Spontaneous dormancy protects Trypanosoma cruzi during extended drug exposure
Figures
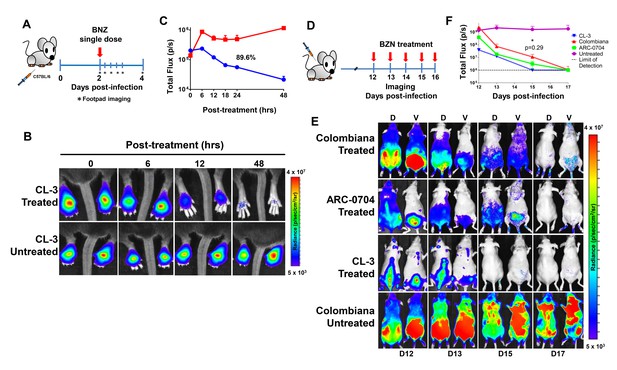
Rapid decrease of parasite load following short-term BZN treatment.
(A) Schematic experimental protocol. C57BL/6 mice were infected in the hind footpads with 2 × 105 luciferase-expressing T. cruzi trypomastigotes of the CL-3 strain. A single oral dose of BZN (100 mg/kg) was administered 2 days post-infection. Cohorts of mice were maintained as untreated controls. Parasite bioluminescence following D-luciferin injection was measured at 6, 12, 18, 24 and 48 hr after BZN dosing. (B) Representative images showing footpad bioluminescent signal 2 days post-infection and at 6, 12 and 48 hr post-treatment. The heat map is on a log10 scale and indicates the intensity of bioluminescence from low (blue) to high (red). (C) Quantification of footpad signal. Each data point represents the mean of 12 footpads bioluminescence from six mice expressed on a logarithmic scale. After subtraction of the background signal, total flux measurements of photons per second (p/s) were quantified. A statistically significant difference (p=0.004) was found between treated (blue) and untreated (red) groups. (D) Protocol to measure systemic parasite load by bioluminescence before and during daily dosing with BZN. Cohorts of six SKH-1/B6 (hairless C57BL/6) mice were infected i.p. with 5 × 105 colombiana, ARC-0704 or CL-3 luciferase-expressing T. cruzi isolates. Oral BZN-treatment (100 mg/kg/day) was initiated on day 12 and continued until day 17 post-infection. (E) Dorsal (D) and ventral (V) images of individual representative infected mice. (F) Quantification of whole animal ventral bioluminescence. Dashed line indicates detection threshold determined as the mean plus two standard deviation of background bioluminescence of uninfected mice. Untreated control animals infected with all the strains showed similar high parasite levels, representative images (E) and data (F) for the group infected with colombiana strain are shown. No statistical differences were observed in the rate of parasite decrease between strains (p=0.29). Results are representative of three independent experiments with six infected animals per parasite strain group.
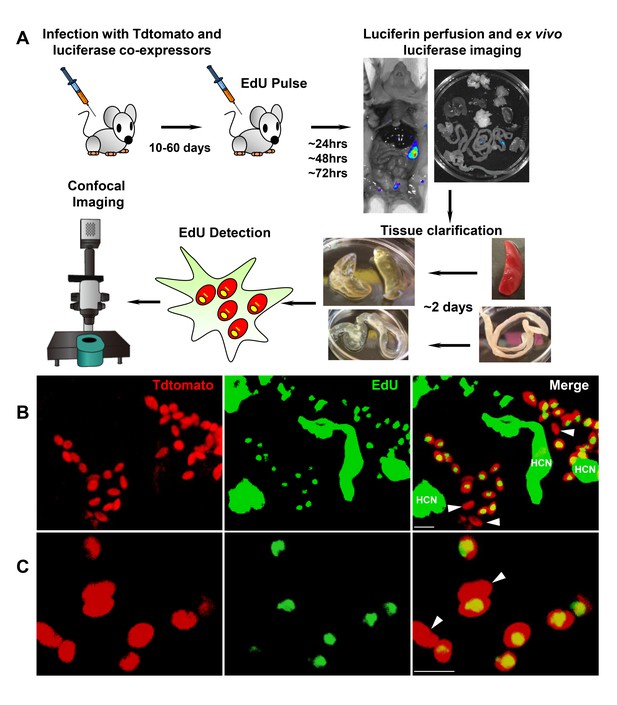
Rare amastigotes fail to incorporate EdU in chronically infected mice.
