A kinase-dependent feedforward loop affects CREBB stability and long term memory formation
Figures
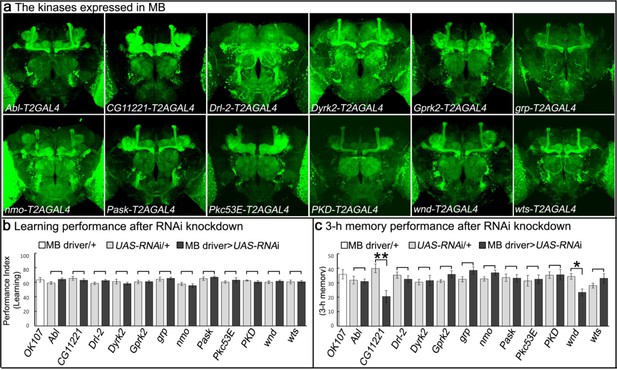
Genes encoding kinases expressed in MB of adult brains and behavioral consequences of RNA interference.
(a) Expression patterns of genes encoding protein kinases in adult brains. The 12 protein kinases shown here are expressed in MB. (b–c) Screening results of behavioral assays by RNAi knockdown. The MB-GAL4 driver, OK107 was used to drive the UAS-RNAi. Flies were raised at 18°C until eclosion, transferred to 25°C for 3 days and behavioral assays were performed to test (b) learning and (c) 3 hr memory. The mean ±SEM is plotted for each genotype; n = 8 for each group. *p<0.05, **p<0.01.
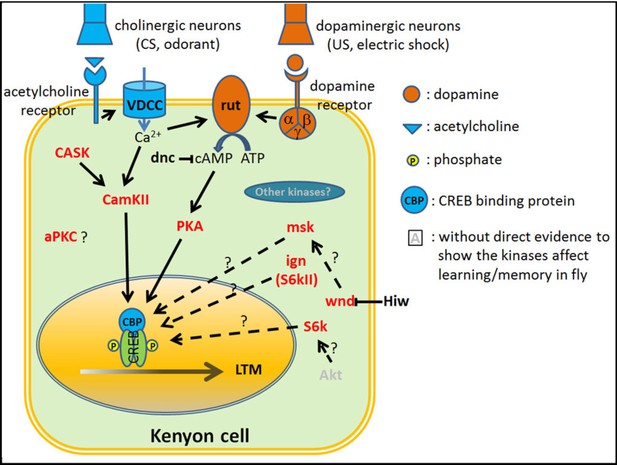
Overview of the proteins required for olfactory aversive learning/memory formation in Drosophila.
In adult flies, the mushroom body (MB) Kenyon cells are responsible for the detection of the conditioned stimulus (CS) and unconditioned stimulus (US), association of CS and US, and memory storage (Dubnau and Tully, 1998). The CS (odorant) signal is delivered by cholinergic projection neurons that cause a Ca2+ influx into Kenyon cells. The US (electric shock) signal is delivered by dopaminergic neurons that activate downstream G protein coupled receptors, triggering Gα (Busto et al., 2010). The adenylyl cyclase rutabaga (rut) can be activated by both Ca2+ and Gα and causes an increase in cAMP (Busto et al., 2010) which activates PKA and CREB through PKA mediated phosphorylation of CREB (Impey et al., 1999). dunce (dnc), a phosphodiesterase regulates cAMP levels (Byers et al., 1981). CREB is a key transcription factor for genes required for long-term memory formation (DeZazzo and Tully, 1995). In addition to the cAMP-PKA-CREB pathway, there are other kinases that have been implicated in memory formation. These include CamKII (Akalal et al., 2010), CASK (Gillespie and Hodge, 2013), msk (MAPK pathway) (Li et al., 2016; Philip et al., 2001; Skoulakis and Davis, 1996), ignorant (ign, S6kII) (Putz et al., 2004), wallenda (wnd) (Huang et al., 2012) and S6K (Fropf et al., 2013). aPKC has been shown to enhance 24 hr memory in Drosophila. However, it enhances 24 hr ARM, but not 24 hr LTM (Drier et al., 2002). Red: protein kinases that have been shown to be involved in learning/memory. Solid line: protein kinases that have been shown to phosphorylate CREBB directly (Horiuchi et al., 2004). Dashed line: There is no evidence to show these genes interact directly.
