Identification of PNG kinase substrates uncovers interactions with the translational repressor TRAL in the oocyte-to-embryo transition
Figures
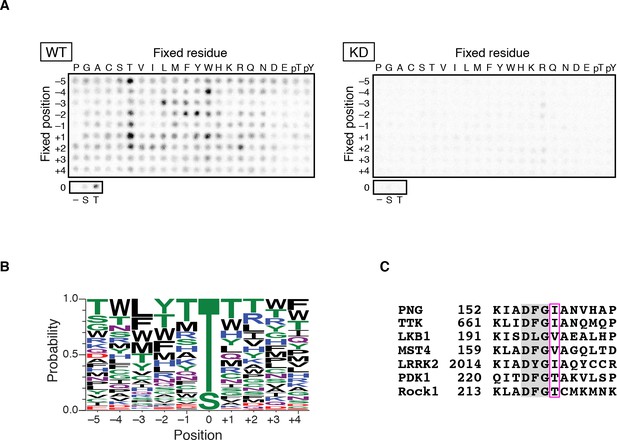
PAN GU (PNG) kinase is a threonine-specific kinase.
(A, B) PNG kinase prefers threonine as a phosphoacceptor site. The peptide library was phosphorylated with PNG kinase wild type (WT) or kinase dead (KD) using radiolabeled ATP (A). The signals were quantified and visualized with WebLogo 3.0. (B) Ten thousand peptide sequences were generated according to the probabilities predicted from the quantified peptide library data. The colors designate classes of amino acids. (C) Alignment of the amino acid sequences near the DFG motif of known threonine-specific kinases and PNG. PNG has a beta-branched residue, isoleucine, immediately downstream of the DFG motif as do other threonine-selective kinases (boxed with magenta (Chen et al., 2014). The peptide library screen with WT was repeated in four replicates.
-
Figure 1—source data 1
Quantification of peptide phosphorylation in Figure 1B.
- https://doi.org/10.7554/eLife.33150.004
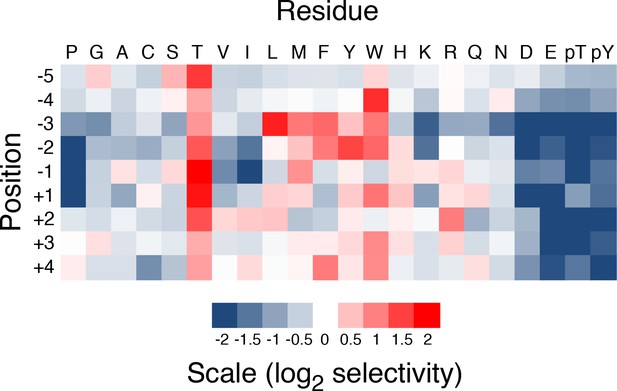
Heat map for quantified selective values of PNG phosphorylation.
Peptide array data were quantified and normalized to an average value of 1 within a position.
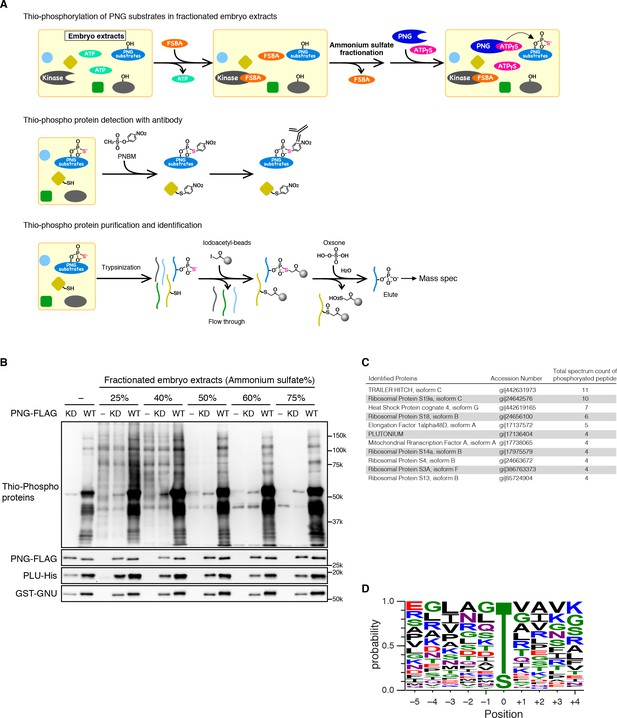
Identification of PNG kinase substrates with a biochemical screen.