(A) Experimental protocol for assessing proliferation of amastigotes in established in vivo infections. C57BL/6 mice were infected with 2.5 × 105 trypomastigotes of colombiana T. cruzi strain co-expressing fluorescent (Tdtomato) and luminescent (luciferase) reporter proteins. Sixty days post-infection, mice were injected i.p. with EdU and sacrificed 24, 48 or 72 hr after injection. Mice were perfused with PBS and luciferin and ex vivo bioluminescence imaging of selected tissues were performed to identify parasite foci. Luciferase-positive thick tissue sections were clarified and Tdtomato + parasites and EdU incorporation was detected by confocal microscopy. Colocalization of Tdtomato (red) and EdU (green) positive signals identifies proliferating amastigotes (yellow nuclei) from (B) skeletal and (C) adipose tissue. Arrows indicate rare red only (non-proliferating) amastigotes. Scale bars, 5 μm. HCN = EdU positive host cell nuclei. Results are representative of three independent experiments using groups of 2–3 mice.
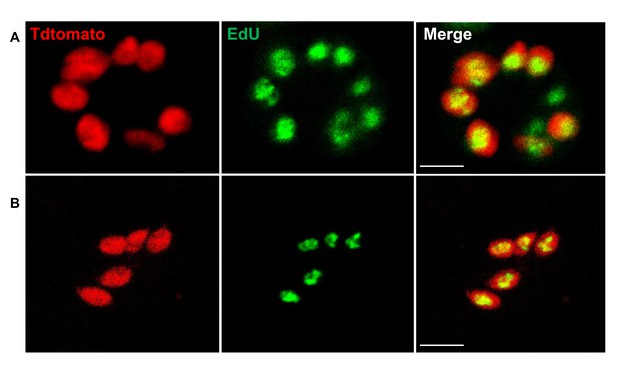
Detection of proliferating amastigotes in acutely infected mice.
(A and B) The protocol used for the detection of proliferating amastigotes in chronic infections (Figure 2A) was standardized in acutely infected IFN-γ knockout mice and in immunocompetent C57BL/6. Mice were infected with 2.5 × 105 trypomastigotes of colombiana strain co-expressing Tdtomato and luciferase reporters. Fifteen days post-infection, mice were EdU-pulsed by ip. injection and 24 hr later, euthanized and transcardially perfused with luciferin. Whole mice ex vivo luciferase imaging was used to identify luciferase-positive thick tissue sections. Consecutive imaging and sectioning was performed to identify the luciferase-bright tissue regions. Tissue sections were clarified using CUBIC protocols and EdU incorporation was detected by click chemistry. Overlay of Td-tomato (red) and EdU (green) signals revealed the proliferating amastigotes (yellow nuclei). Scale bars, 5 μm. Results are representative of two independent experiments with groups of 6 mice.
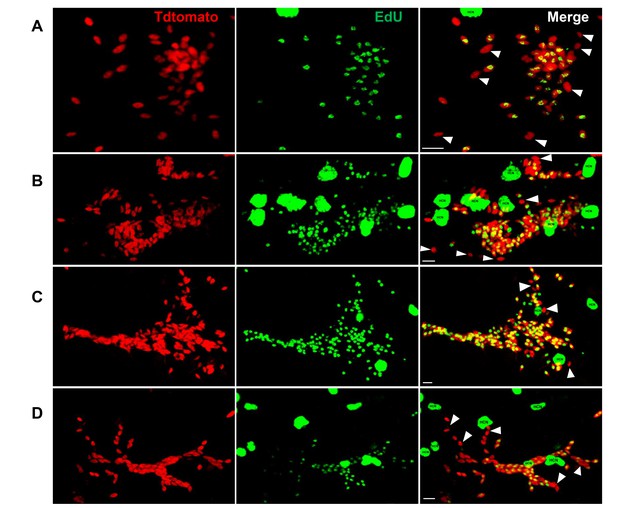
Detection of non-proliferating amastigotes in established infections.