The T2A-GAL4 expression patterns of protein kinases that are not expressed in MB.
Expression patterns of protein kinases in adult brains. The T2AGAL4 lines were crossed with UAS-mCD8::GFP (green: GFP).
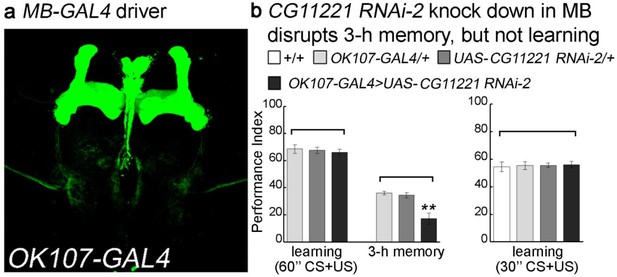
Reducing the levels of CG11221 in MB affects 3 hr memory formation.
(a) MB specific GAL4 driver, OK107-GAL4 was used for RNAi knockdown. The GAL4 driver was crossed with UAS-mCD8::GFP (green: GFP). (b) The second CG11221 RNAi line from VDRC is used to reduce the expression level in MB driven by OK107. Upon knockdown of MP, flies show a normal learning score, but 3 hr memory is impaired. Reducing the number of training sessions from 12x to 6x electric shocks (from 60’ to 30’) reveals that learning is still not affected in the absence of MP. Mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01.

Comparison of protein sequence between human SBK1 and fly MP (CG11221).
The kinase domain of SBK1 is underlined.
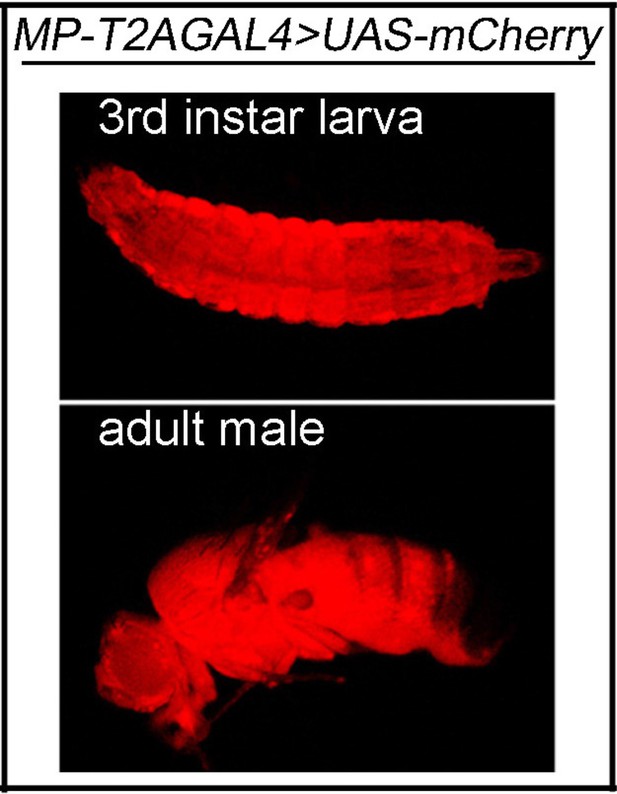
MP is expressed ubiquitously.
The MP expression in third instar larva and adult fly. UAS-mCherry is driven by MP-T2AGAL4 (mCherry: red).
