Ripply2 recruits proteasome complex for Tbx6 degradation to define segment border during murine somitogenesis
Figures
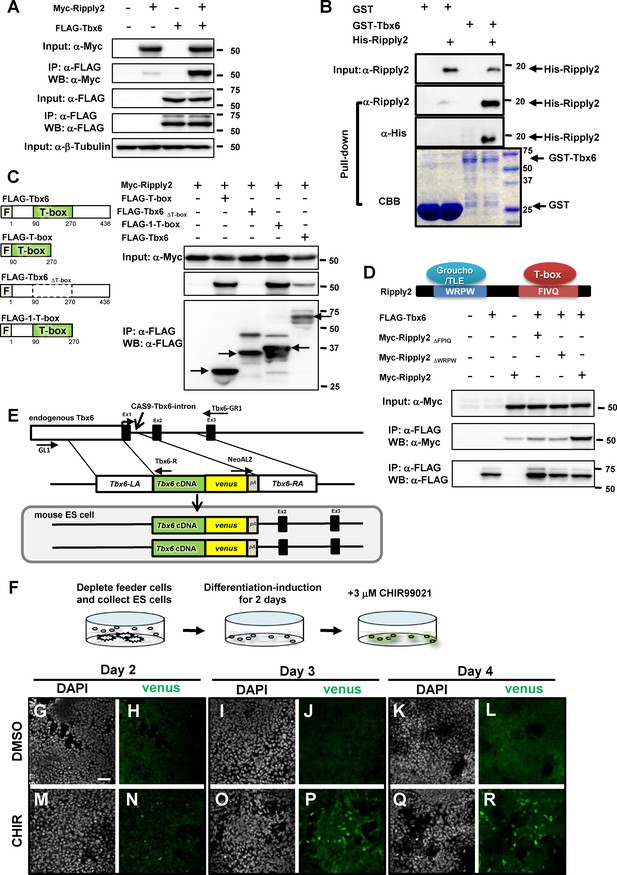
Establishment of the PSM-fated ES cell system.
(A) Immunoprecipitation (IP) for examining Tbx6-Ripply2 interaction. DNA constructs for FLAG-Tbx6 was co-transfected with Myc-Ripply2 into HEK293T cells. IP was conducted using anti-FLAG beads, followed by western blotting for detecting FLAG-Tbx6 and Myc-Ripply2 by anti-FLAG and anti-Myc antibodies (N = 10). (B) GST pull-down assay to examine the direct interaction between Tbx6 and Ripply2. Purified His-Ripply2 protein was mixed with cell lysate of GST-Tbx6. Anti-Ripply2 antibody and anti-His antibody was used to detect His-Ripply2 signal (N = 2). (C) IP-western analyses to determine Tbx6 domain for Ripply2 interaction. DNA constructs for FLAG-Tbx6, FLAG-T-box, FLAG-Tbx6ΔT-box or FLAG-1-T-box, were co-transfected with Myc-Ripply2 into HEK293T cells. IP was conducted using anti-FLAG beads, followed by western blotting for detecting FLAG-Tbx6 and Myc-Ripply2 by anti-FLAG and anti-Myc antibodies (N = 6). Arrows indicate protein bands showing expected molecular size. (D) IP for Tbx6 and mutant Ripply2. FLAG-Tbx6 was co-transfected with Myc-Ripply2ΔFPIQ, Myc-Ripply2ΔWRPW, or Myc-Ripply2 (wild-type) into the HEK293T cultured cells. Cell lysates were incubated with anti-FLAG beads. Western blotting was conducted using anti-FLAG and anti-Myc antibodies (N = 6). (E) Strategy for establishing the Tbx6-venus knock-in (KI) ES cell line. Tbx6 cDNA connected with the venus sequence replaced exon-1 via Cas9-aided homologous recombination. (F) Schematic diagram for the method of PSM differentiation. (G–R) Time course change of Tbx6-venus protein expression in PSM-fated ES cells at 2 (G–N), 3 (I–P), or 4 (K–R) days after the addition of either DMSO or CHIR99021 in culture medium. The Tbx6-venus signals were detected by anti-GFP antibody. (N = 2) Scale bar: 50 µm.
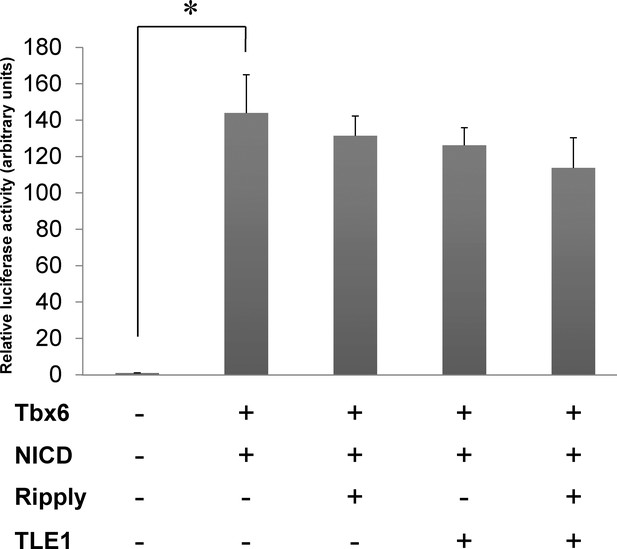
Luciferase reporter assay showing no effect of mouse Ripply2 and TLE1 on mouse Tbx6 transcriptional activity.
