Dual leucine zipper kinase is required for mechanical allodynia and microgliosis after nerve injury
Figures
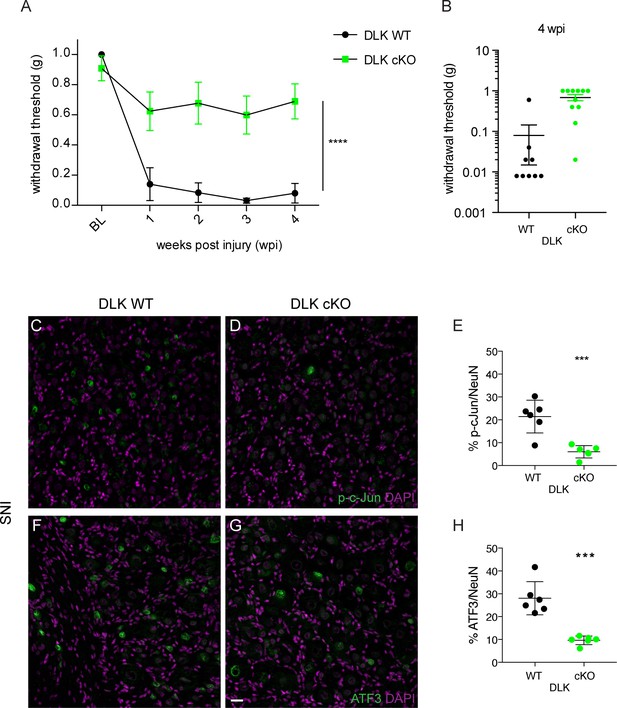
DLK is necessary for the development of mechanical allodynia and induction of injury markers after spared nerve injury (SNI).
(A) DLK deletion is protective in the von Frey behavior assay following SNI at 1, 2, 3 and 4 weeks post injury (wpi). Baseline (BL) mechanical sensitivity thresholds are comparable between DLK WT (n = 9) and DLK cKO (n = 11). ****p<0.0001 by repeated measures ANOVA. (B) Scatter graph to illustrate individual points for DLK WT and DLK cKO at 4 weeks post injury. (C–H) Deletion of DLK reduces injury-induced transcription factors p-c-Jun and ATF3 in DRG following SNI. Representative images of ipsilateral L4 DRG 1 week after SNI stained for p-c-Jun in DLK WT (C) and DLK cKO (D) quantified in (E), and for ATF3 in DLK WT (F) and DLK cKO (G) quantified in (H). n = 6 per genotype. Scale bar in (G) valid for all images in this figure: 20 µm. ***p<0.001 by 2-tailed Student’s t test.
-
Figure 1—source data 1
data plotted in Figure 1A; von Frey behavior testing in DLK WT and cKO mice.
- https://doi.org/10.7554/eLife.33910.006
-
Figure 1—source data 2
data plotted in Figure 1E; p-cJun-positive nuclear counts in ipsilateral L4 DRG 7 days after SNI in DLK WT vs cKO mice.
- https://doi.org/10.7554/eLife.33910.007
-
Figure 1—source data 3
data plotted in Figure 1H; ATF3-positive nuclear counts in ipsilateral L4 DRG 7 days after SNI in DLK WT vs cKO mice.
- https://doi.org/10.7554/eLife.33910.008
-
Figure 1—source data 4
data plotted in Figure 1—figure supplement 1; dynamic brush scores 7 weeks after SNI in DLK WT vs cKO mice.
- https://doi.org/10.7554/eLife.33910.009
-
Figure 1—source data 5
data plotted in Figure 1—figure supplement 2; von Frey testing in DLK WT vs cKO mice at 1, 3, 5 and 7 days post SNI.
- https://doi.org/10.7554/eLife.33910.010
-
Figure 1—source data 6
data plotted in Figure 1E; p-cJun-positive nuclear counts in ipsilateral L4 DRG 48 hr after hind paw formalin injection in DLK WT vs cKO mice.
- https://doi.org/10.7554/eLife.33910.011
-
Figure 1—source data 7
data plotted in Figure 1E; ATF3-positive nuclear counts in ipsilateral L4 DRG 48 hr after hind paw formalin injection in DLK WT vs cKO mice.
