brain samples. (B) qRT-PCR analysis of DAAM2 in primary human glioma samples and associated non-tumor, white matter. (C–H) Expression of DAAM2 in xenograft tumors generated from GBM1 and GBM2 cell lines. (D–E, G–H) high magnification images of in situ hybridization using antisense probes showing expression of Daam2 mRNA in these tumors. (C, F) Low magnification images of xenograft tumors from GBM1 and GBM2 cell lines showing widespread expression of Daam2 in these tumors. Scale bar in E is 50 nm and is applicable to D-E, G-H.
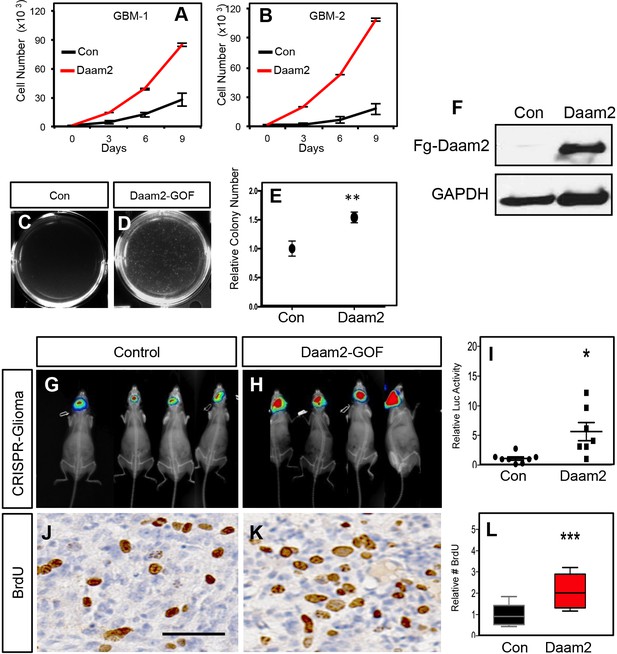
Daam2 accelerates glioma tumorigenesis.
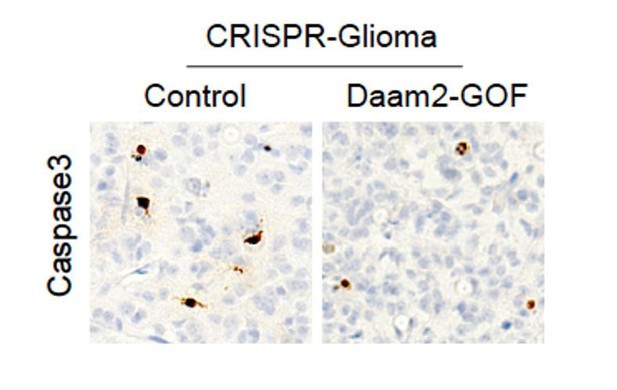
(A–B) Cell proliferation analysis of human GBM1 and GBM2 cell lines infected with lentivirus containing Daam2 or GFP control. (C–D) Soft agar assay with GBM1 cell line infected with lentivirus containing Daam2 or GFP control, images are representative. Each of these in vitro experiments was performed in triplicate and replicated three times; quantification is in E, **p=0.036. (F) Representative immuoblots showing ectopic expression of flag-tagged Daam2 in GBM-1 cell line. (G–I) Representative bioluminescence imaging of mice bearing CRISPR-IUE glioma with Daam2 overexpression or control, imaged at 7 weeks of age. Imaging quantification in I is derived from 7 Daam2-overexpression and eight control mice; *p=0.0079. (J–L) Immunohistochemistry analysis of BrdU expression from mice bearing CRISPR-IUE glioma from each experimental condition. Relative number of BrdU expressing cells is quantified in L and is derived from six total mice, four slides per mouse, from each experimental condition; ***p=0.0019. Scale bar in J is 50 nm. Error bars in E, I and L are ± SEM.