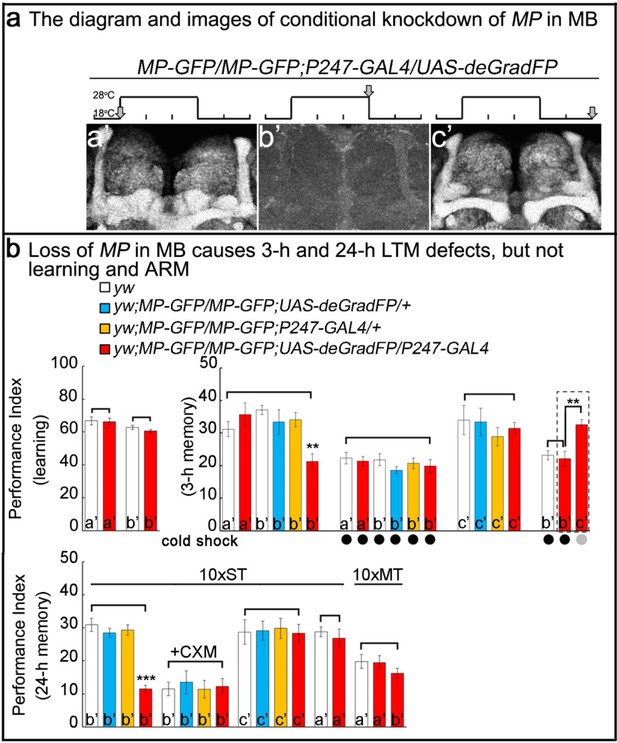
Loss of MP causes a loss of memory.
(a) UAS-deGradFP driven by MB driver, P247-GAL4. a'-c’: scheme for temporal control of deGradFP expression via temperature shift. (a’) 18°C; (b’) 28°C for three days; (c’) 28°C for three days, followed by a shift to 18°C for two days. MP-GFP-MP expression at the time points shown above by arrows. (b) Performance scores of learning, 3 hr memory and 24 hr memory. (a’), (b’) and (c’) are referred as the time points in (a). (b), learning) Learning is normal after knockdown of MP in MB by deGradFP at 28°C for 3 days (b’). Flies raised at 18°C are used as a control (a’). (b, 3 hr memory) 3 hr memory is impaired after knockdown of MP in MB (b’). However, the performance score of ARM is intact in MP knockdown flies which are treated with a cold shock and compared to flies raised at 18°C (3 hr memory a’ and b’+cold shock). In c’ condition, the flies show normal performance score of 3 hr memory. The 3 hr memory impairment (in b’+cold shock, right panel) can be fully rescued when the animals are shifted to 18°C for two days (the groups boxed in dashed line are the exactly same flies). (c), 24 hr memory) 24 hr LTM (10 x ST) is impaired upon knockdown of MP in MB (b’, red). After treatment with 35 mM cycloheximide (CXM), the MP knockdown flies (b’+CXM, red) don’t exhibit a performance that is worse than control flies. In the c’ conditions, the flies exhibit a normal performance score for the 24 hr memory assay. The performance of ARM (10 x MT) is intact. 10 x ST:10 times spaced training. 10 x MT:10 times massed training. The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01. ***p<0.001.
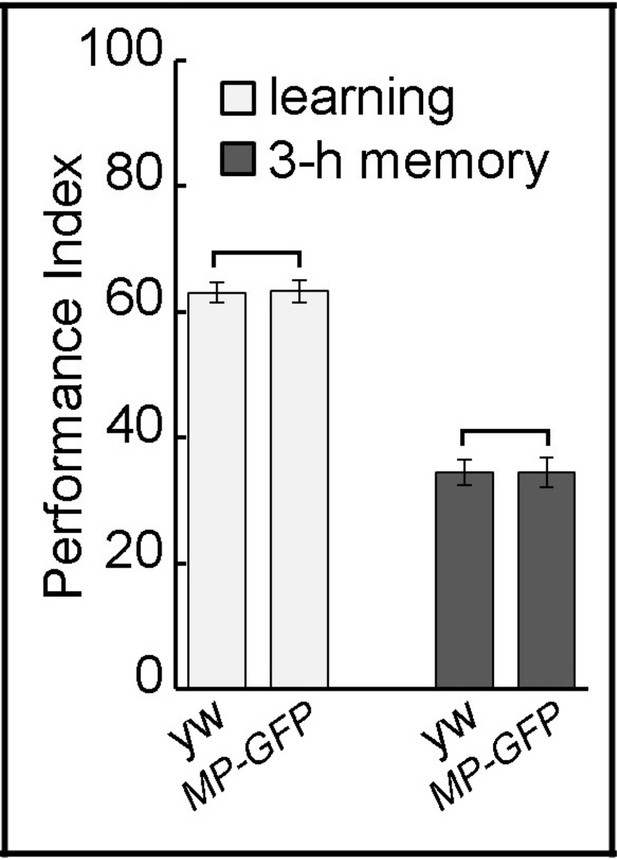
MP-GFP-MP animals have proper learning and memory.