(A) A schematic representation of the substrate screen. Embryo extracts were gel-filtrated to exchange the buffer and treated with 5’-(4-Fluorosulfonylbenzoyl)adenosine (FSBA) to inactivate endogenous kinases, followed ammonium sulfate fractionation. The fractionated extracts were dialyzed to remove ammonium sulfate. The recombinant active PNG kinase complex was added to the FSBA-treated fractions with ATP-γS to thio-phosphorylate PNG kinase substrates. To examine thio-phosphorylated proteins by immunoblot, they were alkylated with p-Nitrobenzyl mesylate (PNBM), and the alkylated thiophosphate was detected by its specific antibody. For identification of the thio-phosphorylated peptides, the PNG kinase-treated fractions were pooled and digested with trypsin. Thio-phosphorylated peptides were purified specifically with iodoacetyl beads and oxsone and identified by mass spectrometry. (B) Detection of thio-phosphoproteins in the fractions by immunoblots. The FSBA-treated fractions were incubated with wild-type (WT) or kinase-dead (KD) PNG kinase complexes. Thio-phosphorylated proteins were detected as shown in (A). The PNG kinase complexes added into the fraction were examined by immunoblots using anti-FLAG (PNG-FLAG), anti-PLU (PLU-His) and anti-GST (GST-GNU) antibodies. (C) A list of PNG substrates identified in the second experiment that were recovered with wild-type but not kinase-dead PNG. The proteins found by mass spectrometry following purification of the thio-phosphorylated peptides are listed in the order of the number of total spectrum count of phosphorylated peptides from the sample treated with the wild-type PNG kinase complex. The top 11 of the 36 substrates identified are shown. (D) Phosphorylation sites analysis. Peptides identified with >95% probability according to Scaffold were used for thio-phosphorylation motif analysis. Phosphopeptides identified in the samples treated with the PNG WT kinase complex but absent from the samples treated with KD kinase were further analyzed. Since elution of the peptides was performed under oxidizing conditions, peptides that differed only in the oxidation state of their methionines were regarded as equal. A total of 112 unique phosphopeptides belonging to 70 different proteins were considered. Using these phosphopeptides, a list of motifs was constructed centered on the phosphosite and including the 5 N-terminal and 4 C-terminal residues found adjacent to the phosphosite on the protein and analyzed with WebLogo 3.0. The screen with the WT kinase was done as a pilot screen, and a second experiment was done in which the results were compared with kinase-dead PNG.
-
Figure 2—source data 1
Lists of proteins and peptides identified by mass spectrometry analyses.
Sheet 1 shows the total spectrum count in each sample for proteins identified in the PNG substrate screens. The substrates identified by phosphopeptides recovered on the iodoacetyl resin are listed for the 25% (F1) and 40–75% (F2) fractions from experiment 1. For experiment 2, the purified thiophosphorylated peptides (elute) and their unbound fractions (flow through) following incubation with wild-type or kinase-dead PNG are listed (see also Figure 2A). Proteins for which at least two peptides were found are shown in sheet 1, but all the spectra found in the analyses are listed in sheet 2. Sheet 3 provides putative targets of PNG based on the analysis of each experiment and the combined analysis.