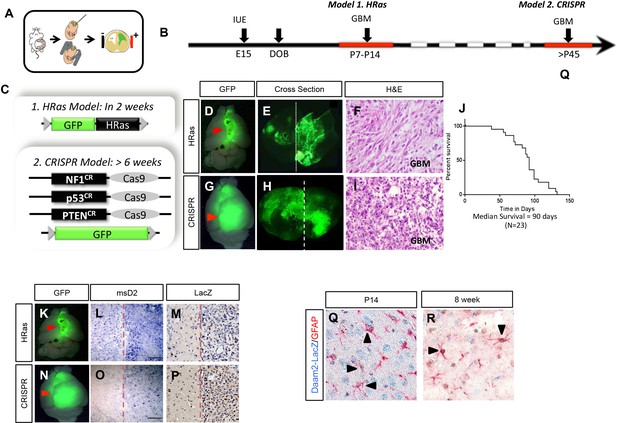
Mouse glioma model systems.
(A) Schematic illustrating the in utero electroporation manipulation used in these model systems to introduce oncogenes or Cas9/guideRNAs for CRISPR gene editing. (B) Timelines illustrating the embryonic electroporation manipulations and the onset of glioma in both the PB-Ras and CRISPR-IUE models. (C) Constructs used to generate glioma in both models. (D–F) Representative images from PB-Ras model at P14; D is whole brain image, E is cross section, and F is H&E staining for histopathological analysis. (G–I) Representative images from CRISPR-IUE model at P70; G is whole brain image, H is cross section, and I is H&E staining for histopathological analysis. (J) Kaplan-Meier Survival Curve for the CRISPR-IUE model. (K–P) Brain cross section demonstrating Daam2 expression in cortical hemispheres using ISH (L,O) and IHC (M,P) in our Ras (K–M) and CRISPR (N–P) mouse models of glioma. Dashed line shows the cortical midline, with the tumor bearing side of the cortex denoted by the red arrow in K and N. (Q–R) IHC showing Daam2-LacZ expression in GFAP-expressing astrocytes in cortex at P14 and adult mice (8 week). Arrows denote co-expression of GFAP and LacZ.
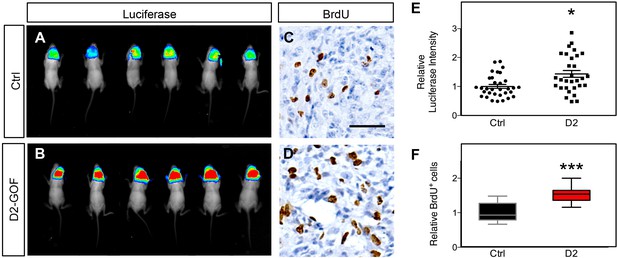
Daam2 accelerates glioma tumorigenesis in PB-Ras model.
(A–B) Representative bioluminescence imaging of mice bearing PB-Ras glioma with Daam2 overexpression or control, imaged at 10 days post-natal. Imaging quantification in E is derived from 17 Daam2-overexpression and 20 control mice; *p=0.0019. (C–D) Immunohistochemistry analysis of BrdU expression from mice bearing PB-Ras glioma from each experimental condition. Relative number of BrdU expressing cells is quantified in F and is derived from six total mice, four slides per mouse, from each experimental condition; ***p<0.0001. Scale bar in C is 50 nm. Error bars in E-F are ± SEM.
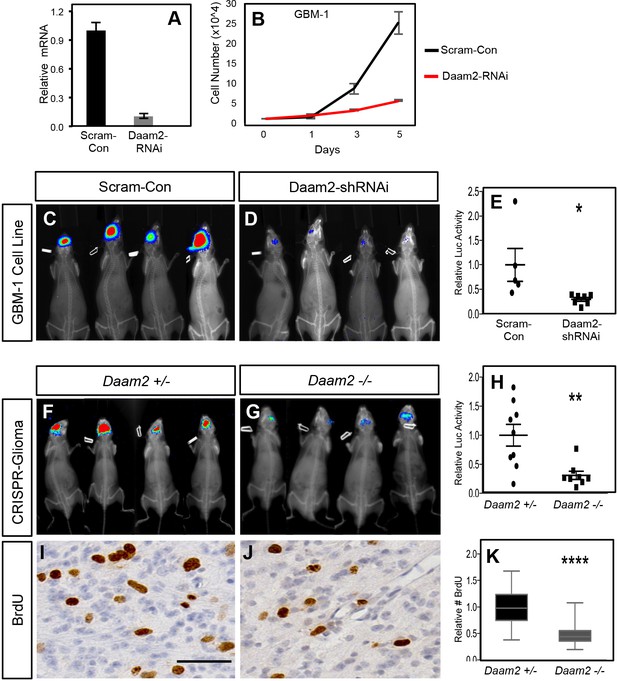
Loss of Daam2 suppresses glioma tumorigenesis.
