Biosynthetic tailoring of existing ascaroside pheromones alters their biological function in C. elegans
Figures
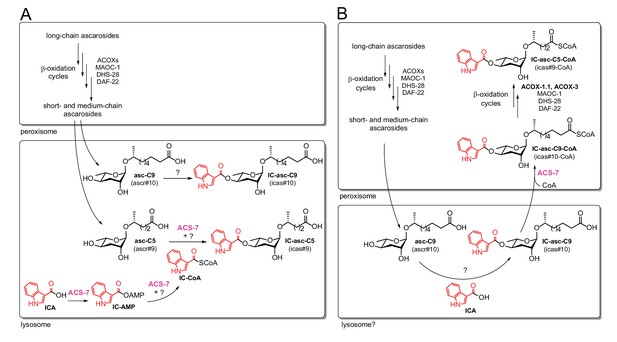
Comparison of the previous model and the model supported by our current data.
In both the previous model (A) and the current model (B), long-chain ascarosides are shortened to medium- and short-chain ascarosides through β-oxidation cycles in the peroxisome. (A) According to the previous model, the final step in the biosynthesis of all of the IC-modified ascarosides is the attachment of the IC head group to the 4’-position of the ascarylose sugar. Short/medium-chain ascarosides, such as asc-C5 and asc-C9, are transported from the peroxisome to the lysosome, where the IC group is attached. According to this model, the acyl-CoA synthetase ACS-7 plays a key role in activating ICA as its corresponding CoA thioester, IC-CoA, for attachment specifically to asc-C5 to make IC-asc-C5. (B) According to our model, the IC head group is only attached to ascarosides with medium-length side chains, such as asc-C9. The biosynthesis of IC-modified ascarosides with shorter side chains, such as IC-asc-C5, requires additional β-oxidation in the peroxisome. In our model, under favorable conditions, the IC head group is attached by unknown enzymes to the medium-chain ascarosides, such as asc-C9, to make aggregation pheromones. This process may occur in the lysosome as it has been shown that the lysosome is required for the biosynthesis of 4’-modified ascarosides (Panda et al., 2017). If conditions decline, such as during starvation, ACS-7 then plays a key role for activating IC-asc-C9 as the corresponding CoA-thioester for further β-oxidation of its side chain in the peroxisome to make shorter-chain IC-ascarosides, such as the dauer pheromone IC-asc-C5.
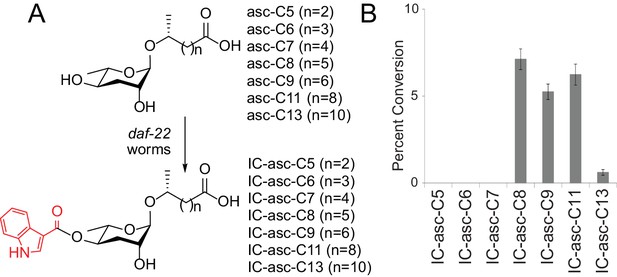
Substrate preference for attachment of the IC head group in biosynthesis of IC-ascarosides.
(A) Possible attachment of the IC group to ascarosides with different side-chain lengths (C5–C13). (B) Percent conversion of each ascaroside to the corresponding IC-ascaroside by daf-22(m130) worms. Data represent the mean ± SD of three independent experiments.