The graph shows the representative results of luciferase activity of Mesp2 enhancer in the presence or absence of either NICD, Tbx6, Ripply2, or TLE1 in Cos7 cells. The Mesp2-luciferase reporter activity was strongly induced by the addition of Tbx6+ NICD, but did not demonstrate any significant change in the presence of Ripply2 and TLE1. Values represent means ± SD. Asterisks indicate p<0.001; paired t-test with Tbx6 +NICD samples. (N = 2).
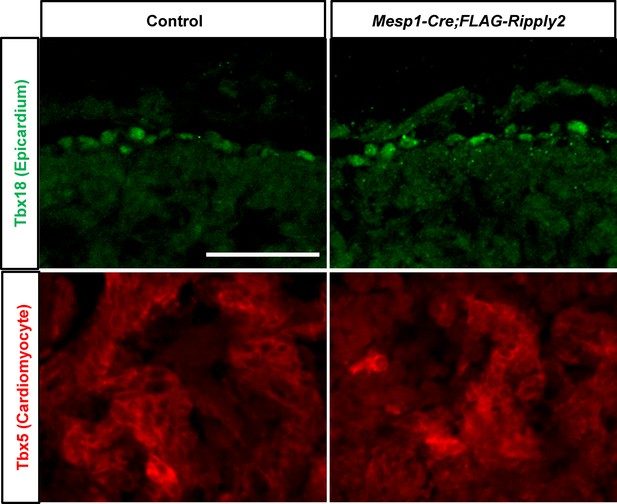
Ectopic expression of Ripply2 in the heart had no influence on the heart development.
The expression of Tbx18 (Epicardium) and Tbx5 (Cardiomyocyte) in the control (N = 5) and Mesp1-Cre;CAG-floxed-lynmRFP-FLAG-Ripply2 (N = 5) mouse hearts at E10.5. Scale bar: 100 µm.
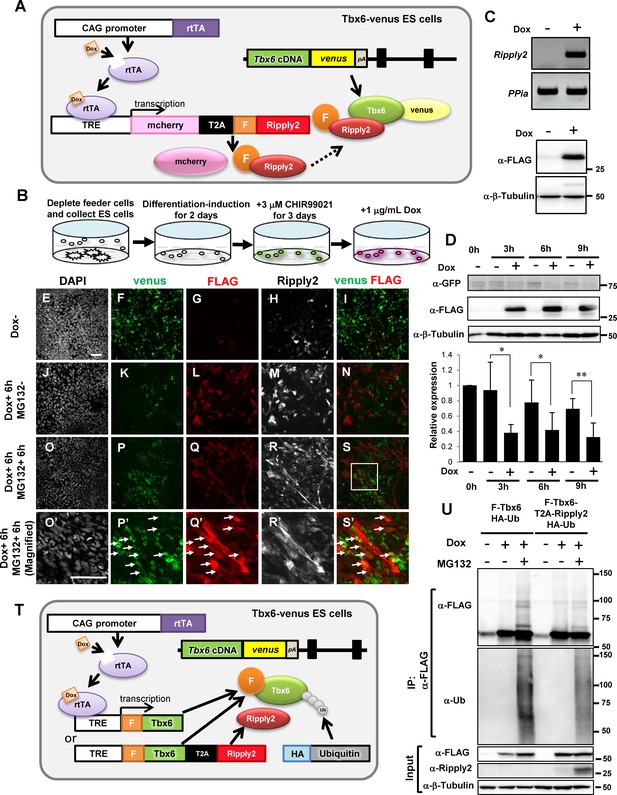
Ripply2 repressed Tbx6 protein expression in PSM-fated ES cells.