C57B/6 mice were infected with 2.5 × 105 trypomastigotes of the colombiana T. cruzi strain co-expressing Tdtomato and luciferase reporter proteins. Sixty days post-infection, mice were injected with EdU and sacrificed (A) 24, (B) 48 or (C and D) 72 hr after injection. Mice were perfused with PBS and luciferin. Ex vivo tissue-specific bioluminescence imaging identified foci of persisting parasites and luciferase-positive thick tissue sections were clarified and EdU incorporation was detected by confocal microscopy in (A and B) skeletal muscle and (C and D) adipose tissue. Yellow nuclei revealed the co-localization of Tdtomato and EdU signals on replicating amastigotes. Arrows indicate non-proliferating amastigotes (red). Scale bars, 5 μm. HCN, host cell nuclei. Results are representative of three independent experiments using groups of 2–3 mice.
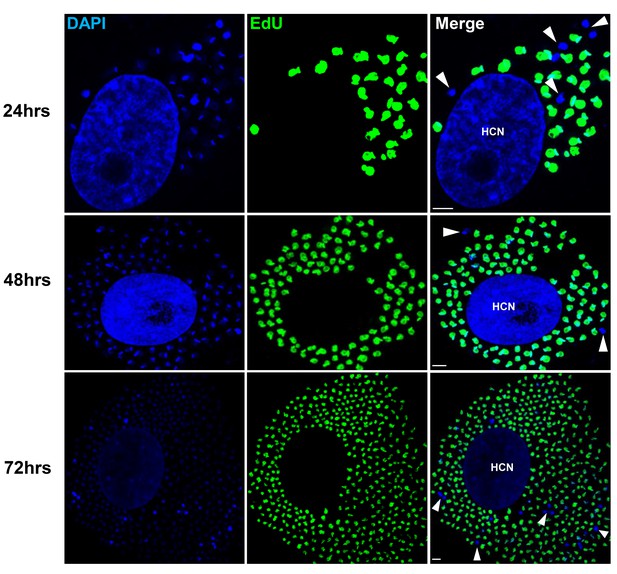
Non-replicating amastigotes are also evident in vitro.
Nearly confluent monolayers of Vero cells were infected with trypomastigotes of colombiana, CL, Brazil or ARC-0704 T. cruzi strain. Twelve hours after infection, cultures were washed and incubated with EdU (100 µM) diluted in fresh RPMI medium. EdU was detected after an additional 24, 48 or 72 hr culture period. DAPI staining (blue) allows the identification of EdU-negative amastigotes (arrows) from the EdU-positive (green). HCN, host cell nuclei. Scale bars, 5 μm. Results are representative of three independent experiments.
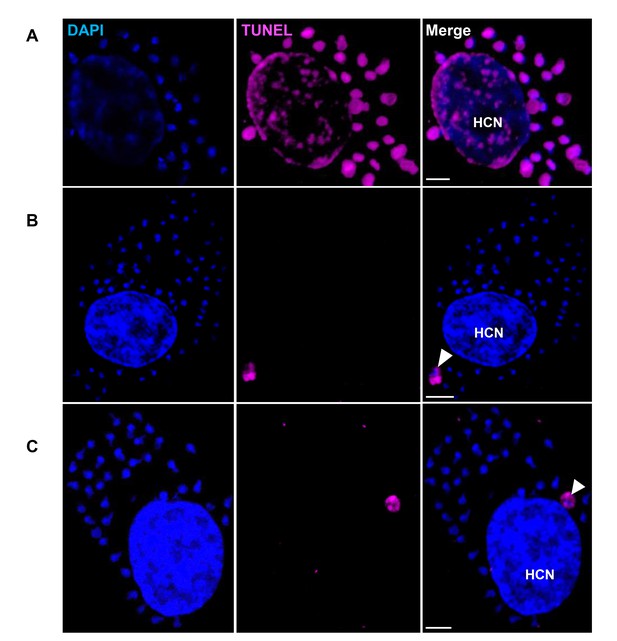
Rare apoptosis of amastigotes in host cells.