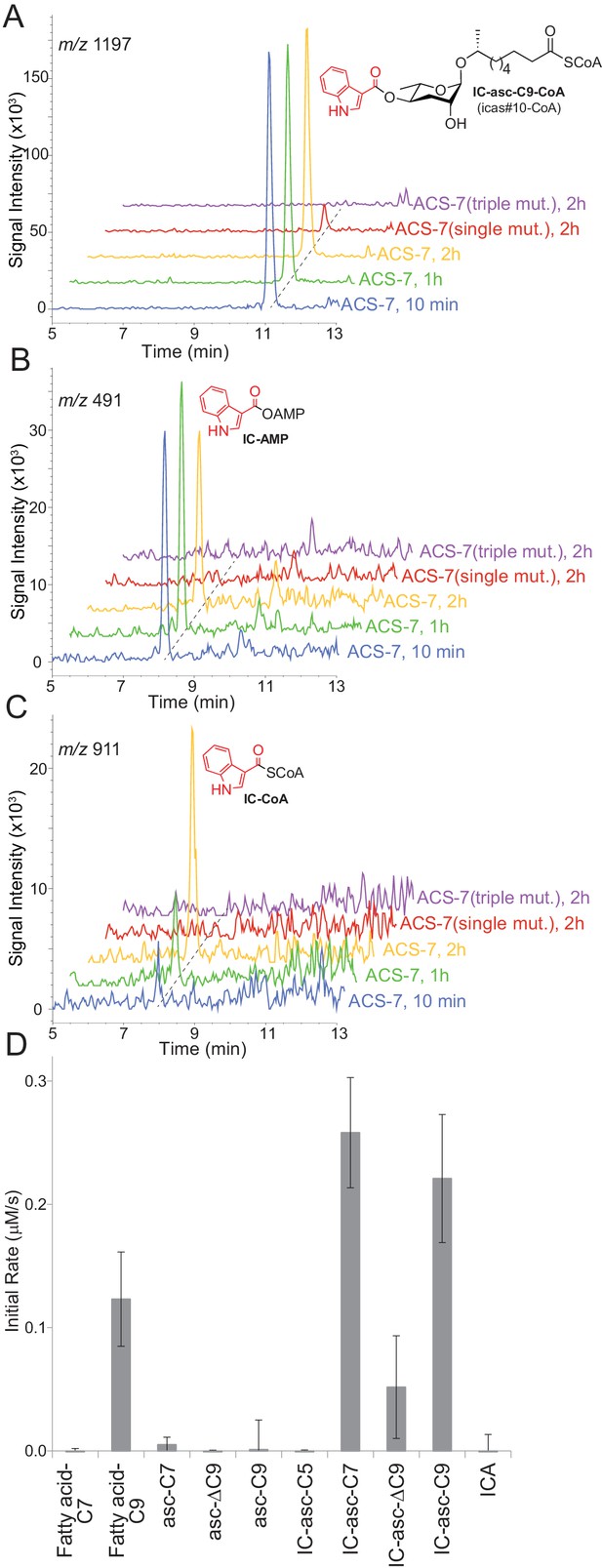
In vitro activity of ACS-7 towards ascaroside and ICA substrates.
(A) LC-MS traces of the reaction of ACS-7, ACS-7(E339A) single mutant, or ACS-7(E339A, S186A, S187A) triple mutant with IC-asc-C9, detecting IC-asc-C9-CoA. (B–C) LC-MS traces of the reaction of ACS-7, ACS-7 (E339A), or ACS-7 (E339A, S186A, S187A) with ICA, detecting IC-AMP (B) or IC-CoA (C). (D) Activity of ACS-7 in a coupled enzyme assay against a panel of substrates, including fatty acids (fatty acid-C7 and fatty acid-C9), ascarosides (asc-C7, asc-ΔC9, and asc-C9), IC-modified ascarosides (IC-asc-C5, IC-asc-C7, IC-asc-ΔC9, and IC-asc-C9), and ICA. Data represent the mean ± SD of three independent experiments.
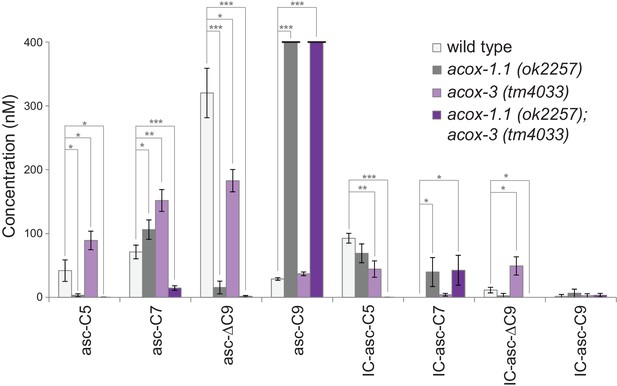
Ascaroside production in deletion mutants of acyl-CoA oxidase genes acox-1.1 and acox-3.