(A) Schematic representation of Tet-inducible Ripply2 in Tbx6-venus ES cells. (B) Schematic diagram for ES culture. Tbx6Tbx6-venus/Tbx6-venus;mcherry-T2A-FLAG-Ripply2 ES cells were cultured under feeder-free conditions for 2 days, followed by culture with 3 μM CHIR99021 for 3 days, and were then treated with 1 μg/ml Dox with or without 10 μM MG132. (C) The expression of FLAG-Ripply2 mRNA (up) and protein (down) in Tbx6Tbx6-venus/Tbx6-venus ES cells with Dox treatment for 12 hr without PSM-induction. (D) Western blot analyses to monitor Tbx6-venus expression with or without FLAG-Ripply2 expression in PSM-fated ES cells (N = 5). The histogram shows the quantitation of GFP signal normalized by β-Tubulin signal. Asterisks indicate p<0.05, Double asterisks indicate p<0.01; paired t-test. (E–S) Immunofluorescence of PSM-fated ES cells for the detection of Tbx6-venus and FLAG-Ripply2 after 6 hr incubation without Dox (E–I), with Dox (J–N), or with Dox and MG132 (O–S, O’–S’). (O’–S’) are the magnified rectangle regions in (O–S). Arrows indicate GFP-FLAG double-positive cells. (N = 3) Scale bar: 50 µm. (T) Schematic representation of Tet-inducible FLAG-Tbx6;HA-Ubiquitin or FLAG-Tbx6-T2A-Ripply2;HA-Ubiquitin in Tbx6Tbx6-venus/Tbx6-venus ES cells. (U) IP-western analyses of FLAG-Tbx6 immunoprecipitated with anti-FLAG beads from PSM-fated ES cells that expressed FLAG-tagged Tbx6 or FLAG-tagged Tbx6-T2A-Ripply2 with HA-tagged ubiquitin in the absence or presence of MG132 (N = 3).
-
Figure 2—source data 1
Quantification of Tbx6-venus protein in the absence or presence of FLAG-Ripply2 in PSM-fated ES cells.
Relative expression of Tbx6-venus to b-tubulin protein was quantified using Analyze Gel function of ImageJ. then normalized 0hr sample as 1.
- https://doi.org/10.7554/eLife.33068.010
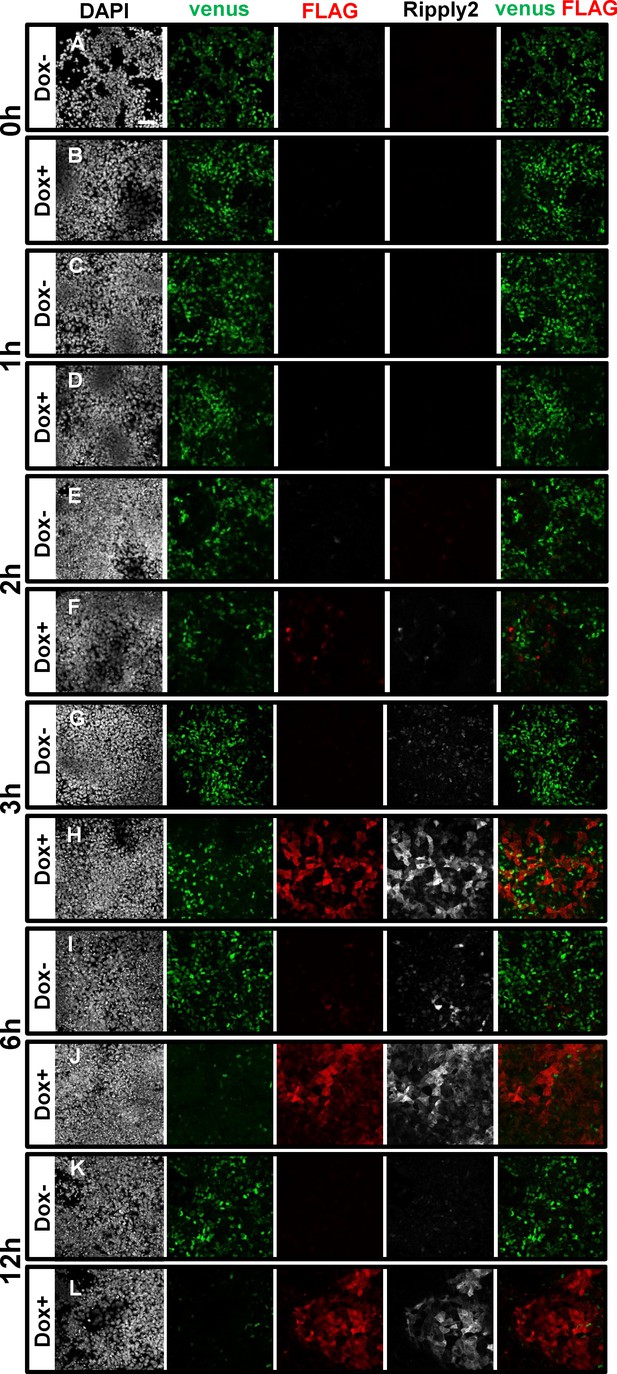
Decreased Tbx6 protein induced by Ripply2 in the PSM-fated ES cell system.
Immunostaining using anti-GFP, anti-FLAG, and anti-Ripply2 antibodies for the detection of Tbx6-venus and FLAG-Ripply2 after 0 hr (A–B), 1 hr (C–D), 2 hr (E–F), 3 hr (G–H), 6 hr (I–J) and 12 hr (K–L) without (A,C,E,G,I) or with (B,D,F,H,J) Dox treatment. (N = 3) Scale bar: 50 µm.