- https://doi.org/10.7554/eLife.33910.012
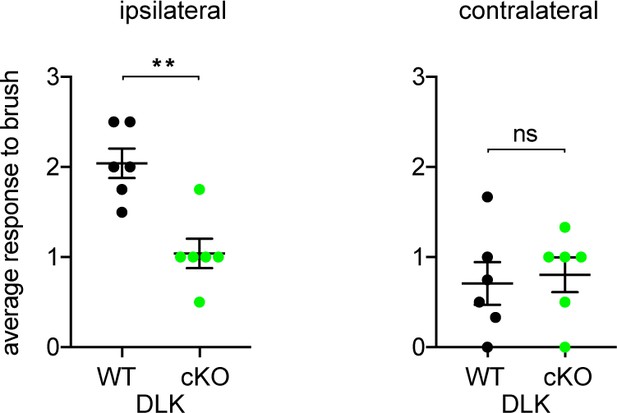
DLK deletion prevents allodynia to dynamic mechanical stimulus after spared nerve injury (SNI).
Dynamic brush testing at 7 weeks post injury showing that DLK cKO mice display less nocifensive and protective behaviors compared with DLK WT littermates (n = 6 per genotype group; 3 males and three females). Each data point represents the average of 4 trials per animal. Score scale: 0 – rapid paw lifting; 1 – paw guarding; 2 – strong paw lifting/kicking; 3 – paw licking. **p<0.01 by 2-tailed Student’s t test. ns: not significant.
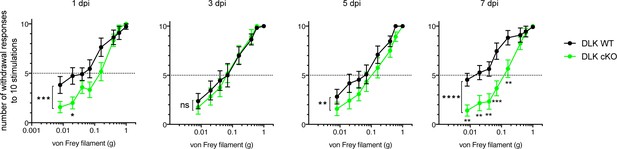
DLK is necessary for the development of mechanical allodynia after spared nerve injury (SNI).
Von Frey testing at acute time points 1, 3, 5, and 7 days post injury (dpi) in DLK cKO (n = 11, 5 females and 6 males) and DLK WT littermates (n = 12, 6 per sex). DLK deletion is protective in the von Frey behavior assay following SNI at 1, 5, and 7 days post injury (dpi) by ordinary two-way ANOVA with filament size and genotype as factors. P values for a genotype effect were 0.0002 (1 dpi), 0.0038 (5 dpi), and p<0.0001 (7 dpi). Asterisks under green data points represent significant p values for differences between genotypes at individual filaments by Sidak’s multiple comparisons post test. *p<0.05, **p<0.01, ***p<0.001. The apparent hypersensitivity of the DLK WT group was reduced at 3 and 5 dpi, perhaps due to the aversive nature of paw withdrawal causing additional pain due to movement during this acute recovery period 1–5 days after injury.
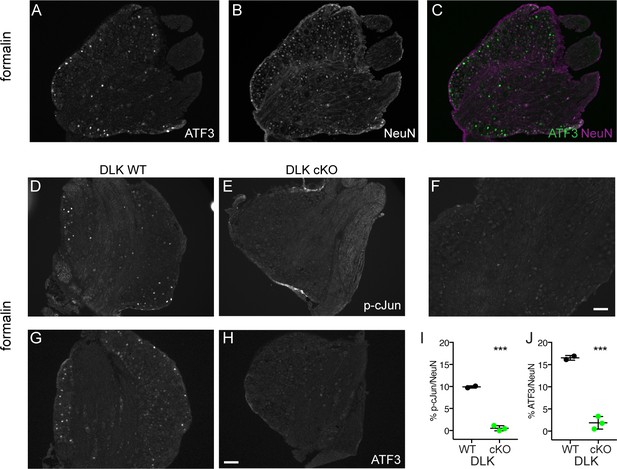
DLK deletion reduces injury markers in DRG in the formalin model of neuropathic pain.
(A–C) Example images of co-immunostaining for p-c-Jun (injury marker) and NeuN (all neuronal nuclei) in a wild type DRG to illustrate the % NeuN-positive neuron counts that were done for the data shown in Figure 1C–H and I–J in this figure. (D–J) Deletion of DLK reduces injury-induced transcription factors p-c-Jun and ATF3 in DRG following nerve injury by intraplantar formalin injection. Representative images of ipsilateral L4 DRG 48 hr after formalin treatment stained for p-c-Jun in DLK WT (D) and DLK cKO (E) quantified in (I), and for ATF3 in DLK WT (G) and DLK cKO (H) quantified in (J). (DLK WT n = 2; DLK cKO n = 3). (F) Representative image of p-cJun staining in a contralateral L4 DRG showing that p-cJun is normally low; compare with injury-induced p-cJun in D). Scale bar in (H) 100 µm, valid for all images in this figure except in (F) where scalebar = 50 µm). ***p<0.001 by two-tailed Student’s t test.