(A) qRT-PCR demonstrating effective knockdown of human Daam2 mRNA expression in human GBM1 cell line infected with lentivirus containing Daam2-shRNAi or scrambled control. (B) Cell proliferation analysis of human GBM1 cell line infected with human Daam2-shRNAi or scrambled control lentivirus. (C–E) Representative bioluminescence imaging of mice transplanted with GBM1 cell lines transfected with Daam2-shRNAi or scrambled control, imaged 6 weeks post-transplantation. Imaging quantification is derived from seven mice transplanted with GBM1/Daam2-shRNAi and 5 GBM1/scrambled controls; *p=0.0325. (F–H) Representative bioluminescence imaging of mice bearing CRISPR-IUE glioma generated in Daam2 ± or Daam2-/- mice, imaged at 8 weeks of age. Imaging quantification is derived from 8 Daam2-/- and 9 Daam2 ± mice; **p=0.0049. (I–K) Immunohistochemistry analysis of BrdU expression from mice bearing CRISPR-IUE glioma from each experimental condition. Relative number of BrdU expressing cells is quantified in K and is derived from six total mice, four slides per mouse, from each experimental condition; ****p=0.00001. Scale bar in I is 50 nm. Error bars in E, H and K are ± SEM.
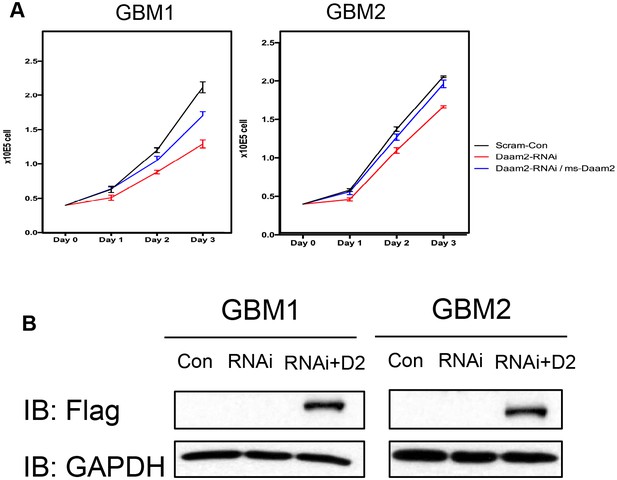
Rescue of Daam2-shRNAi growth defects in GBM cell lines.
(A) Rescue of growth defects manifest in Daam2-shRNAi knockdown cell lines via overexpression of mouse Daam2. In both cases overexpression of mouse Daam2 in the presence of the Daam2-shRNAi (blue line) rescued the growth defects present with Daam2-shRNAi alone (red line). (B) Immunoblot showing ectopic expression of flag-tagged Daam2 in GBM1/2 cell lines expressing the Daam2-shRNAi.
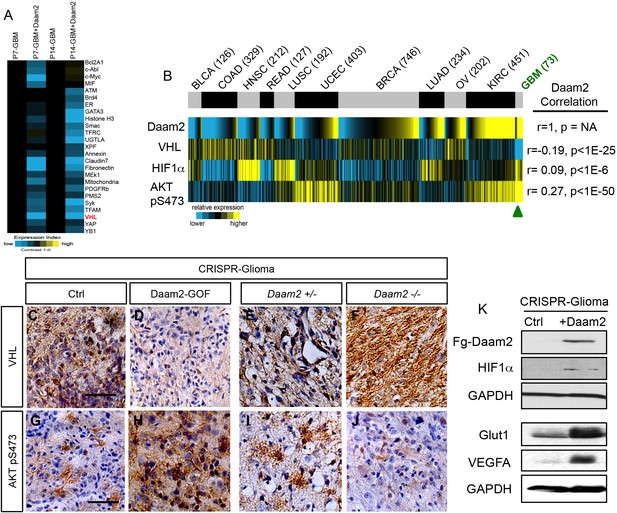
Daam2 suppresses VHL expression.