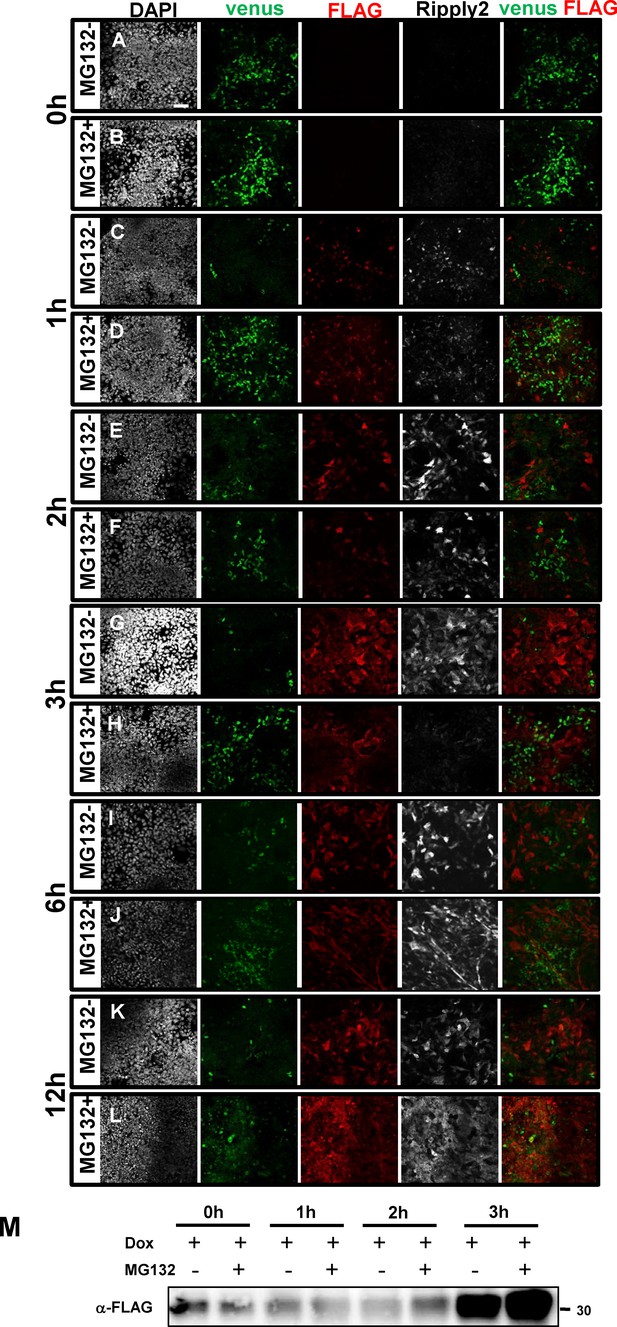
Proteasome inhibitor interrupted Ripply2-dependant Tbx6 protein degradation in the PSM-fated ES cell system.
Immunostaining using anti-GFP, anti-FLAG, and anti-Ripply2 antibody for the detection of Tbx6-venus and FLAG-Ripply2 after treatment with Dox together without (A,C,E,G,I) or with (B,D,F,H,J) MG132 for 0 hr (A–B), 1 hr (C–D), 2 hr (E–F), 3 hr (G–H), 6 hr (I–J) and 12 hr (K–L). (N = 3) (M) The expression level of Ripply2 with or without MG132 for 0 hr, 1 hr, 2 hr, and 3 hr. Scale bar: 50 µm.
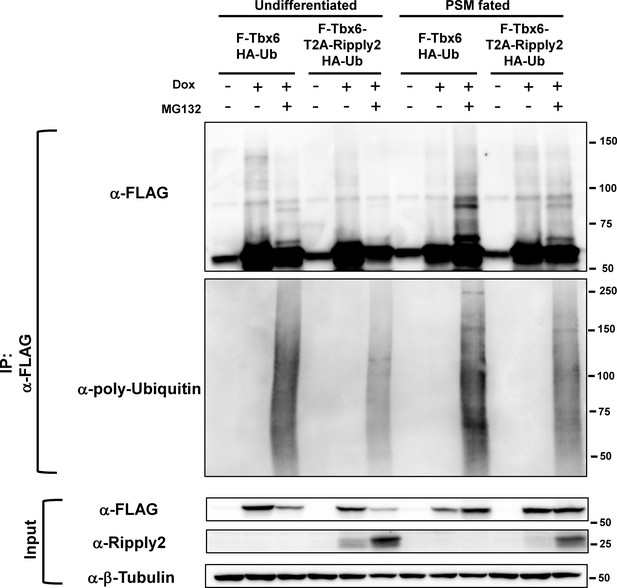
Tbx6 is ubiquitinated in the PSM fated cells.