Comparison of ascaroside and corresponding IC-ascaroside production in wild type, the acox-1.1(ok2257) deletion mutant, the acox-3(tm4033) deletion mutant, and the acox-1.1(ok2257);acox-3(tm4033) double deletion mutant. Data represent the mean ± SD of three independent experiments. Two-tailed, unpaired t-tests were used to determine statistical significance (*p≤0.05, **p≤0.01, ***p≤0.001).
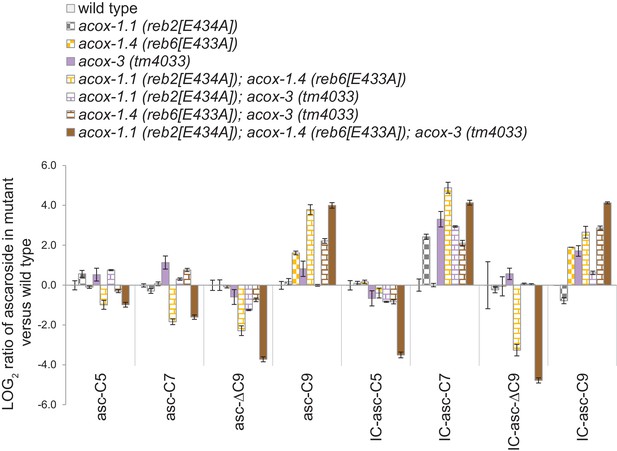
Ascaroside production in wild type, the acox-1.1(reb2[E434A]) catalytic mutant, the acox-1.4(reb6[E433A]) catalytic mutant, the acox-3(tm4033) deletion mutant, and double mutant and triple mutants.
Log ratio of ascaroside production in wild type and mutants, relative to wild type. For ascarosides that could not be detected, their peak area was set at the detection limit of the LC-MS. Data represent the mean ± SD of three independent experiments.
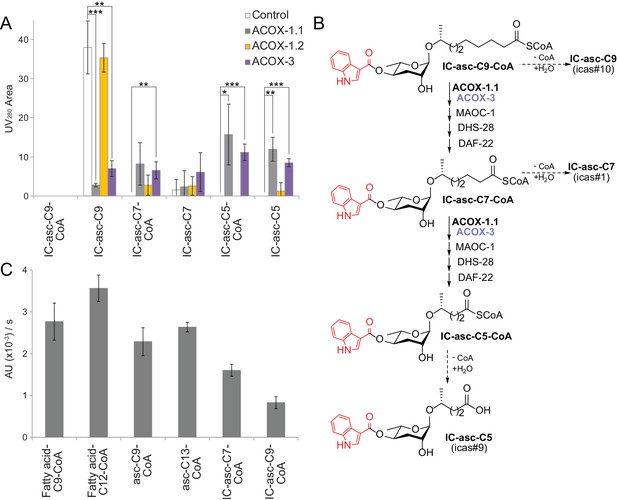
In vitro activity of ACOX-1.1, ACOX-1.2, and ACOX-3.
(A) Reaction of IC-asc-C9-CoA with ACOX-1.1, ACOX-1.2, or ACOX-3, in the presence of the additional β-oxidation enzymes, MAOC-1, DHS-28, and DAF-22, as monitored by LC-MS. Chemical structures are shown in (B). Data represent the mean ± SD of three independent experiments. Two-tailed, unpaired t-tests were used to determine statistical significance (*p≤0.05, **p≤0.01, ***p≤0.001). (B) Proposed role for ACOX-1.1 and ACOX-3 in the β-oxidation of IC-asc-C9-CoA to IC-asc-C5-CoA. (C) Activity of ACOX-1.1 in a coupled enzyme assay against the CoA-thioesters of fatty acids (fatty acid-C9-CoA and fatty acid-C12-CoA), ascarosides (asc-C9-CoA and asc-C13-CoA), and IC-modified ascarosides (IC-asc-C7-CoA and IC-asc-C9-CoA). Data represent the mean ± SD of three independent experiments.