(A) Heatmap analysis of RPPA data showing the core cohort of proteins downregulated by overexpression of Daam2 in the PB-Ras mouse glioma model. (B) Correlation of Daam2 mRNA expression with VHL and Akt pS473 protein expression data and HIF1α mRNA expression. Protein and mRNA expression data from samples was obtained from The Cancer Proteome Atlas (TCPA) and correlations with Daam2 mRNA was performed using Pearson’s. Abbreviated cancer type is listed on top panel and number of tumors analyzed in the TCPA dataset for each cancer type is listed in parenthesis; GBM is denoted by green text. Cancer type abbreviations: BLCA: bladder urothelial carcinoma, COAD: Colon Adenocarcinoma, HNSC: head and neck squamous cell carcinoma, READ: rectal carcinoma, LUSC: lung squamous cell carcinoma, UCEC: uterine corpus endometrial carcinoma, BRCA: breast adenocarcinoma, LUAD: lung adenocarcinoma, OV: ovarian serous carcinoma, KIRC: kidney renal clear cell carcinoma, GBM: glioblastoma multiforme. (C–F) Representative immunohistochemical analysis of VHL expression in CRISPR-IUE glioma tumors derived from Daam2-overexpression (D) or knockout of Daam2 (F) and associated controls. (G–J) Representative immunohistochemical analysis of Akt pS473 expression in CRISPR-IUE glioma tumors derived from Daam2-overexpression (H) or knockout of Daam2 (J) and associated controls. Images are representative of analysis performed on six independent tumors for each experimental condition. (K) Immunoblot with antibodies against HIF1α, Glut1, and VEGFA from protein lysates derived from CRISPR-IUE glioma overexpressing Daam2 or control. Scale bars in C and G are 50 nm.
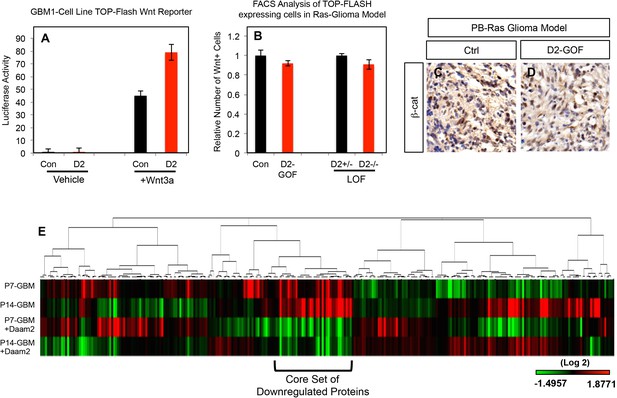
Daam2 effect on Wnt signaling in glioma and RPPA analysis.
(A) Luciferase reporter assay using Wnt reporter (TOP-Flash) in GBM1 cell lines. Cell line was transfected with Daam2, GFP control, with and without Wnt3a, and the TOP-FLASH reporter to measure Wnt activity. Note that Daam2 alone does not activate Wnt reporter; activation by Daam2 requires presence of canonical Wnt ligand, Wnt3a. Activation by Daam2 in the context of Wnt3a is very mild ~ 1.8 fold. Each experiment was performed in triplicate, with three replicates for each group. (B) FACS analysis of the relative number of TOP-FLASH-BFP expressing cells in our PB-Ras model. We performed IUE with the PB-Ras model and included Daam2 overexpression (GOF) along with the TOP-FLASH-BFP construct. In addition, we generated PB-Ras glioma in our Daam2 ± and Daam2-/- mice (LOF) that were also electroporated with the TOP-FLASH-BFP reporter. At post-natal day 12, tumors were harvested, dissociated and subjected to FACS analysis, where we assessed the relative number of BFP+/GFP + cells within the tumor as an index of Wnt activity across the respective experimental conditions in both the GOF and LOF studies. (C–D) Representative immunohistochemistry staining of PB-Ras, Daam2-GOF tumors with β-catenin antibodies. This analysis was performed on six different tumors for each condition, four slides per tumor. (E) Heatmap representation of median centered unsupervised hierarchical clustering of RPPA protein expression analysis from P7 and P14 PB-Ras glioma subjected to Daam2-GOF. Please see Supplementary file 1 for the raw data and the methods section for additional details. Note that the bracket denotes a core set of proteins that are downregulated in the presence of Daam2. These proteins are featured in Figure 4a.