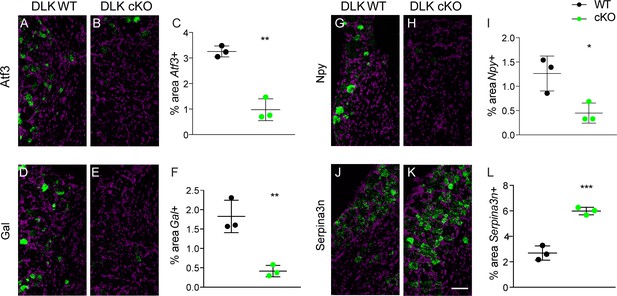
DLK is upstream of injury-associated genesimplicated in the establishment of neuropathic pain: Atf3, Gal, Npy, and Serpina3n.
Representative images and quantification of expression of 4 genes whose expression changes following sciatic nerve axotomy: Atf3 (A–C), Gal (D–F), Npy (G–I), and Serpina3n (J–L) by in situ hybridization in DLK WT vs DLK cKO ipsilateral L3 DRG 1 week after SNI (7 dpi). Scale bar 100 µm. *p<0.05, **p<0.01, ***p<0.001 by 2-tailed Student’s t test. Averaged data for n = 4–6 sections quantified in n = 3 male mice per genotype.
-
Figure 2—source data 1
data plotted in Figure 2; quantification of in situ hybridization for Atf3, Gal, Npy and Serpina3n in DLK WT vs cKO in ipsilateral L4 DRG 7 days post SNI.
- https://doi.org/10.7554/eLife.33910.016
-
Figure 2—source data 2
data plotted in Figure 2—figure supplement 1; quantification of in situ hybridization for Cckbr, Ecel1, Gpr151, Nts, Sox11, and Sprr1a in DLK WT vs cKO in ipsilateral L4 DRG 7 days post SNI.
- https://doi.org/10.7554/eLife.33910.017
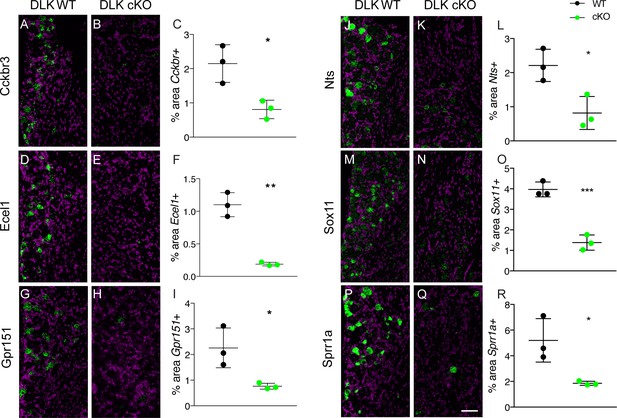
DLK is upstream of multiple genes implicated in the establishment of neuropathic pain.
(A–R) Representative images and quantification of expression of 6 genes whose expression changes following sciatic nerve axotomy: Cckbr (A–C), Ecel1 (D–F), Gpr151 (G–I), Nts (J–L), Sox11 (M–O), Sprr1a (P–R) by in situ hybridization in DLK WT vs DLK cKO L3 DRG 1 week after SNI. Scale bar 100 µm. *p<0.05 and **p<0.01 by 2-tailed Student’s t test. Averaged data for n = 4–6 sections quantified for n = 3 male mice per genotype.