MP-GFP-MP transgenic animals show normal learning and 3 hr memory showing that the GFP-tag insertion does not affect protein function. The mean ±SEM is plotted for each genotype; n = 8 for each group.
P247-GAL4 was used for protein knockdown.
The GAL4 driver was crossed with UAS-mCD8::GFP (green: GFP).
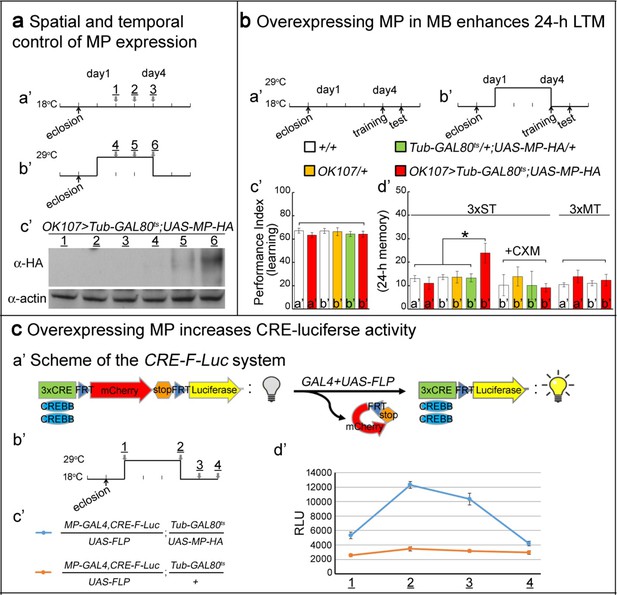
Overexpressing MP enhances 24 hr LTM and increases CREBB activity.
(a) Spatial and temporal control of MP expression was achieved with a MB-GAL4 driver and Tub-GAL80ts. Schemes are shown in (a’) and (b’). MP expression was assessed by Western blot with α-HA antibody in (c’). (b) Temporal overexpression of MP in MB enhances 24 hr LTM. Diagrams for MP overexpression, training and testing of behavior are shown in (a’) and (b’). The learning scores are normal after MP overexpression in (c’) (red column, b’). 24 hr LTM is significantly enhanced after 3x ST training in (d’) (red column, (b’). However, memory enhancement can be erased by feeding flies with cycloheximide. The 24 hr ARM is unaltered after MP overexpression in (d’) (red column, (b’), a’=no overexpression, b’=overexpression, and 3x ST (spaced training) or 3x MT (massed training). The mean ±SEM is plotted for each genotype; n = 8–12 for each group with 200 animals per group. *p<0.05. (c) Overexpressing MP increases CRE-luciferase activity. The CRE-F-Luc system is shown in (a’). The diagram is shown in (b’). The fly genotypes are shown in (c’). blue curve: MP-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/UAS-MP-HA. orange curve: MP-GAL4,CRE-F-Luc/UAS-FLP;Tub-GAL80ts/+. In (d’), the luciferase activity of the two genotypes of flies at the following time points: 1, 2, 3 and 4 which are shown in (b’). (1) at 18°C for one day after eclosion. (2) at 29°C for three days. (3) at 29°C for three days, then shift to 18°C for one day. (4) at 29°C for three days then shift to 18°C for two days. 10 fly heads (five males/five females) were collected for a single assay. RLU: relative luminescence unit. The mean ±SEM is plotted for each point; n = 6.