- https://doi.org/10.7554/eLife.33150.008
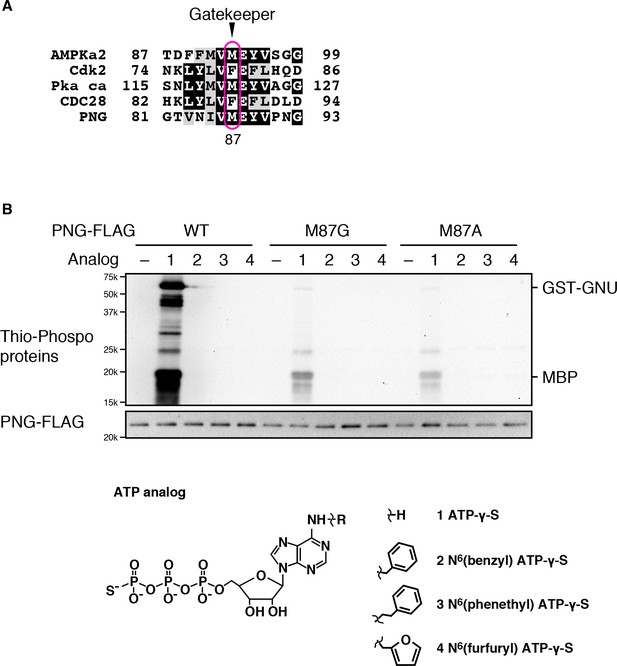
Mutation of the gatekeeper residue of PNG inactivates its kinase activity.
(A) Alignment of the gatekeeper residue with its surrounding sequence in PNG and other kinases whose analog-sensitive mutants have been reported (Banko et al., 2011; Niswender et al., 2002; Polson et al., 2001; Ubersax et al., 2003). The gatekeeper residues of the kinases are boxed in magenta. Methionine 87 of PNG is likely the gatekeeper residue. (B) Substitution of the gatekeeper residue to small amino acids reduced PNG kinase activity. Methionine 87 of PNG was changed to the small amino acid glycine or alanine (M87G or M87A). Kinase activity of the mutants and wild-type (WT) PNG kinase with GST-GNU was examined using myelin basic protein (MBP) as an in vitro substrate in the presence of ATP-γS or its analogs as indicated. Thio-phosphorylated proteins were alkylated and detected by immunoblot using anti alkylated thiophosphate antibody. Protein levels of PNG-FLAG also were examined using anti-FLAG.
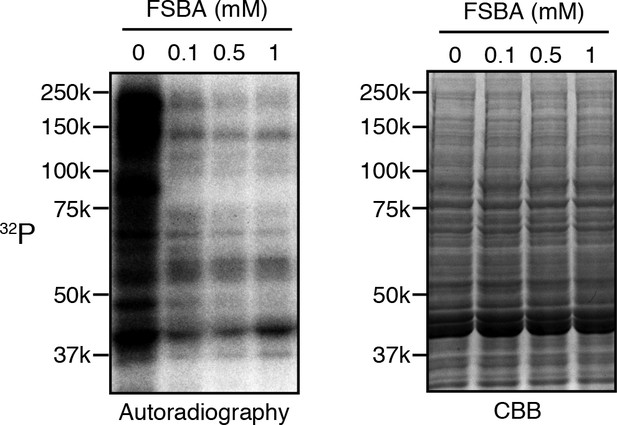
5’-(4-Fluorosulfonylbenzoyl)adenosine (FSBA) inactivates endogenous kinases in the embryo extracts.
Extracts made from embryos collected for 2 hr were dialyzed to remove ATP and treated with the indicated concentration of FSBA. After removal of free FSBA, the extracts were incubated with radioactive ATP. Radioactivity incorporated into proteins was detected by autoradiography (left panel). Proteins in the extracts were examined by Coomassie staining (right panel. CBB).
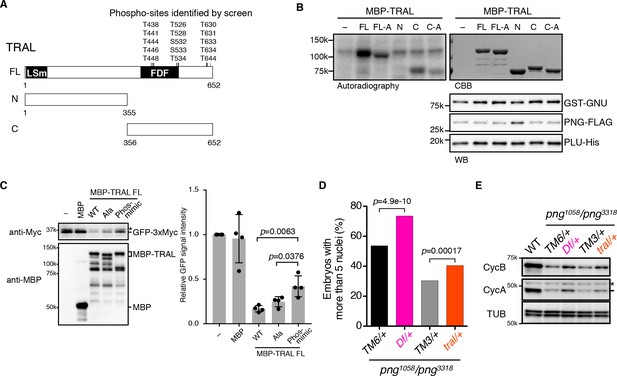
PNG kinase directly phosphorylates Trailer hitch (TRAL) and can suppress its repressor function.