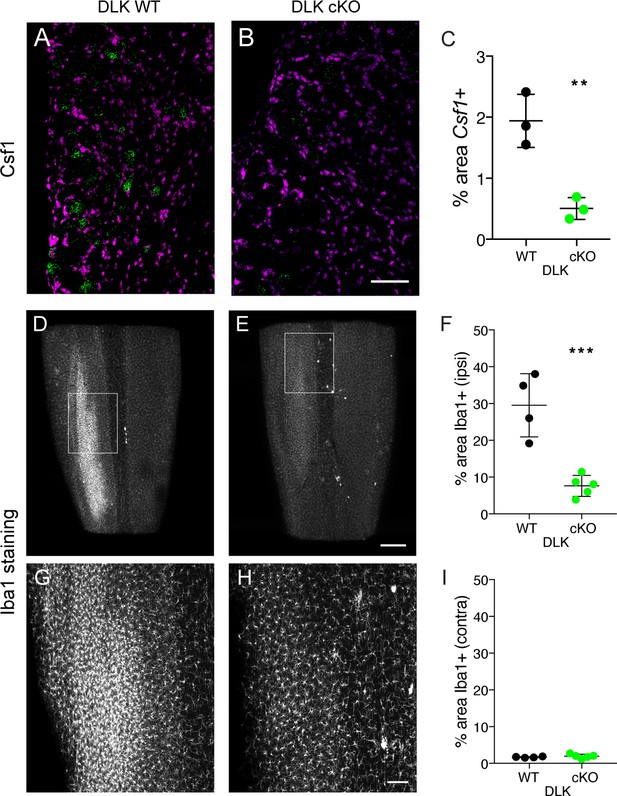
DLK is necessary for Csf1 upregulation in DRG neurons and for spinal cord microgliosis elicited by SNI.
(A–C) Representative images and quantification of Csf1 gene expression changes following SNI by in situ hybridization in DLK WT vs DLK cKO L3 DRG 7 days post SNI. Scale bar 100 µm. **p<0.01, by 2-tailed Student’s t test. Averaged data for n = 4–6 sections quantified in n = 3 mice per genotype. (D–I) Representative images and quantification of microgliosis in DLK WT vs DLK cKO spinal cords 8 days post-SNI. Iba1 staining in cleared spinal cords (top view, longitudinal plane) showing the SNI-induced microgliosis in the ipsilateral (left) dorsal horn of DLK WT spinal cord (D, G) that does not occur in the DLK cKO (E, H). (G) and (H) are higher magnification images of boxed areas in (D) and (E) respectively. Quantification of dorsal horn microgliosis in ipsilateral (F) and contralateral (I) spinal cord of DLK WT (n = 4) vs DLK cKO (n = 5). Scale bar in (D, E) 400 µm. Scale bar in (G, H) 100 µm. ***p=0.0010 by 2-tailed Student’s t test.
-
Figure 3—source data 1
data plotted in Figure 3C; quantification of in situ hybridization for Csf1 in DLK WT vs cKO in ipsilateral L4 DRG 7 days post SNI.
- https://doi.org/10.7554/eLife.33910.019
-
Figure 3—source data 2
data plotted in Figure 3F; quantification of Iba1-positive signal in DLK WT vs cKO dorsal spinal cord ipsilateral to injury at 8 dpi.
- https://doi.org/10.7554/eLife.33910.020
-
Figure 3—source data 3
data plotted in Figure 3I; quantification of Iba1-positive signal in DLK WT vs cKO dorsal spinal cord contralateral to injury at 8 dpi.
- https://doi.org/10.7554/eLife.33910.021
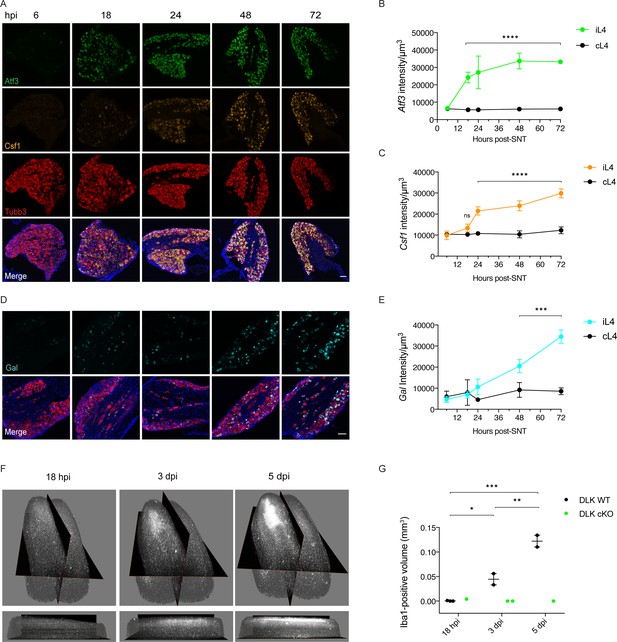
Time course of DLK-dependent gene expression in DRG neurons and corresponding spinal cord microgliosis.