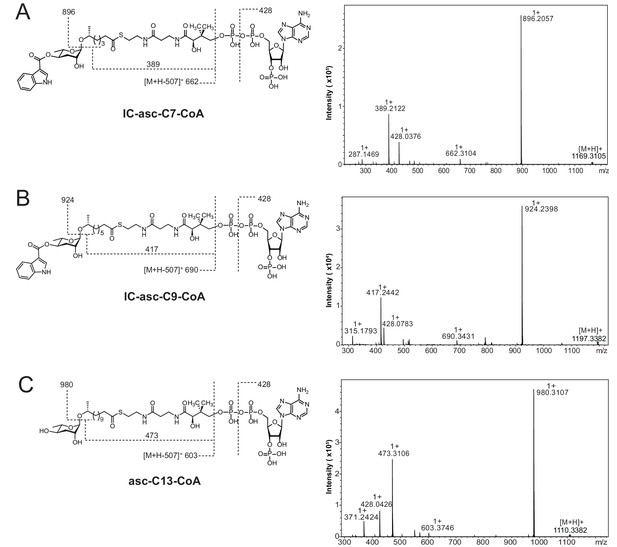
High-resolution LC-MS/MS analysis of CoA-thioesters of ascarosides produced chemoenzymatically and used as substrates in Figure 5A,C.
Structures with key product ions indicated (left) and MS-MS spectrum (right) for IC-asc-C7-CoA (A), IC-asc-C9-CoA (B), and asc-C13-CoA (C).
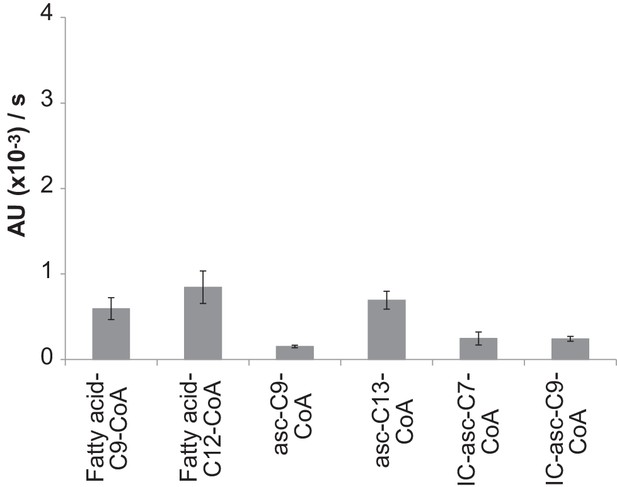
Activity of ACOX-3 towards different substrates.
Activity was monitored using the peroxidase-coupled in vitro activity assay. Data represent the mean ± SD of three independent experiments.
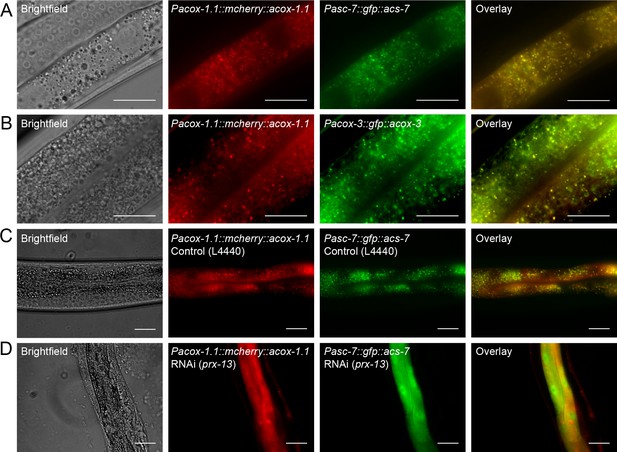
Peroxisomal co-localization of ACS-7, ACOX-1.1, and ACOX-3.