FLAG-Tbx6 was immunoprecipitated with anti-FLAG beads from undifferentiated and PSM-fated ES cells that expressed FLAG-tagged Tbx6 with HA-tagged ubiquitin in the presence or absence of Ripply2 and MG132. Western analyses were conducted with anti-FLAG and anti-ubiquitin antibodies.
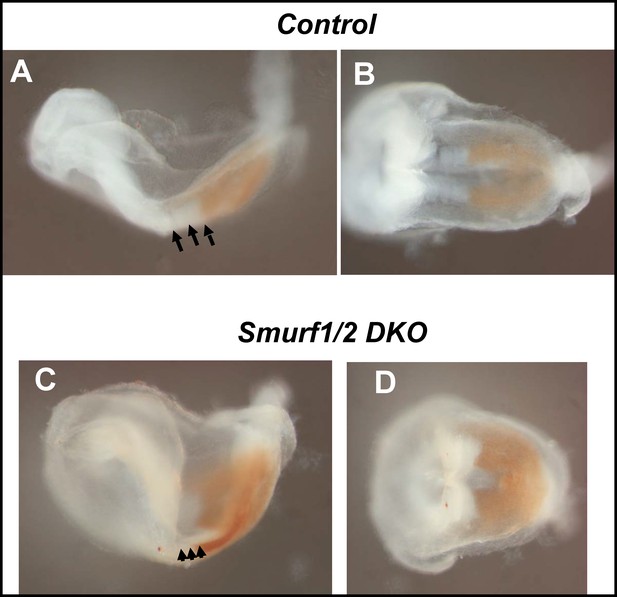
Smurf1/2 are not involved in the Tbx6 degradation mechanism.
Comparison of the expression patterns for Tbx6 protein between wild-type (A, B) and Smurf1/2 DKO mice (C, D). In Smurf1/2 DKO mice, the Tbx6 protein domain was not expanded anteriorly but had a clear boundary (C, D) similar with wild-type embryos (A, B). Embryos were fixed at E8.0. (A, C) are lateral view and (B), D) are dorsal view. Arrows indicate segmented somites.
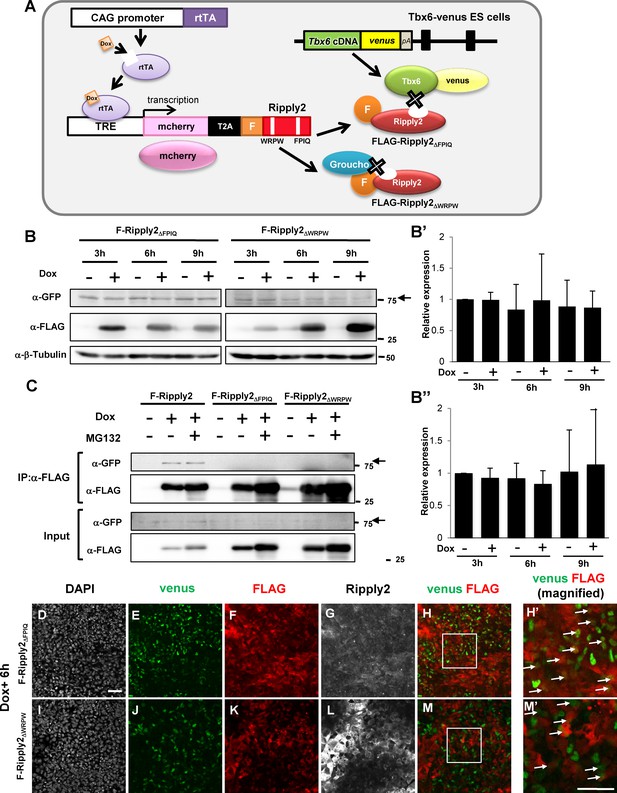
Ripply2-mutants lacking interaction with Tbx6 were defective for Tbx6 protein degradation.
(A) Schematic presentation of Tet-inducible Ripply2ΔFPIQ or Ripply2ΔWRPW in Tbx6Tbx6-venus/Tbx6-venus ES cells. (B) Western blotting of Tbx6-venus in the absence or presence of FLAG-Ripply2ΔFPIQ or FLAG-Ripply2ΔWRPW in PSM-fated ES cells. Arrow: Tbx6-venus. The histogram shows the quantitation of GFP signal in Ripply2ΔFPIQ induced cells (B’:N = 5) and in FLAG-Ripply2ΔWRPW induced cells (B’’:N = 6), normalized by β-Tubulin signal. (C) Co-immunoprecipitation of Tbx6-venus using the anti-FLAG antibody for FLAG-Ripply2 and the mutants. Tbx6-venus signal was detected using an anti-GFP antibody. Arrows: Tbx6-venus. (N = 5). (D–M) Immunofluorescence of PSM-fated ES cells for the detection of Tbx6-venus and FLAG-Ripply2ΔFPIQ or FLAG-Ripply2ΔWRPW after 6 hr incubation with Dox. (H’ and M’) are the magnified rectangle regions in (H and M). Arrows indicate GFP-FLAG double-positive cells. (N = 3) Scale bar: 50 µm.