(A–C) Representative images and quantification of gene expression changes in ipsilateral L4 DRGs of WT mice 6, 18, 24, 48 and 72 hr post-SNT (sciatic nerve transection). (A) and (D) Representative images of Atf3 (green), Csf1 (orange), Gal (cyan) and Tubb3 (red) in situ hybridizations in iL4 DRGs after SNT. Scale bars 100 μm. Quantification of Atf3 (B), Csf1 (C), and Gal (E) expression changes in L4 DRGs ipsilateral (green, orange, cyan iL4) vs contralateral (black, cL4) to injury 6–72 hr post-SNT. n = 4 (2 males, 2 females) for 6, 18, 24, and 48 hr groups, n = 2 (1 male, 1 female) for 72 hr group. ns: not significant; ***p<0.001, ****p<0.0001 by 2-way ANOVA followed by Sidak’s multiple comparisons post test comparing values between ipsilateral and contralateral L4 DRG at each time point. Note in (D), in representative images at 48 and 72 hr, that Gal comes on in fewer cells than for Atf3 and Csf1 (A), and these include a mixture of large and small diameter neurons. (F–G) Iba1 immunolabeling in cleared lumbar spinal cord shows no region of microglial reactivity at 18 hr post-SNI. Microgliosis becomes evident by 3 dpi and robust by 5 dpi. Representative images of 3D views from the top and the ipsilateral side (F) and quantification (G). DLK cKO prevents microgliosis in lumbar spinal cord 3 and 5 days post-SNI, as observed previously at 7 dpi (Figure 3D–H). *p<0.05, **p<0.01, ***p<0.001 by one-way ANOVA followed by Tukey’s multiple comparisons.
-
Figure 4—source data 1
data plotted in Figure 4B,C and E; quantification of time course of expression of Atf3, Csf1 and Gal in ipsilateral L4 DRGs normalized by total neuronal volume sampled as labeled by Tubb3.
- https://doi.org/10.7554/eLife.33910.024
-
Figure 4—source data 2
data plotted in Figure 4G; quantification of Iba1-positive volume in dorsal spinal cord ipsilateral to injury at the indicated time points.
DLK cKO samples are provided for comparison.
- https://doi.org/10.7554/eLife.33910.025
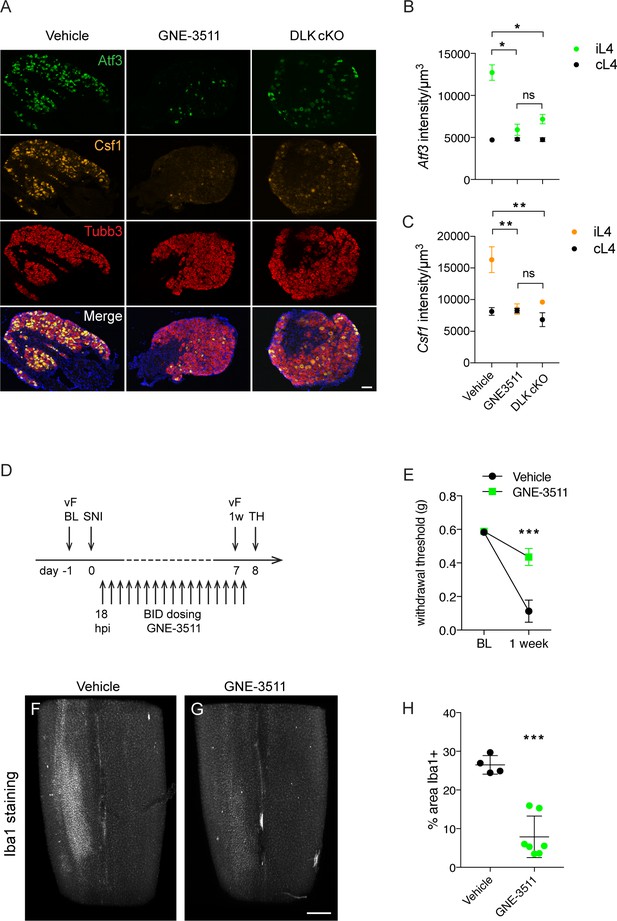
DLK inhibition prevents DLK-dependent gene upregulation, mechanical allodynia, and microgliosis after SNI.