(A) Schematic representation of Trailer hitch (TRAL) protein. Drosophila melanogaster TRAL consists of 652 amino acids and has conserved domains: the LSm (Like Sm) domain is essential for P-body localization of TRAL; the FDF domain binds to ME31B, a translational repressor (Marnef et al., 2009). The phosphorylation sites of TRAL identified in the screen map exclusively to the C-terminus. (B) PNG kinase phosphorylates the C-terminus of TRAL in vitro. Maltose-binding protein (MBP)-fused full length (FL) and N- and C-terminal fragments (N: 1–355 and C: 356–652, respectively) of TRAL were expressed and purified from bacteria (A). The MBP-fused TRAL proteins were incubated with the active PNG kinase complex in the presence of radioactive ATP. Incorporated radioactivity in the TRAL proteins was detected by autoradiography. The levels of the MBP-fused TRAL proteins were examined by coomassie staining (CBB). The active PNG kinase complex in the reactions also was examined by immunoblot using anti-GST (GST-GNU), anti-FLAG (PNG-FLAG) and anti-PLU (PLU-His) antibodies. Substitution of the phosphosites in TRAL to alanine (FL-A and C-A) reduced phosphorylation by PNG. The kinase assay was repeated three times. Representative results are shown. (C) Phosphomimetic mutation of the PNG phosphorylation sites in TRAL suppresses translational repression activity of TRAL in vitro. In vitro transcribed GFP-3xMyc mRNA was translated in rabbit reticulocyte lysate with or without MBP-fused TRAL wild-type (WT) or MBP-TRAL mutant proteins, which have alanine or aspartic acid in the PNG phosphorylation sites (Ala or Phos-mimic). MBP was used as a control. Translation of GFP-3xMyc mRNA was examined by GFP-3xMyc protein levels on an immunoblot using anti-Myc antibody. MBP and MBP-fused TRAL protein levels were examined by immunoblot using anti-MBP antibody. GFP-3xMyc protein levels were quantified and normalized to its levels in the control reaction. Error bars represent standard deviation (n = 4, unpaired t-test; mean ± SD). Representative blots are shown. (D, E) tral dominantly suppresses png phenotypes. (D) Embryos from females whose genotype were png1058/png3318 with Df(3L)ED4483/+ (Df/+) or tral1/+ (tral/+) were collected, fixed, and DNA stained with DAPI. Nuclear numbers in the embryos were quantified by fluorescent microscopy. TM6C, cu1 Sb1/+ (TM6/+) and TM3, Sb1 Ser1/+ (TM3/+) were used as controls for Df/+ and tral/+, respectively. The results are the sum of three experiments, and their significance was tested with Fisher’s exact test. (E) Cyclin A and B (CycA, CycB) protein levels of the embryos from the females with the indicated genotypes were examined by immunoblot. Alpha-tubulin (TUB) was used as a loading control. The asterisk shows a non-specific band.
-
Figure 3—source data 1
Quantification of GFP protein levels in Figure 3C.
- https://doi.org/10.7554/eLife.33150.013
-
Figure 3—source data 2
Raw data of embryo numbers for Figure 3D.
- https://doi.org/10.7554/eLife.33150.014
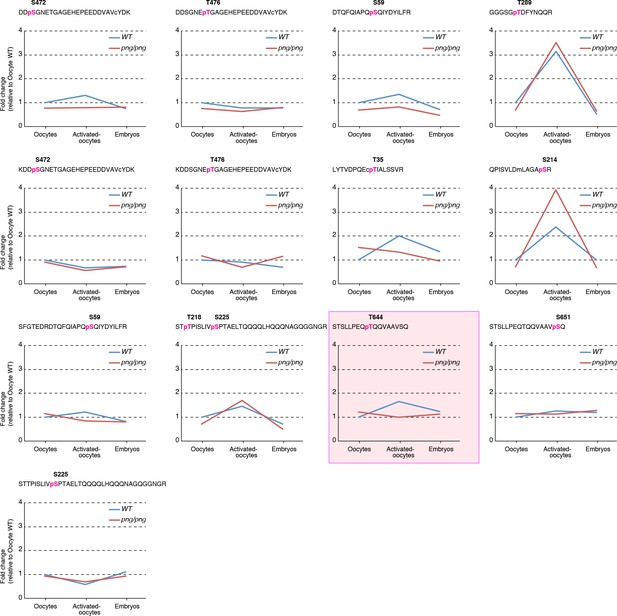
TRAL phosphorylation analysis by quantitative mass spectrometry.