-
Figure 3—source data 1
Quantification of Tbx6-venus protein in the absence or presence of FLAG-Ripply2DFPIQ or FLAG-Ripply2DWRPW in PSM-fated ES cells.
Relative expression of Tbx6-venus to b-tubulin protein was quantified using Analyze Gel function of ImageJ. then normalized 3hr Dox- sample as 1.
- https://doi.org/10.7554/eLife.33068.013
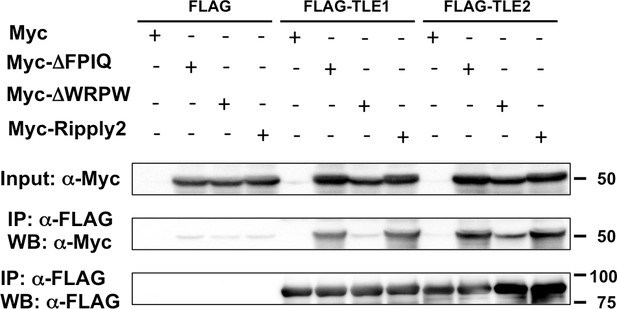
Ripply2 is able to interact with TLE1/2.
IP-western analyses for the interaction between Ripply2 and TLE1/2. Each Myc-tagged construct was transfected with the DNA construct expressing FLAG, FLAG-TLE1, or FLAG-TLE2 into HEK293T cells. The deletion of the tetrapeptide WRPW but not FPIQ in the Ripply2 protein destroyed its ability to bind with TLE1/2. (N = 3).
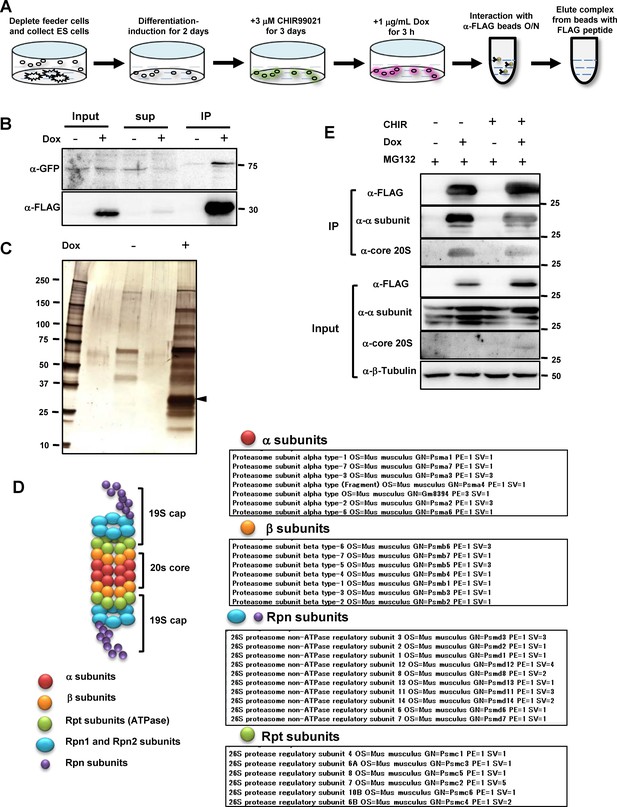
MASS-spec analysis of Tbx6-Ripply2 degradation complex using immunoprecipitation of FLAG-Ripply2.
(A) Schematic diagram of PSM-fated ES culture used for immunoprecipitation. PSM-fated ES cells containing Tbx6Tbx6-venus/Tbx6-venus;mcherry-T2A-FLAG-Ripply2 were incubated with Dox for 3 hr and the cell lysate was subjected to immunoprecipitation. (B) Western blotting showing Tbx6-venus and FLAG-Ripply2 in input (left), supernatant after reaction with beads (middle), and after elution with 3xFLAG peptide (right) with anti-GFP and anti-FLAG antibodies. (N = 4) (C) Silver staining of eluates from beads reacted with control and Tet-induced ES cells lysates. Arrowhead: band for FLAG-Ripply2 protein. (D) Proteasome subunit components specifically detected as Ripply2 interacting proteins. (E) ES cells cultured under several conditions according to the protocol shown in (A) were subjected to immunoprecipitation using anti-FLAG antibody, and interacting proteins were detected with anti-FLAG (Ripply2), anti-proteasome 20 S-1, 2, 3, 5, 6 and 7 subunits, and anti-proteasome 20S core subunit. (N = 2).