(A–C) GNE-3511 treatment prevents upregulation of DLK-dependent genes in the DRG after SNI. (A) Representative images of Atf3 (green), Csf1 (orange), and Tubb3 (red) in situ hybridizations in ipsilateral L4 DRGs from vehicle-treated, GNE3511-treated, and DLK cKO mice 5 days post-SNI. Scale bar 100 μm. (B, C) Quantification of neuronal gene expression intensity in ipsilateral L4 DRGs at 5 dpi showing reduced expression of Atf3 (B) and Csf1 (C) in DLK cKO and GNE-3511 vs vehicle treated mice (n = 2 per group). *p<0.05, **p<0.01 by Tukey’s multiple comparisons test. (D) DLK inhibitor study design. vF: von Frey; BL: baseline; SNI: spared nerve injury; 1 w: 1 week post SNI; TH: tissue harvest. Twice daily dosing of GNE-3511 began at 18 hr post injury (hpi). (E) DLK inhibitor GNE-3511 prevents mechanical allodynia measured 1 week post injury. Vehicle n = 12; GNE-3511 n = 17. ***p=0.0004 for genotype by 2-way ANOVA. (F–H) GNE-3511 prevents the spinal cord microgliosis elicited by SNI as shown by Iba1 staining in cleared immunostained lumbar spinal cord. Representative images from vehicle (F) vs GNE-3511 treated mice (G) and quantification of Iba1-positive signal in ipsilateral spinal cord (H). Vehicle n = 4; GNE-3511 n = 7. ***p=0.0001 by 2-tailed Student’s t test. Scale bar in (F), (G) 400 µm.
-
Figure 5—source data 1
data plotted in Figure 5B,C; quantification of in situ signal intensity at 5 dpi for Atf3 and Csf1 in ipsilateral and contralateral L4 DRGs normalized by total neuronal volume sampled as labeled by Tubb3.
- https://doi.org/10.7554/eLife.33910.027
-
Figure 5—source data 2
data plotted in Figure 5E; von Frey thresholds (>5 responses out of 10 stimulations) in vehicle vs GNE-3511-treated at 7 dpi.
- https://doi.org/10.7554/eLife.33910.028
-
Figure 5—source data 3
data plotted in Figure 5H; quantification of Iba1-positive signal in vehicle vs GNE-3511-treated dorsal spinal cord ipsilateral to injury at 8 dpi.
- https://doi.org/10.7554/eLife.33910.029
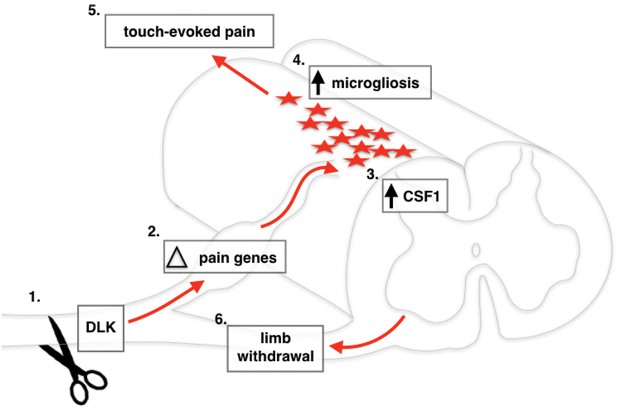
Schematic of DLK activation in DRG neurons following spared nerve injury.
Nerve injury activates DLK (Ji and Strichartz, 2004) which induces a transcriptional response in DRG neurons (Basbaum et al., 2009), including Csf1 upregulation (von Hehn et al., 2012) which leads to microgliosis in the spinal cord (Decosterd and Woolf, 2000). Overall, the transcriptional changes induced by DLK lead to the establishment of touch-evoked pain (Bourquin et al., 2006) as evidenced by the paw withdrawal reflex (Svensson et al., 1993).