Trypomastigotes from the T. cruzi colombiana strain were used to infect monolayers of Vero cells. Infected cells were fixed, permeabilized and the TUNEL assay was used to detect DNA fragmentation in intracellular amastigotes 72 hr post-infection. DAPI staining was used to identify DNA-containing organelles (nuclei and kinetoplast). (A) DNase-treated cells were used as TUNEL-positive controls. (B and C) Arrows show examples of the 2–3 apoptotic amastigotes detected upon scanning approximately 1 × 106 infected host cells. Scale bars, 5 μm. HCN, host cell nuclei. Results are representative of two independent experiments.
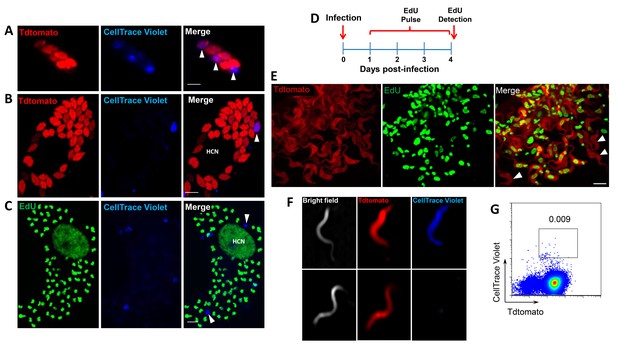
Non-proliferating amastigotes can transition to trypomastigotes.
(A, B) Td-tomato expressing T. cruzi trypomastigotes were labeled with CellTrace Violet and used to infect Vero cells. Amastigote replication was monitored for 72–96 hr post-infection by live imaging (A) and confocal microscopy (B). Note the violet signal dilution in proliferating amastigotes and the retention of the dye in the non-replicating amastigote (arrows). (C) Vero cell cultures infected with CellTrace Violet-labeled T. cruzi trypomastigotes were incubated with EdU (100 µM) for 72 hr before fixation and detection of EdU incorporation. CellTrace Violet-positive amastigotes were also EdU-negative (arrow). Experiment was performed three times. (D) Schematic of the experimental protocol. T. cruzi colombiana strain trypomastigotes expressing Tdtomato were used to infect nearly confluent monolayers of Vero cells. Twelve hours after infection, the dishes were washed and incubated with EdU (100 µM) for 72 hr. Cultures were fixed, permeabilized and EdU (green) detection was performed just prior to trypomastigote release from host cells (~96 hr post-infection). (E) Arrowheads indicate EdU-negative trypomastigotes (red only) within host cells filled with EdU-positive trypomastigotes (green). (F, G) Violet-labeled Td-tomato-expressing trypomastigotes (colombiana or CL strains) were used to infect Vero cell cultures. Trypomastigotes released 92 hr post-infection were harvested by centrifugation and flow-imaged by ImageStream and sorted via fluorescence-activated flow sorting. Only a small subpopulation of trypomastigotes retained substantial CellTrace Violet after a single round of host cell infection. Similar results were obtained with CFSE staining of trypomastigotes (not shown). Scale bars, 5 μm. Experiment was performed a minimum of three times.
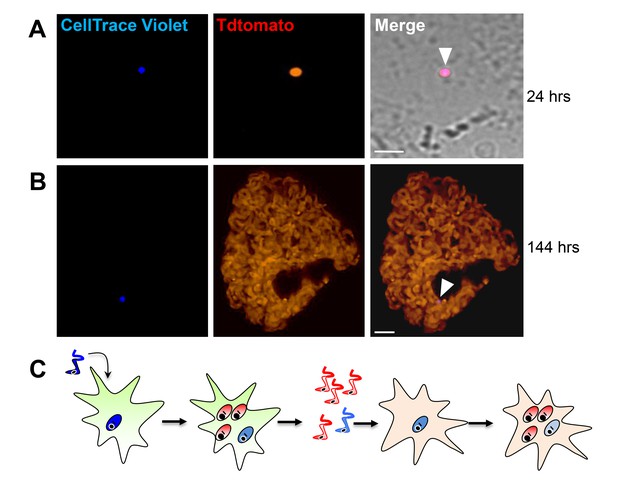
Trypomastigotes originating from non-replicative amastigotes are able to infect host cells and produce both replicating and dormant progeny.