(A) Co-injection of plasmids containing Pacox-1.1::mcherry::acox-1.1 and Pacs-7::gfp::acs-7 into wild-type worms shows that ACOX-1.1 and ACS-7 co-localize in a punctate pattern in the intestine. (B) Co-injection of plasmids containing Pacox-1.1::mcherry::acox-1.1 and Pacox-3::gfp::acox-3 into wild-type worms shows that ACOX-1.1 and ACOX-3 co-localize in a punctate pattern in the intestine. (C) RNAi using the control plasmid L4440 in Pacox-1.1::mcherry::acox-1.1;Pacs-7::gfp::acs-7 worms gives a punctate pattern of expression in the intestine. (D) RNAi against prx-13 in Pacox-1.1::mcherry::acox-1.1; Pacs-7::gfp::acs-7 worms gives a diffuse pattern of expression in the intestine. In (A–D), L4 to young adult stage worms were imaged. Scale bar = 20 μm.
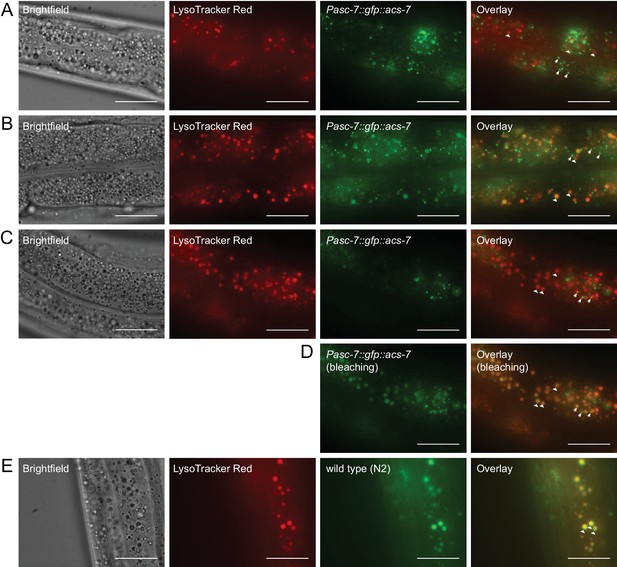
ACS-7 does not localize primarily to the lysosome.
(A) Staining of Pacs-7::gfp::acs-7 L4 worms with LysoTracker Red dye. (B) Staining of Pacs-7::gfp::acs-7 adult worms with LysoTracker Red dye. (C,D) Staining of Pacs-7::gfp::acs-7 adult worms with LysoTracker Red dye, before (C) and after (D) bleaching the GFP signal under the fluorescent microscope. Bleaching in (C) selectively eliminates peroxisomal GFP while making lysosomal autofluoresence more prominent. (E) Staining of wild-type (N2) worms with LysoTracker Red dye. In (A–E), peroxisomes are indicated with arrows and lysosomes are indicated with triangles. In (A), L3 worms are imaged, and in (B–E), L4 to adult worms are imaged. Scale bar = 20 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (C. elegans) | acox-1.1 | WBGene00008564 | Other names: CELE_F08A8.1, acox-1 | |
| acox-1.2 | WBGene00008565 | Other names: CELE_F08A8.2, acox-2, drd-100 | ||
| acox-1.3 | WBGene00008566 | Other names: CELE_F08A8.3, acox-3 | ||
| acox-1.4 | WBGene00008567 | Other names: CELE_F08A8.4, acox-4 | ||
| acox-1.5 | WBGene00008167 | Other names: CELE_C48B4.1, acox-5, drd-51 | ||
| acox-1.6 | WBGene00010336 | Other names: CELE_F59F4.1, acox-6 | ||
| acox-3 | WBGene00019060 | Other names: CELE_F58F9.7, acox-6 | ||
| acs-7 | WBGene00007228 | Other name: CELE_C01G6.7 | ||
| maoc-1 | WBGene00017123 | Other name: CELE_E04F6.3 | ||
| dhs-28 | WBGene00000991 | Other name: CELE_M03A8.1 | ||
| daf-22 | WBGene00013284 | Other name: CELE_Y57A10C.6 | ||
| Gene (M. tuberculosis) | fadD6 | Gene ID: 887549 | ||
| Strains (C. elegans) | N2 | CGC | RRID:WB-STRAIN:N2_(ancestral) | wild type (Bristol) |
| RAB1 | doi: 10.1021/acschembio.7b01021 | backcrossed VC1785 acox-1.1(ok2257)I | ||
| RAB21 | doi: 10.1021/acschembio.7b01021 | backcrossed acox-1.4(tm6415)I | ||
| VC40449 | CGC | RRID:WB-STRAIN:VC40449 | acox-1.4(gk892586)I | |