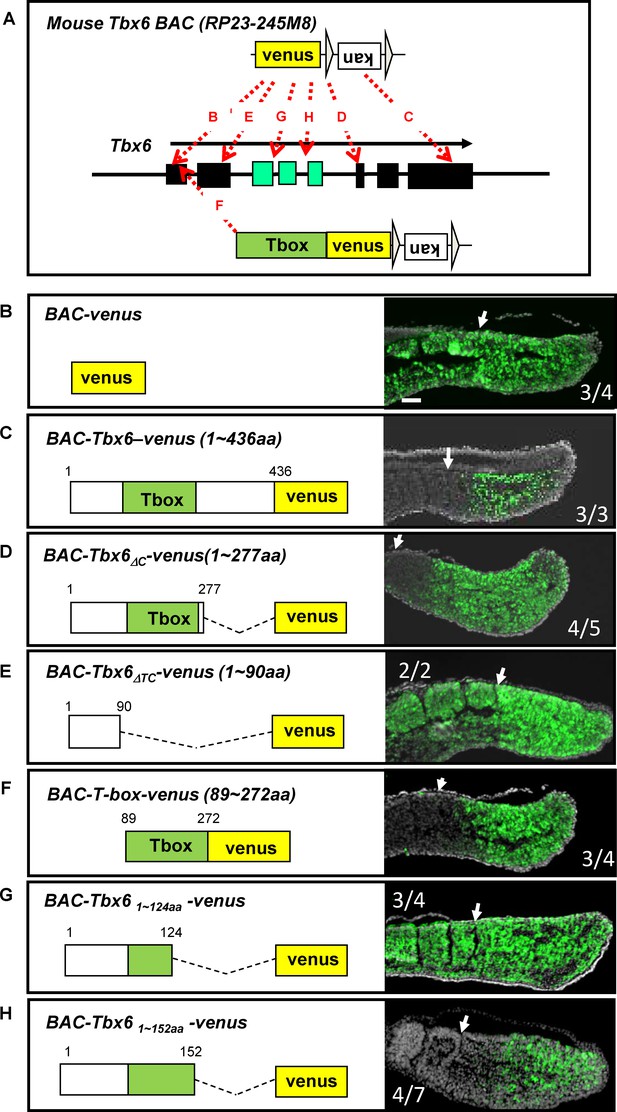
The T-box is essential and sufficient for Tbx6 destabilization in vivo.
(A) Strategies to generate BAC constructs with venus-tag at different positions in the Tbx6 protein. The venus-tag was introduced in frame with either the translational initiation site (B, BAC-venus), translational termination site (C, BAC-Tbx6-venus), after T-box (D, BAC-Tbx6ΔC-venus) or before the T-box (E, BAC-Tbx6ΔTC-venus), after amino acid 124 within the T-box (G, BAC-Tbx61~124aa -venus, amino acid 152 (H, BAC-Tbx61~152aa -venus). A similar method was also used to generate a construct containing only the T-box with venus (F, BAC-T-box-venus). Black and green boxes indicate exons in the Tbx6 locus. Green ones correspond to the T-box region. (B–H) Sections of E10.5 transgenic mouse embryos harboring each BAC construct were stained with anti-GFP antibody. Construct names and amino acid sequences included in the Tbx6-venus fusion-proteins are indicated in each panel. Green; venus signal. Gray; DAPI staining. Newly formed somite borders are indicated by white arrows. Numbers of GFP-positive embryos among transgenic embryos recovered are shown within each box. Scale bar: 100 µm.
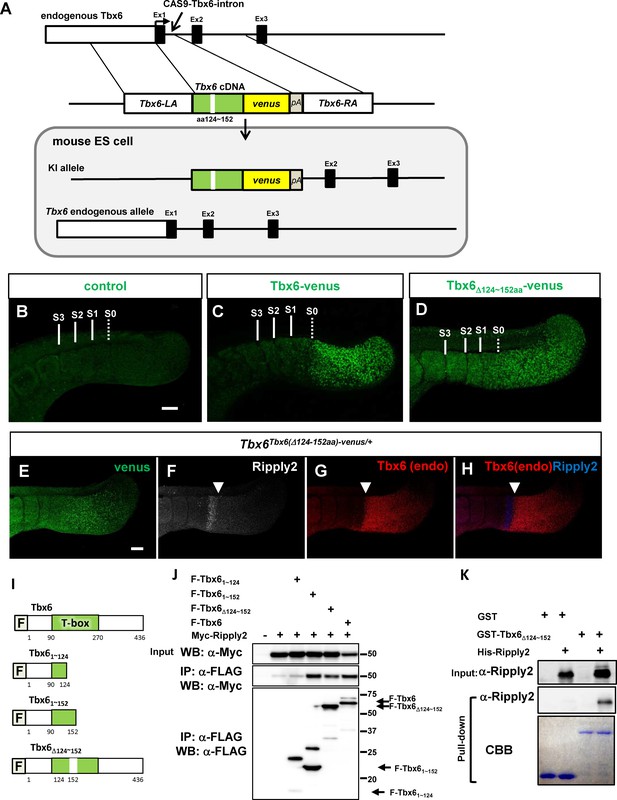
Identification of T-box subdomain required for the degradation of Tbx6 via Ripply2.