TRAL was immunoprecipitated from extracts of oocytes, in vitro activated oocytes and embryos of OrR (WT, blue line) or png1058/png1058 (png/png, red line). The precipitated TRAL proteins were analyzed by mass spectrometry. Identified phosphopeptides were manually validated. Threonine 644 is one of the phosphorylation sites identified in the biochemical PNG substrate screen (boxed in magenta). This was done one time. The Y-axes designate a relative value for peptide abundance.
-
Figure 3—figure supplement 1—source data 1
Quantification of phosphopeptide recovery.
- https://doi.org/10.7554/eLife.33150.011
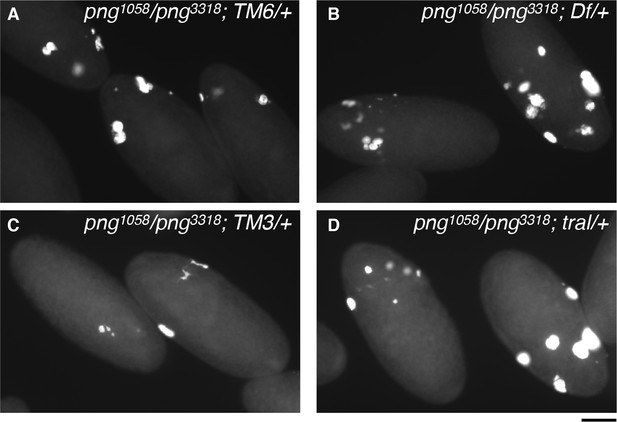
Examples of DAPI-stained embryos scored in Figure 3D.
(A) Three embryos from png1058/png3318; TM6/+ sibling control mothers are shown. The embryo in the middle has six nuclei and the two flanking have five or less nuclei. (B) Two embryos from png1058/png3318; Df(3L)ED4483/+ (Df/+) experimental mothers. These have greater than five nuclei, and mitotic figures can be seen in the embryo on the left. (C) Two embryos with five or less nuclei from png1058/png3318; TM3/+ sibling control mothers. (D) Two embryos from png1058/png3318; tral1/+ (tral/+) experimental mothers. The images show the range of ploidy in the nuclei and apparent bridges from failed mitoses, even with high ploidy. All panels are at the same magnification, and the scale bar is 100 μm.
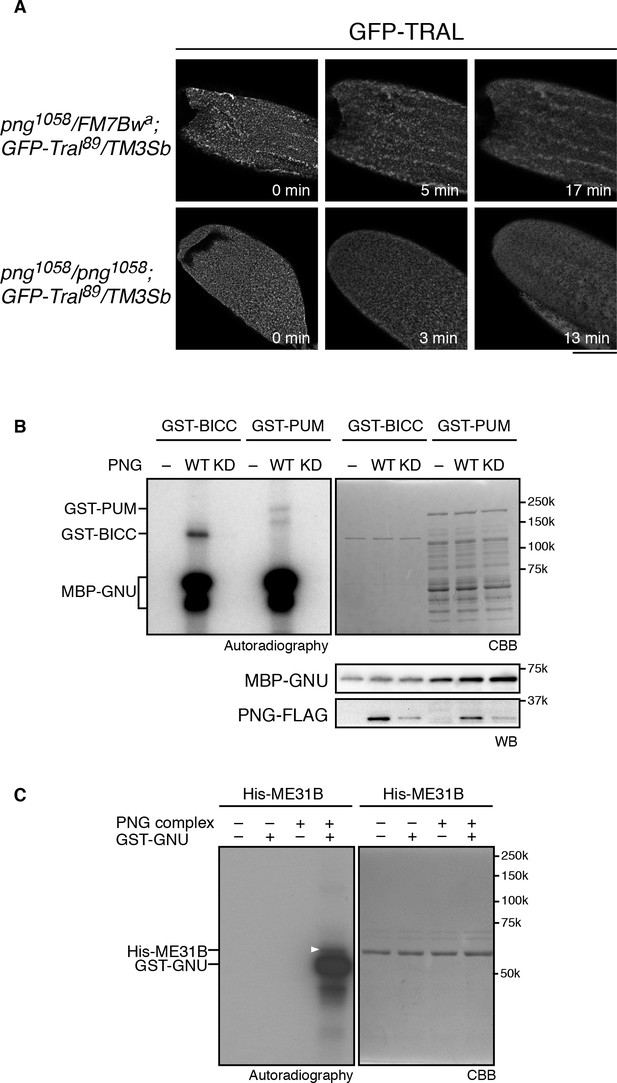
PNG kinase phosphorylates translational repressors in vitro.