| RAB22 | doi: 10.1021/acschembio.7b01021 | backcrossed acox-3(tm4033)IV | ||
| DR476 | CGC | RRID:WB-STRAIN:DR476 | daf-22(m130)II | |
| RAB24 | doi: 10.1021/acschembio.7b01021 | acox-1.1(reb2[E434A])I | ||
| RAB28 | doi: 10.1021/acschembio.7b01021 | acox-1.4(reb6[E433A])I | ||
| RAB30 | doi: 10.1021/acschembio.7b01021 | acox-1.1(ok2257)I; acox-3(tm4033)IV | ||
| RAB31 | doi: 10.1021/acschembio.7b01021 | acox-1.1(reb2[E434A])I; acox-1.4(reb6[E433A])I | ||
| RAB32 | doi: 10.1021/acschembio.7b01021 | acox-1.1(reb2[E434A])I; acox-3(tm4033)IV | ||
| RAB35 | this paper | acox-1.4(reb6[E433A])I; acox-3(tm4033)IV | ||
| RAB36 | this paper | acox-1.1(reb2[E434A])I; acox-1.4(reb6[E433A])I; acox-3(tm4033)IV | ||
| RAB37 | this paper | rebEx11 (Pacs-7::gfp::acs-7) | ||
| RAB38 | this paper | rebEx12 (Pacox-1.1::mcherry::acox-1.1; Pacs-7::gfp::acs-7) | ||
| RAB39 | this paper | rebEx13 (Pacox-1.1::mcherry::acox-1.1; Pacox-3::gfp::acox-3) | ||
| RAB40 | this paper | rebEx14 (Pacox-1.1::mcherry::acox-1.1; Pacs-7::gfp::acs-7) | ||
| Plasmids for protein expression | pET-22b-acs-7 | this paper | Cloning described in 'ACS-7 expression and mutagenesis' | |
| pET-22b-acs-7(E339A) | this paper | Cloning described in 'ACS-7 expression and mutagenesis' | ||
| pET-22b-acs-7(E339A,S186A,S187A) | this paper | Cloning described in 'ACS-7 expression and mutagenesis' | ||
| pET-16b-acox-1.1a | doi: 10.1073/pnas.1423951112 | |||
| pET-16b-acox-1.2 | doi: 10.1073/pnas.1423951112 | |||
| pET-16b-acox-3 | this paper | Cloning described in 'β-oxidation enzyme expression' | ||
| pACYCDuet-1-maoc-1 | this paper | Cloning described in 'β-oxidation enzyme expression' | ||
| pACYCDuet-1-dhs-28Δscp-2 | this paper | Cloning described in 'β-oxidation enzyme expression' | ||
| pET-16b-daf-22 | this paper | Cloning described in 'β-oxidation enzyme expression' | ||
| pET-28a-fadD6 | doi: 10.1021/acschembio.7b01021 | Cloning described in 'Synthesis of CoA-thioesters' | ||
| Plasmids for generating transgenic strains | pPD114.108-Pacox-1.1::mcherry::acox-1.1 | this paper | Cloning described in 'ACOX-1.1, ACOX-3, and ACS-7 localization and RNAi' | |
| pPD114.108-Pacs-7::gfp::acs-7 | this paper | Cloning described in 'ACOX-1.1, ACOX-3, and ACS-7 localization and RNAi' | ||
| pPD114.108-Pacox-3::gfp::acox-3 | this paper | Cloning described in 'ACOX-1.1, ACOX-3, and ACS-7 localization and RNAi' | ||
| Commercial kit | Q5 Site-Directed Mutagenesis Kit | New England Biolabs | E0554S | |
| Commercial enzymes | adenylate kinase | Sigma | M3003 | |
| pyruvate kinase and lactate dehydrogenase | Sigma | P0294 | ||
| Commercial compounds | LysoTracker Red (Deep Red) | Thermo Fisher | L12492 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33286.013