(A) Violet-positive trypomastigotes obtained as shown in Figure 4G were used to infect a fresh culture of Vero cells. Infected cells with CellTrace Violet-positive amastigotes were observed 24 hr post-infection (arrow). (B) Six days post-infection, a non-dividing CellTrace Violet-positive progeny was identified (arrow) in cells filled with dye-negative trypomastigotes. (C) Experiment summary: CellTrace Violet-positive trypomastigotes can produce both dormant (violet+) and actively replicating (violet-) progeny, both of which can convert to trypomastigotes. The trypomastigotes from previously dormant amastigotes can infect new host cells and repeat the process of generating both dormant and actively replicating progeny. Scale bars, 5 um. Experiments repeated twice.
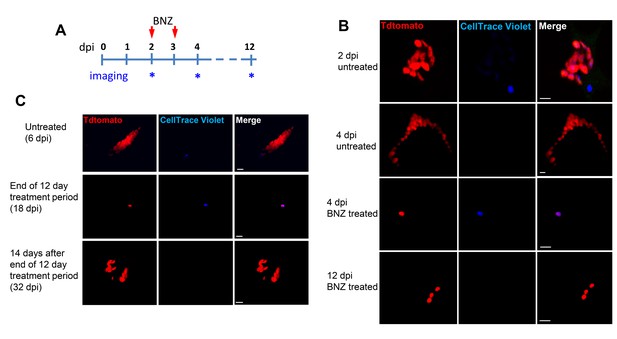
Only dormant amastigotes are resistant to short-term in vivo BZN treatment.
(A) Schematic of experimental protocol. (B) C57BL/6 mice were infected i.p. with 5 × 106 Td-Tomato-expressing and CellTrace Violet-labeled trypomastigotes and orally treated with BZN (100 mg/kg/day) on day 2 and 3 post-infection or left untreated. At the indicated time points, mice were euthanized and adipose tissue was excised, fixed and processed for confocal imaging. Images are representative of three independent experiments with 3–4 animals per group. (C) Infection with 1.5 × 107 Td-Tomato-expressing and CellTrace Violet-labeled trypomastigotes was established for 6 days in IFN-g deficient mice before initiation of daily treatment with BZN (100 mg/kg) for 12 days. Peritoneal adipose tissue was harvested from mice on day 6 (prior to treatment), at the end of the 12 day treatment period on day 18 post-infection, or 14 days after the end of treatment (32 dpi). The peritoneal adipose tissue sample from each mouse is ~0.3 g of tissue spread over a surface of ~1.6 cm2 when mounted for microscopic analysis. In tissue harvested on day 6 of infection (prior to treatment), infected cells can be observed within 15 min of scanning. Following 12 days of treatment this same amount of tissue must be exhaustively scanned for up to 3 hr per sample to detect between 4 and 5 infected cells per sample. Images are representative of two independent experiments with four animals each.
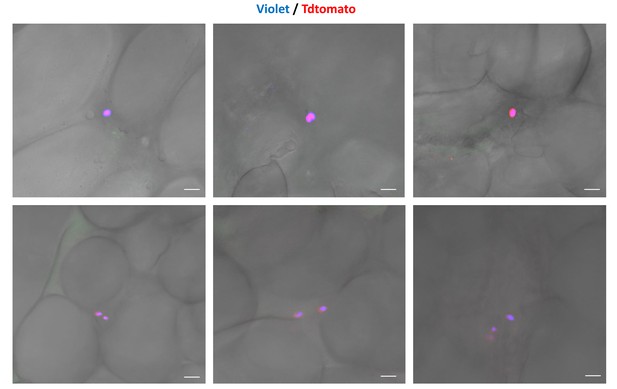
Residual CellTrace Violet-labeled amastigotes after BZN treatment.
C57BL/6 mice were infected with CellTrace Violet-labeled trypomastigotes of the colombiana strain. Mice were BZN-treated (by the oral route, 100 mg/kg/day) on day 2 and 3 post-infection. On day four post-infection, mice were euthanized and adipose tissue was collected, fixed and processed for confocal imaging. Representative images of CellTrace Violet-positive amastigotes within adipocytes detected after BZN treatment. Images are representative of three independent experiments with 3–4 animals per group.