Videos
untreated spinal cord, 5 dpi
https://doi.org/10.7554/eLife.33910.022GNE-3511-treated spinal cord, 5 dpi
https://doi.org/10.7554/eLife.33910.030Tables
Top 25 genes with altered expression 7 days following sciatic nerve transection from (Guan et al., 2016)
Genes are sorted by significance, then by fold change. The genes we examined for DLK-dependence by in situ hybridization are indicated by an asterisk *
| Gene | log2 fold_change | |
|---|---|---|
| 1 | Sprr1a * | 6.96879 |
| 2 | Npy * | 5.89862 |
| 3 | Gpr151 * | 5.72999 |
| 4 | Cckbr * | 5.35581 |
| 5 | Ecel1 * | 4.97284 |
| 6 | Atf3 * | 4.95333 |
| 7 | Gal * | 4.77653 |
| 8 | Nts * | 4.60215 |
| 9 | Hrk | 4.55133 |
| 10 | Fst | 4.09785 |
| 11 | Lmo7 | 3.76662 |
| 12 | Car3 | 3.48156 |
| 13 | Sox11 * | 3.37483 |
| 14 | Sema6a | 3.32589 |
| 15 | Mmp16 | 3.31338 |
| 16 | Flrt3 | 3.28661 |
| 17 | Loxl2 | 3.25419 |
| 18 | Flnc | 3.21128 |
| 19 | Sez6l | 3.20287 |
| 20 | Adam8 | 2.98222 |
| 21 | Stmn4 | 2.89975 |
| 22 | Xdh | 2.77339 |
| 23 | Gadd45a | 2.66746 |
| 24 | Sdc1 | 2.59248 |
| 25 | Csf1 * | 2.51895 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay/kit- RNAscope | RNAscope Fluorescent Multiplex Detection Reagents | Advanced Cell Diagnostics | ACD:320851 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Atf3; Atf3 | Advanced Cell Diagnostics | ACD:426891 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Serpina3n; Serpina3n | Advanced Cell Diagnostics | ACD:430191 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Csf1; Csf1 | Advanced Cell Diagnostics | ACD:315621 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Tubb3; Tubb3 | Advanced Cell Diagnostics | ACD:423391 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Gal; Galanin; Gal | Advanced Cell Diagnostics | ACD:400961 | |
| Commercial Assay/kit- RNAscope | RNAscope Probe-Mm-Npy; Npy | Advanced Cell Diagnostics | ACD:313321 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Cckbr; Cckbr | Advanced Cell Diagnostics | ACD:439121 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Ecel1; Ecel1 | Advanced Cell Diagnostics | ACD:475331 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Gpr151; Gpr151 | Advanced Cell Diagnostics | ACD:317321 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Nts; Nts | Advanced Cell Diagnostics | ACD:420441 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Sox11; Sox11 | Advanced Cell Diagnostics | ACD:440811 | |
| Commercial assay/kit- RNAscope | RNAscope Probe-Mm-Sprr1a; Sprr1a | Advanced Cell Diagnostics | ACD:426871 | |
| Antibody | anti-phospho-c-Jun S63 (rabbit polyclonal) | Cell Signaling | Cell Signaling:9261; RRID:AB_2130162 | 1:500 |
| Antibody | anti-NeuN (mouse monoclonal) | Millipore | Millipore:MAB377; RRID:AB_2298772 | 1:300 |
| Antibody | anti-ATF3 (rabbit polyclonal) | Santa Cruz | Santa Cruz:sc-188; RRID:AB_2258513 | 1:3000 |
| Antibody | anti-Iba1 (rabbit polyclonal) | Wako | Wako:019–19741; RRID:AB_2665520 | 1:200; 1:600 |
| Antibody | Donkey anti-rabbit AlexaFluor 647 | ThermoFisher | ThermoFisher:A31573; RRID:AB_2536183 | 1:500 for immunostaining; 1:600 for iDisco |
| Antibody | Donkey anti-rabbit AlexaFluor 594 | ThermoFisher | ThermoFisher:A21207; RRID:AB_141637 | 1:500 for immunostaining; 1:600 for iDisco |
| Antibody | Donkey anti-mouse AlexaFluor 488 | ThermoFisher | ThermoFisher:A21202; RRID:AB_141607 | 1:500 for immunostaining; 1:600 for iDisco |
| Strain, strain background (mus musculus) | Inbred strain C57BL/6J (mus musculus) | The Jackson Laboratory | Jax:000664; RRID:IMSR_JAX:000664 | |
| Strain, strain background (mus musculus) | STOCK Tg(CAG-cre/Esr1*) 5Amc/J, C57BL/6J (mus musculus) | The Jackson Laboratory | Jax:004453; RRID:IMSR_JAX:004453 | |
| Strain, strain background (mus musculus) | Map3k12 flox mouse, C57BL/6J (mus musculus) | Genentech, by MTA; PMID 24166713 | ||
| Chemical compound, drug | small molecule GNE-3511 (DLK inhibitor) | Genentech, by MTA; PMID 25341110 | ||
| Software, algorithm | FIJI | NIH | RRID:SCR_002285 | |
| Software, algorithm | arivis Vision 4D | Arivis |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33910.032