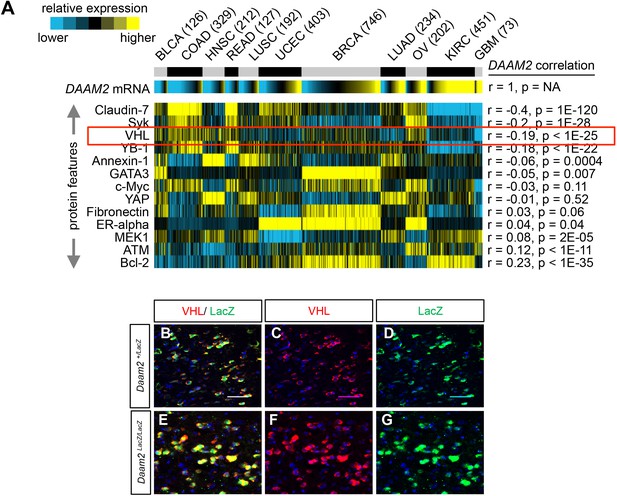
Correlation between Daam2 and proteins suppressed in glioma.
Correlation of Daam2 mRNA expression with the core set of proteins downregulated in the PB-Ras Daam2-GOF RPPA analysis. Please also see Figure 4a and Figure 4—figure supplement 1, panel E. Across a broad spectrum of cancers. Protein and mRNA expression data across this broad spectrum of cancer was derived from samples obtained from The Cancer Proteome Atlas (TCPA) and correlations with Daam2 mRNA was performed using Pearson’s. Abbreviated cancer type is listed on top panel and number of tumors analyzed in the TCPA dataset for each cancer type is listed in parenthesis. VHL is highlighted in red box. (B–G) Immunostaining showing Daam2-LacZ and VHL expression in the adult cortex from Daam2LacZ/+ and Daam2LacZ/LacZ mice. Note that the levels of VHL that are co-expressed with Daam2-LacZ are increased in the Daam2LacZ/LacZ mice compared to Daam2LacZ/+ mice (C v. (F).
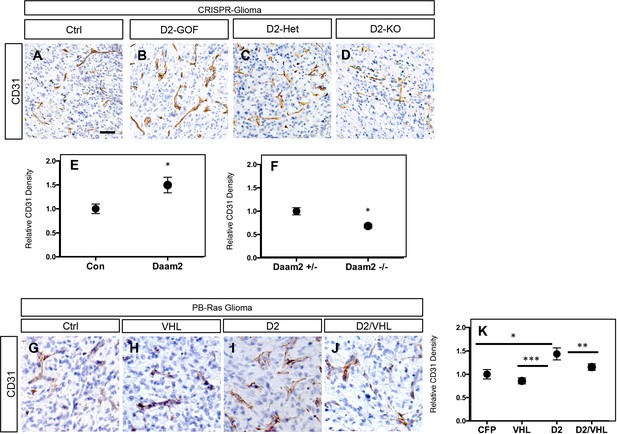
Manipulation of Daam2 expression influences angiogenesis.
Representative immunohistochemistry staining of the vascular marker, CD31, in our glioma model, across a series of experimental manipulations. (A–B) CRISPR-IUE glioma in the context of Daam2 GOF and associated control, (C–D) CRISPR-IUE glioma generated in Daam2 ± and Daam2-/- mice; quantification in E-F. In E, *p=0.033. In F, p=0.0026. (G–J) PB-Ras glioma in the context of VHL-GOF, Daam2-GOF, Daam2 + VHL, and control. This analysis was performed on six different tumors for each condition, four slides per tumor. Quantification is in K: *p=0.034, **p=0.044, and ***p=0.023. In E, F, and K error bars are ± SEM.
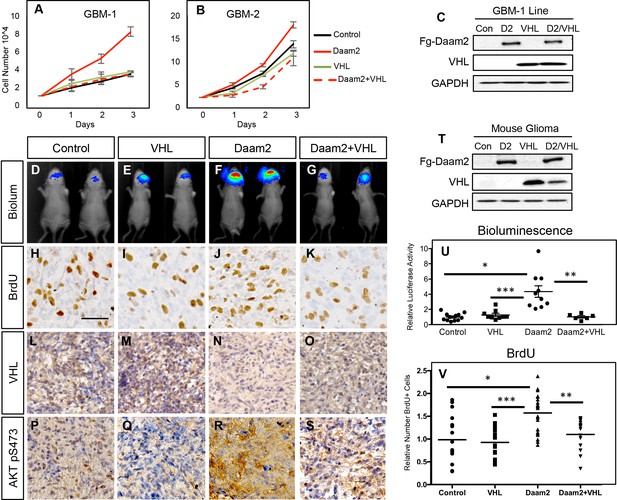
Daam2 promotes gliogenesis via suppression of VHL.