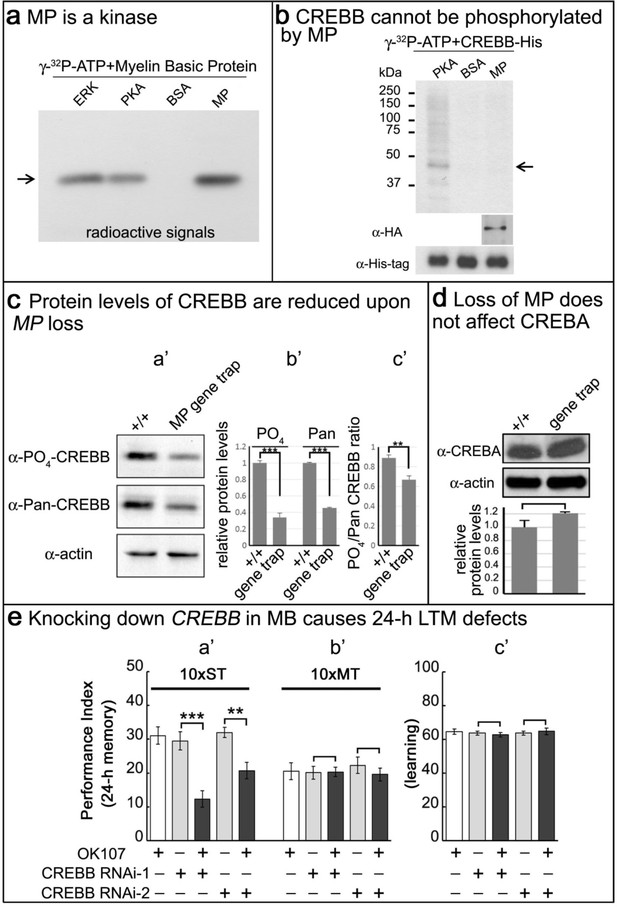
MP is a kinase, and loss of MP affects CREBB protein levels in MP gene trap animals.
(a) MP can phosphorylate myelin basic protein similar to ERK and PKA. Arrow points to MBP. (b) CREBB is not phosphorylated by MP kinase. As a positive control, we used PKA to phosphorylate the CREBB-His tag protein. MP failed to phosphorylate CREBB under these conditions. BSA was used as a negative control. Arrow points to CREBB. (c) In (a’–b’), protein levels of phospho-CREBB and total CREBB in the absence of MP are decreased, and also the phosphorylation ratio of phospho-CREBB/total CREBB in (c’). Total CREBB was detected by anti-Pan-CREBB, phosphorylated CREBB was detected by anti-PO4-CREBB (**p<0.01. ***p<0.001). (d) Protein levels of CREBA are not affected in MP gene-trap flies. CREBA protein was detected by anti-CREBA. The protein levels were normalized to actin. (Student’s t test, n = 3). (e) Knocking down CREBB in MB with CREBB RNAi causes 24 hr LTM defects (10x ST, a’), but not ARM (10x MT, b’) and learning in (c’). 10x ST = 10 times spaced training. 10x MT = 10 times massed training. The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01. ***p<0.001.