(A) TRAL granules in oocytes diminished after egg activation independently of png. Heterozygous (png1058/FM7B wa) or homozygous png (png1058/png1058) oocytes expressing GFP-Tral (GFP-Tral89/TM3 Sb) were activated in vitro. GFP-TRAL signal in the oocytes was observed by confocal microscopy. Time in the images indicates time after egg activation. Bar indicates 100 μm. Representative oocytes images, taken with the same exposure settings, are shown (WT: n = 8, png: n = 3) (B) PNG kinase phosphorylates Bicaudal C (BICC) and Pumilio (PUM). GST-fused BICC and PUM were incubated with or without wild-type (WT) or kinase-dead (KD) PNG kinase activated with MBP-GNU in the presence of radioactive ATP. Radioactivity incorporated into proteins was detected by autoradiography (left panel). The substrate protein levels were examined by coomassie staining (right panel, CBB). MBP-GNU and PNG protein levels were examined by immunoblot using anti-MBP (MBP-GNU) and anti-FLAG (PNG-FLAG) (bottom panels, WB). The kinase assay was repeated in two replicates. Representative results are shown. (C) PNG kinase phosphorylates ME31B in vitro. Recombinant His-ME31B was incubated with or without PNG kinase complex in the presence of radioactive ATP. Because PNG kinase complex requires additional recombinant GNU (GST-GNU) for its full kinase activation, His-ME31B was phosphorylated with or without GST-GNU. Radioactivity incorporated into His-ME31B was detected by autoradiography (left panel). Protein levels of His-ME31B were examined by coomassie staining (right panel, CBB). The white arrowhead indicates phosphorylated His-ME31B protein. The kinase assay was repeated in two replicates. Representative results are shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (Drosophila melanogaster) | Bicaudal C (BicC) | NA | FLYB: FBgn0000182 | |
| gene (D. melanogaster) | giant nuclei (gnu) | NA | FLYB: FBgn0001120 | |
| gene (D. melanogaster) | maternal expression at 31B (me31B) | NA | FLYB: FBgn0004419 | |
| gene (D. melanogaster) | pan gu (png) | NA | FLYB: FBgn0000826 | |
| gene (D. melanogaster) | plutonium (plu) | NA | FLYB: FBgn0003114 | |
| gene (D. melanogaster) | pumilio (pum) | NA | FLYB: FBgn0003165 | |
| gene (D. melanogaster) | trailer hitch (tral) | NA | FLYB: FBgn0041775 | |
| strain, strain background (D. melanogaster) | WT: OregonR | NA | ||
| genetic reagent (D. melanogaster) | Df(3L)ED4483 | Bloomington Drosophila Stock Center | BDSC:8070; RRID:BDSC_8070; FLYB:FBst0008070 | FlyBase symbol: Df(3L)ED4483; Genotype: w[1118]; Df(3L)ED4483, P{w[+mW. Scer\FRT.hs3]=3'.RS5+3.3'} ED4483/TM6C, cu[1] Sb[1] |
| genetic reagent (D. melanogaster) | gfp-tral89 | The Flytrap Project; (Morin et al., 2001); PMID:11742088 | Flytrap:G00089; DGRC:110584; RRID:DGGR_110584 | Genotype: w[*]; P{w[+mC]=PTT-un1}G00089 |
| genetic reagent (D. melanogaster) | png1058 | (Shamanski and Orr-Weaver, 1991); PMID:1913810 | ||