(A–B) Cell proliferation analysis of human GBM1 and GBM2 cell lines infected with lentivirus containing GFP control, Daam2, VHL, or Daam2 +VHL. Associated immunoblots are in C. (D–G, U) Representative bioluminescence imaging of mice bearing PB-Ras glioma with Daam2 overexpression, VHL overexpression, Daam2 +VHL overexpression, or control, imaged at 10 days post-natal. Imaging quantification in U is derived from 10 Daam2-overexpression, 9 VHL overexpression, 6 Daam2 +VHL, and 12 control mice; *p=0.0001, **p=0.0046, ***p=0.0016 (H–K, V) Immunohistochemistry analysis of BrdU expression from mice bearing PB-Ras glioma from each experimental condition. Relative number of BrdU expressing cells is quantified in V and is derived from six total mice, four slides per mouse, from each experimental condition; *p=0.0006, **p=0.0009, ***p<0.0001. (L–O) Representative immunohistochemical analysis of VHL expression in PB-Ras glioma from described experimental conditions and associated controls. (P–S) Representative immunohistochemical analysis of Akt pS473 expression in PB-Ras glioma tumors derived from the described experimental conditions and associated controls. (T) Representative immunoblot showing overexpression of Daam2 and VHL in the various experimental conditions. Images in L-O are representative of analysis performed on six independent tumors for each experimental condition. Statistics derived by one-way analysis of variance (ANOVA) and followed by Tukey’s test for between-group comparisons. Scale bar in F is 50 nm. Error bars in U and V are ± SEM.
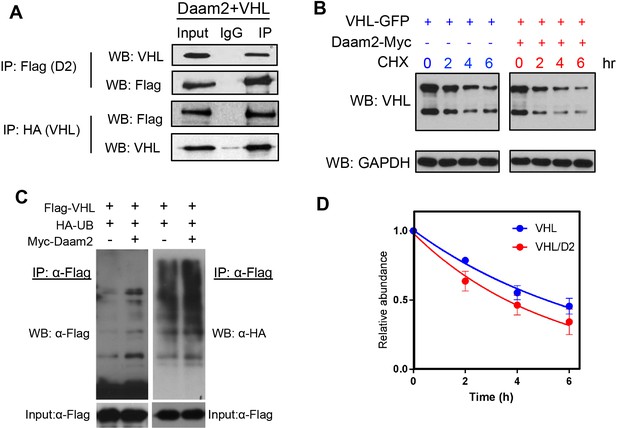
Daam2 mediates VHL degradation and ubiquitination.
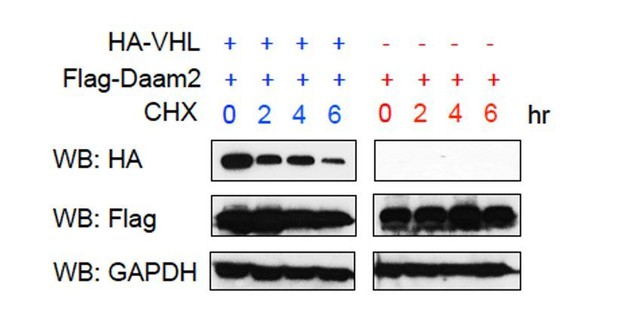
(A) Immunoprecipitation of Flag-Daam2, followed by immunoblot with VHL on lysates derived from PB-Ras glioma overexpressing Flag-Daam2. Detection of endogenous VHL in the ‘IP’ lane, indicates Daam2 associates with VHL in mouse glioma. The reverse IP was also performed using HA-VHL to pulldown Flag-Daam2. (B,D) 293 cells were transfected with VHL and Daam2 (or control). 24 hr after transfection, cells were treated with 40 μg/ml cycloheximide (proteasome inhibitor) and incubated for the indicated time before harvest. Western blotting was performed to monitor VHL levels using anti-VHL antibody. Quantification in D shows the effect of Daam2 on VHL stability. Half-life of VHL (5.082 hr) was significantly decreased in the presence of Daam2 (3.679 hr). Error bars represent mean ± SEM (n = 3). Half-life was determined via nonlinear, one-phase exponential decay analysis (half-life parameter, K, is significantly different in two conditions: p=0.0081). Error bars represent mean ± SEM (n = 3). (C) Daam2 enhances VHL ubiquitination. 293 T cells were co-transfected with Flag-VHL, HA-UB(ubiquitin), and Myc-Daam2. Cells were treated with MG132 (10 ug/ml) 6 hr before harvest. Whole-cell lysate were immunoprecipitated with anti-Flag M2 beads then analyzed by western blotting with Flag and HA antibodies.