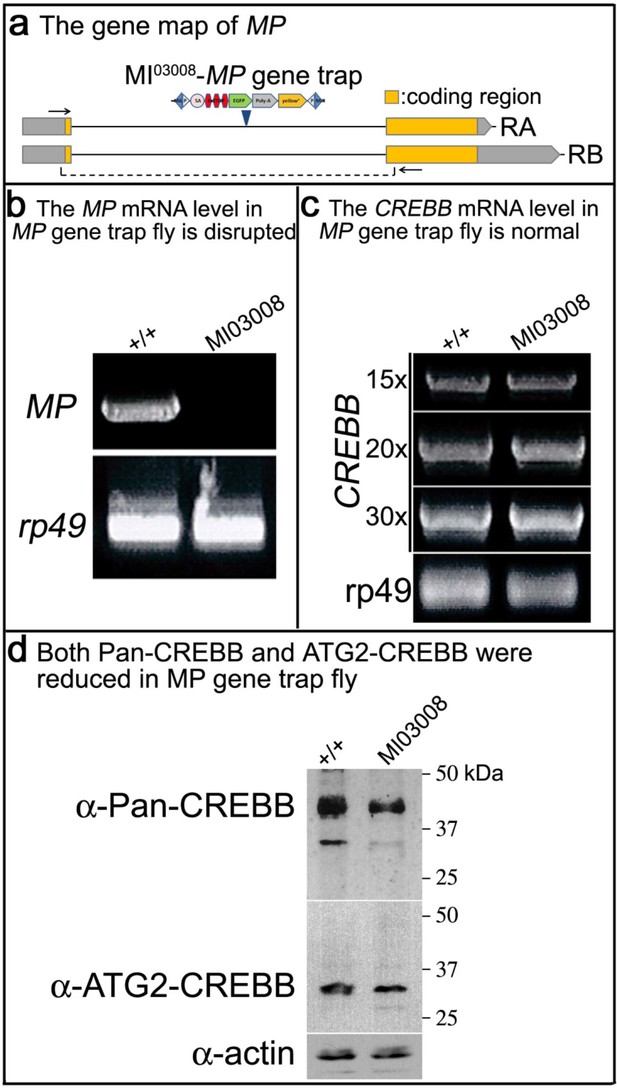
CREBB mRNA level is not affected in MP gene trap fly, but CREBB proteins are reduced.
(a) MI03008 is a gene-trap and produces no transcript. The MiMIC construct is described in Venken et al., 2011. This MiMIC insertion disrupts both RA and RB isoforms of MP. RT-PCR was performed using specific primers for (b) MP and (c) CREBB. For MP, the primer set (arrows) is shown in (a). For CREBB, the number of PCR cycles is shown in (c). rp49 was used as an internal control. (d) Protein levels of Pan- and ATG2-CREBBs are reduced in MP mutant flies. Pan- and ATG2-CREBB protein levels were assessed with anti-Pan- and anti-ATG2-CREBB antibodies. Actin was used as an internal control.
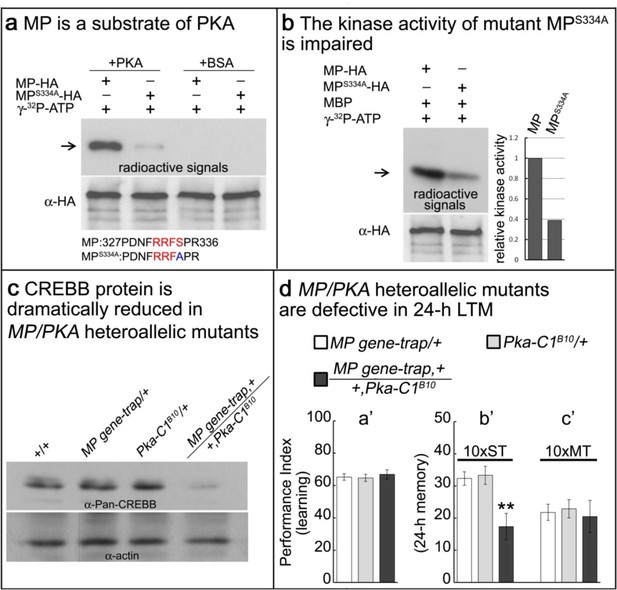
MP kinase is regulated by PKA.