| genetic reagent (D. melanogaster) | png3318 | (Shamanski and Orr-Weaver, 1991); PMID:1913810 | ||
| genetic reagent (D. melanogaster) | tral1 | Bloomington Drosophila Stock Center; (Wilhelm et al., 2005); PMID: 16256742 | BDSC:14933; RRID:BDSC_14933; FLYB:FBst0014933 | Genotype: y[1]; P{y[+mDint2] w[BR.E.BR]= SUPorP}tral[KG08052] ry[506]/TM3, Sb[1] Ser[1] |
| antibody | alkylated thiophosphate antibody (Rabbit monoclonal) | Abcam | Abcam:ab92570; RRID:AB_10562142 | Anti-Thiophosphate ester antibody [51-8] (1/2000 in 5% Skim milk TBS-T) |
| antibody | anti-PNG (Rabbit polyclonal) | (Hara et al., 2017); PMID: 28555567 | (1/1000 in Hikari solution A) | |
| antibody | anti-PLU (Rabbit polyclonal) | (Elfring et al., 1997); PMID: 9247640 | Affinity-purified (1/200 in Hikari solution A) | |
| antibody | anti-GNU (Guinea pig polyclonal) | (Lee et al., 2003); PMID: 14665672 | (1/5000 in TBS-T) | |
| antibody | anti-TRAL (Rat polyclonal) | (Tritschler et al., 2008); PMID:18765641 | ||
| antibody | anti-FLAG (Mouse monoclonal) | Sigma-Aldrich | Sigma-Aldrich:F1804; RRID:AB_262044 | (1/2000 in TBS-T) |
| antibody | anti-MBP (Rat monoclonal) | Sigma-Aldrich | Sigma-Aldrich:SAB4200082 | (1/2000 in Hikari solution A) |
| antibody | anti-Myc (Mouse monoclonal) | Covance | Covance:MMS-150R-1000; RRID:AB_291325 | 9E10; (1/2000 in TBS-T) |
| antibody | anti-GST (Mouse monoclonal) | MBL | MBL:PM013-7; RRID:AB_10598029 | Anti-GST-tag pAb-HRP-DirecT; (1/5000 in Hikari solution A) |
| antibody | HRP-conjugated anti-rabbit IgG | Jackson Immuno Research | Jackson ImmunoResearch: 711-035-152; RRID:AB_10015282 | (1/10000 in TBS-T or Hikari solution B) |
| antibody | HRP-conjugated anti-guinea pig IgG | Jackson Immuno Research | Jackson Immuno Research:706-035-148: RRID:AB_2340447 | (1/50000 in TBS-T) |
| antibody | HRP-conjugated anti-mouse IgG | Jackson Immuno Research | Jackson Immuno Research:115-035-164: RRID:AB_2338510 | (1/20000 in TBS-T) |
| antibody | HRP-conjugated anti-rat IgG | Jackson Immuno Research | Jackson Immuno Research:112-035-062: RRID:AB_2338133 | (1/5000 in TBS-T) |
| antibody | mouse monoclonalanti-Cyclin A | Developmental Studies Hybridoma Bank | DSHB Cat #A12RRID:AB_528188 | 1/100 in Hikari Solution A |
| antibody | mouse monoclonalanti-Cyclin B | Developmental StudiesHybridoma Bank | DSHB Cat#F2F4RRID:AB_528189 | 1/200 in Hikari Solution B |
| recombinant DNA reagent | pFastBac Dual | Thermo Fisher | Thermo Fisher:10712024 | |
| recombinant DNA reagent | pFastBac1 | Thermo Fisher | Thermo Fihser:10359016 | |
| recombinant DNA reagent | pGEX-6P-1 | GE Healthcare | GE Healthcare:28954648 | |
| recombinant DNA reagent | pMAL-c2X | New England Biolabs | New England Biolabs:N8076S | |
| recombinant DNA reagent | pSP64 Poly(A) | Promega | Promega:P1241 | |
| recombinant DNA reagent | pET28b | Merck | Merck:69865-3 | |