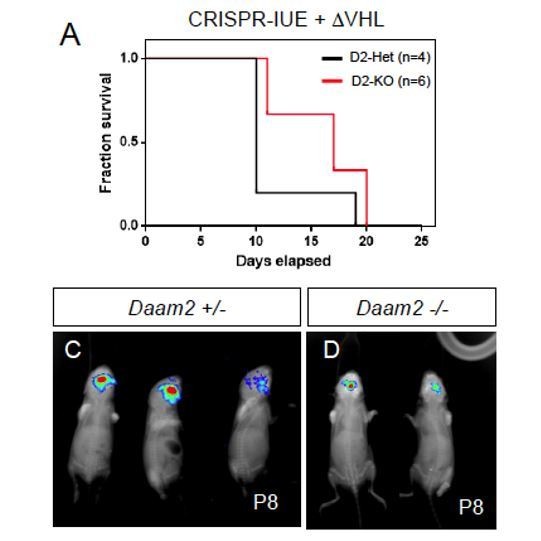
CRISPR-IUE tumor model, combined with CRISPR deletion of VHL, generated in a Daam2+/- or Daam2-/- background.
Deletion of VHL accelerates glioma formation in this model. (A) Kaplan-Meyer survival curve. (C-D) Bioluminescence imaging of tumors at P8.
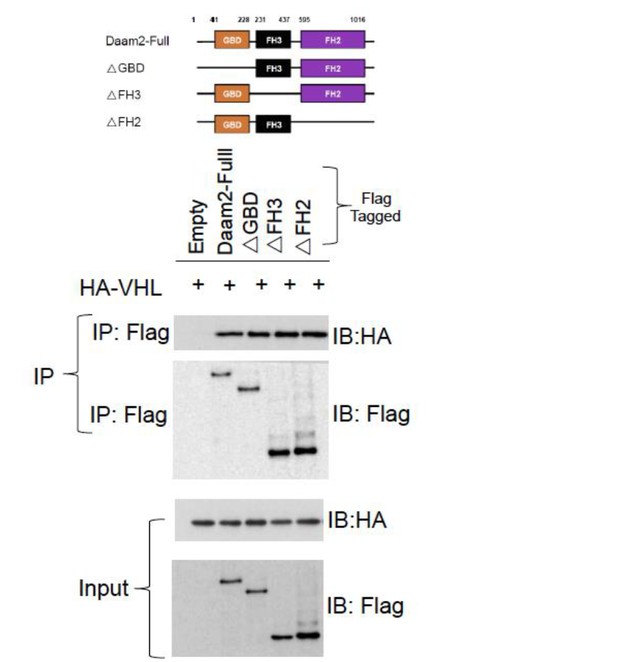
Co-IP of Daam2 truncation mutants (see Lee, et al. 2012) and VHL.
These data show that each of the Daam2 truncation mutants is capable of associating with Daam2.
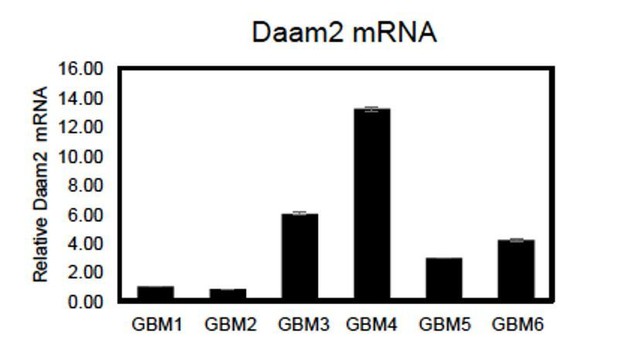
qRT-PCR showing relative expression of Daam2 across 6 primary GBM cell lines.
Additional files
-
Supplementary file 1
RPPA data analysis.
- https://doi.org/10.7554/eLife.31926.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31926.017