(A) The strategy for generating Tbx6Tbx6(Δ124-152aa)-venus/+ + cells. (B–D) Whole-mount immunofluorescent image for venus signal of WT (B) and chimera embryos produced with Tbx6Tbx6-venus/Tbx6-venus (C), and Tbx6 Tbx6(Δ124-152aa)-venus/+ (D) ES cells at E10.5. (E–H) Whole-mount triple-immunostaining for the chimera embryo produced with Tbx6Tbx6(Δ124-152aa)-venus/+ cells showing anti-GFP (E), anti-Ripply2 (F), and anti-Tbx6 (G), and the merged (H, Tbx6 and Ripply2) signals. Expanded GFP signal but not endogenous Tbx6 signal was observed in the chimera embryo with Tbx6Tbx6(Δ124-152aa)-venus/+ cells. Arrow heads: anterior limit of the endogenous Tbx6/posterior limit of Ripply2. Scale bar: 100 µm. (I) Schematic presentation of Tbx6 constructs used for binding assays shown in (J) and (K). (J) IP-western analyses. Each FLAG-tagged Tbx6 construct was co-transfected with Myc-Ripply2 into HEK293T cells. The lysates were subjected to IP with anti-FLAG antibody, followed by western blot analyses. (N = 5) (K) GST-pull down assay. GST or GST-Tbx6Δ124-152aa was incubated with purified His-Ripply2 and pulled down with Glutathione Sepharose, and then subjected to western blotting. His-Ripply2 was detected by anti-Ripply2 antibody. CBB: Coomassie Brilliant Blue staining. (N = 2).
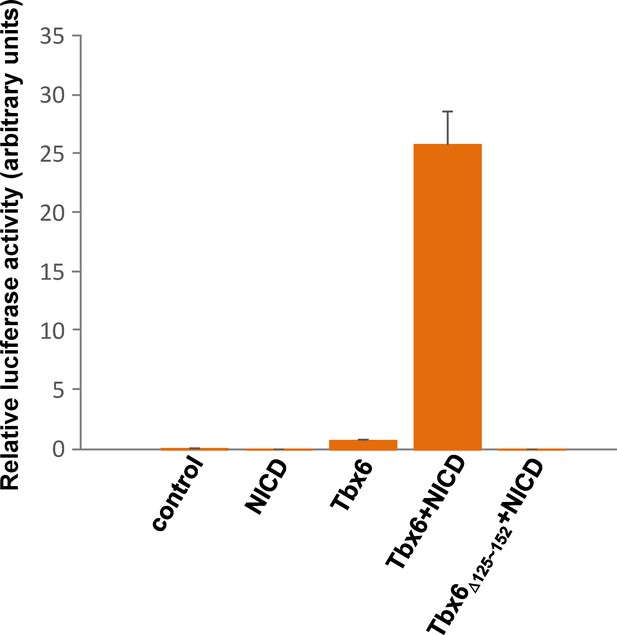
Tbx6Δ124~152aa has no transcriptional activity.
Graph of luciferase activity driven by Mesp2 enhancer when nothing, NICD, Tbx6, Tbx6 +NICD, or Tbx6Δ124~152aa + NICD was transfected into Cos7 cells. High luciferase activity was detected in the sample for Tbx6 +NICD but not in that for Tbx6Δ124~152aa + NICD. Values represent means ± SD.
-
Figure 6—figure supplement 1—source data 1
Quantification of Tbx6-venus protein in the absence or presence of FLAG-Ripply2 in PSM-fated ES cells.
Relative expression of Tbx6-venus to b-tubulin protein was quantified using Analyze Gel function of ImageJ. then normalized 0hr sample as 1.
- https://doi.org/10.7554/eLife.33068.018
Additional files
-
Supplementary file 1
Ripply2-interecting proteins identified by Mass Spectrometry.
Identified proteins, which spectrum counts are higher in Flag IP than control, are listed. Ripply2 is highlighted with yellow, Proteasome subunits are highlighted with green, and Tbx6 is highlighted with pink.
- https://doi.org/10.7554/eLife.33068.019
-
Supplementary file 2
Vector information used for each construct.
Vector information of cDNA constructs used for immunoprecipitation experiments are indicated. The corresponding Figures obtained by using each construct are also listed.
- https://doi.org/10.7554/eLife.33068.020
-
Supplementary file 3
Strategies to generate mutant constructs.
Primer information and the cloning strategies are listed for each mutant cDNA constructs.
- https://doi.org/10.7554/eLife.33068.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33068.022