| recombinant DNA reagent | pFastBac Dual PNG/PLU | (Hara et al., 2017); PMID: 28555567 | ||
| recombinant DNA reagent | pFastBac Dual PNG172/PLU | This paper | png172:kinase dead mutant (G157E) | |
| recombinant DNA reagent | pFastBac1 GNU | (Lee et al., 2003); PMID: 14665672 | ||
| recombinant DNA reagent | pFastBac Dual PNG M87G/PLU | This paper | PNG M87G: gatekeeper mutant | |
| recombinant DNA reagent | pFastBac Dual PNG M87A/PLU | This paper | PNG M87A: gatekeeper mutant | |
| recombinant DNA reagent | pGEX-6P-1 GNU | (Hara et al., 2017); PMID: 28555567 | ||
| recombinant DNA reagent | pGEX-6P-1 BICC | This paper | ||
| recombinant DNA reagent | pGEX-6P-1 PUMILIO | This paper | ||
| recombinant DNA reagent | pMAL-c2X GNU | This paper | ||
| recombinant DNA reagent | pMAL-c2X TRAL FL | This paper | ||
| recombinant DNA reagent | pMAL-c2X TRAL N | This paper | TRAL 1–355 | |
| recombinant DNA reagent | pMAL-c2X TRAL C | This paper | TRAL 356–652 | |
| recombinant DNA reagent | pMAL-c2X TRAL FL A | This paper | T438A, T441A, T444A, T446A, T448A, T526A, T528A, S532A, S533A, T534A, T630A, T631A, T633A, T634A, T644A | |
| recombinant DNA reagent | pMAL-c2X TRAL C A | This paper | T438A, T441A, T444A, T446A, T448A, T526A, T528A, S532A, S533A, T534A, T630A, T631A, T633A, T634A, T644A | |
| recombinant DNA reagent | pMAL-c2X TRAL FL Phos-mimic | This paper | T438D, T441D, T444D, T446D, T448D, T526D, T528D, S532D, S533D, T534D, T630D, T631D, T633D, T634D, T644D | |
| recombinant DNA reagent | pSP64 Poly(A) EGFP-3xMyc | This paper | ||
| recombinant DNA reagent | pET28b ME31B-3xMyc | This paper | ||
| commercial assay or kit | mMESSAGE mMACHINE SP6 Transcription Kit | Thermo Fisher | Thermo Fisher:AM1340 | |
| commercial assay or kit | Rabbit Reticulocyte Lysate System, Nuclease Treated | Promega | Promega: L4960 | |
| commercial assay or kit | TMTsixplex Isobaric Label Reagent Set | Thermo Fisher | Thermo Fisher:90061 | |
| chemical compound, drug | FSBA | SIGMA-Aldrich | SIGMA-Aldrich:F9128 | 5’-(4-fluorosulphonylbenzoyl)adenosine |
| chemical compound, drug | PNBM | Abcam | Abcam:ab138910 | p-Nitrobenzyl mesylate |
| chemical compound, drug | N6(benzyl) ATP-γS | Axxora | Axxora:BLG-B072 | |
| chemical compound, drug | N6(phenethyl) ATP-γS | Axxora | Axxora:BLG-P026 | N6(Phenylethyl) ATP-γ-S |
| chemical compound, drug | N6(furfuryl) ATP-γS | Axxora | Axxora:BLG-F008 | |
| chemical compound, drug | HIKARI signal enhancer | Nacalai | Nacalai:02270–81 | Signal Enhancer HIKARI for Western Blotting and ELISA |
| software, algorithm | WebLogo | (Crooks et al., 2004); PMID:15173120 | RRID:SCR_010236 | |
| software, algorithm | Proteome Discoverer | Thermo Fisher | RRID:SCR_014477 | |
| software, algorithm | Mascot | Matrix Science | RRID:SCR_014322 | |
| software, algorithm | CAMV | (Curran et al., 2013); PMID:23500044 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33150.016