(a) The potential PKA phosphorylation site of MP was altered to S334A. MP can be phosphorylated by PKA, but MPS334A is severely impaired. Arrow points to wild-type and mutant MP. (b) MPS334A has severely reduced kinase activity compared to wild-type MP. Arrow points to MBP. (c) CREBB protein levels are dramatically reduced in MP gene-trap +/+Pka-C1B10 heteroallelic flies. CREBB protein levels are probed with anti-Pan-CREBB antibody. (d) MP gene-trap +/+Pka-C1B10 heteroallelic flies have intact learning in (a’) and ARM in (c’), but show 24 hr LTM impairment in (b’). The mean ±SEM is plotted for each genotype; n = 8 for each group. **p<0.01.
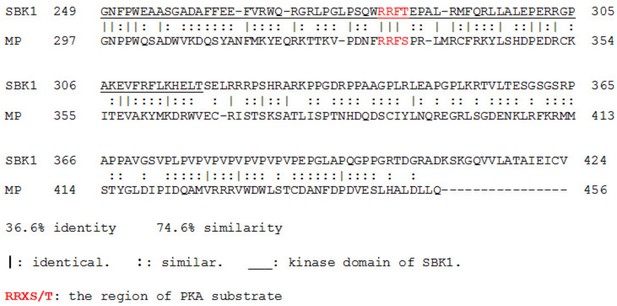
Human SBK1 and fly MP (CG11221) both contain a PKA phosphorylation site.
The kinase domain of SBK1 is underlined. Both proteins contain a PKA phosphorylation site marked in red.
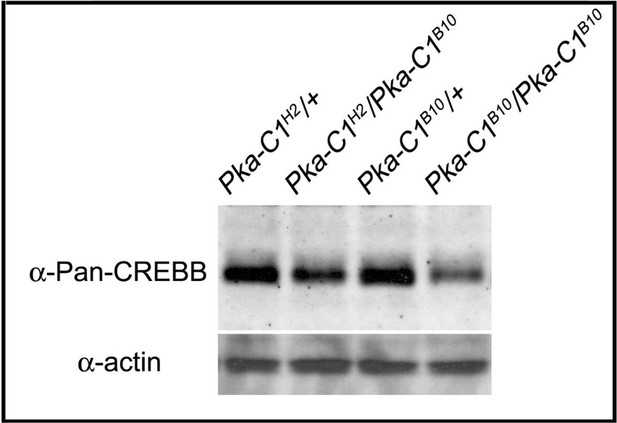
Protein levels of CREBB are reduced in Pka-C1 mutants.
Total CREBB protein levels of adult heads are reduced in Pka-C1H2/Pka-C1B10 heteroallelic flies and Pka-C1B10/Pka-C1B10 mutant flies. Total CREBB protein levels are probed with anti-Pan-CREBB antibody. Actin was used as an internal control.
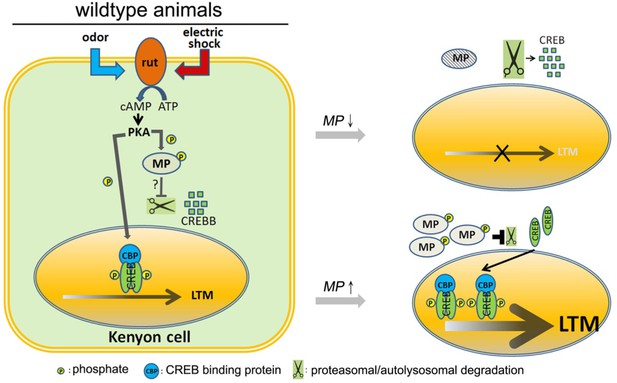
Model.
In wild-type animals, rutabaga (rut) as a coincidence detector can receive the odor and electric shock signals then activate cAMP-PKA signaling. MP is phosphorylated by PKA which maintains the CREBB levels, possibly by inhibiting proteasomal or autolysosomal degradation. It permits CREB-dependent LTM to form. Knocking down MP will unlock this inhibition and facilitate CREBB degradation thereby disrupting LTM. In contrast, overexpression of MP promotes LTM formation.
Additional files
-
Supplementary file 1
Fly stocks and antibodies information.
- https://doi.org/10.7554/eLife.33007.018
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33007.019