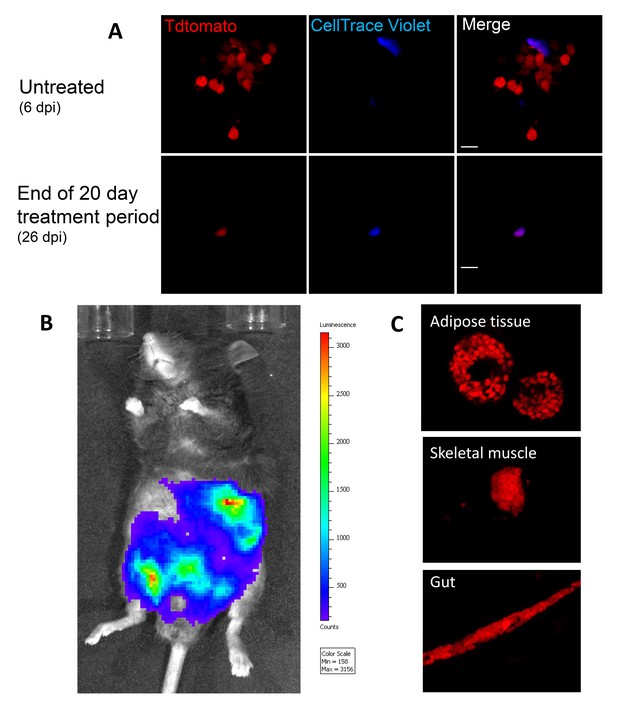
Dormant amastigotes resist up to 20 days of BNZ treatment in vivo and are able to rebound after treatment cessation.
IFN-g deficient mice were infected with 1.5 × 107 Tdtomato-expressing and CellTrace Violet-labeled colombiana strain trypomastigotes and orally treated with BZN (100 mg/kg/day) starting at 6 days post-infection for 12 or 20 days. (A) Adipose tissue cells with abundant amastigotes, including non-replicating (CellTrace Violet+) are evident on day 6 (prior to treatment) while only a minimal number of only CellTrace Violet + amastigotes are detected at the end of the 20 day treatment period. (B) Robust systemic spread of parasites remaining after 12 days of BNZ treatment detected by whole animal in vivo live imaging (luciferase) and confocal microscopy (Tdtomato) of adipose tissue, skeletal muscle and gut (C) at 28 days after the end of treatment. Images are representative of three independent experiments with 3–4 animals per group.
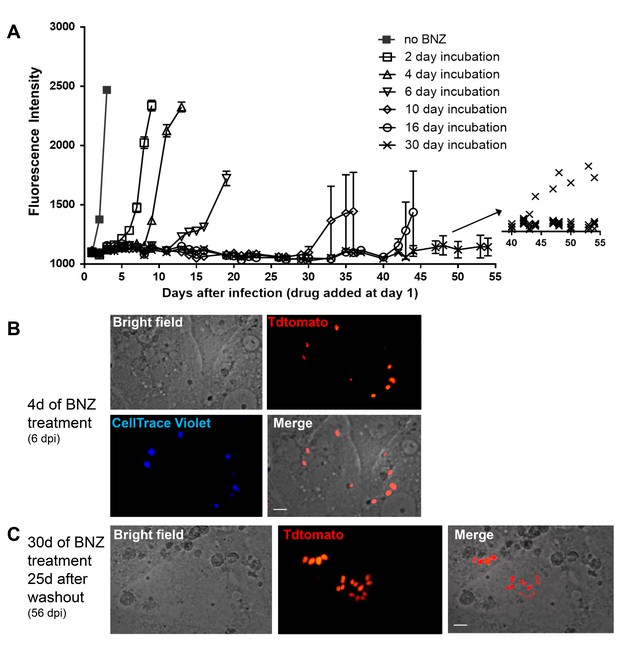
Dormant amastigotes are resistant to extended BNZ treatment in vitro.
Vero cell cultures were infected with Tdtomato-expressing trypomastigotes of colombiana or CL strains (5:1 ratio parasites:cells) 24 hr prior to addition of BNZ (10 uM). At various times post-treatment, BNZ was removed from the cultures and the subsequent rebound of parasites surviving the drug treatment was determined by whole well fluorescence intensity reading (A) and in vitro live imaging (B, C). The retention of the CellTrace Violet label during the course of BNZ treatment indicates that the BNZ-resistant parasites are dormant (B) and capable of replication after drug washout (C). Results are representative of three independent experiments with six replicates per condition. Arrow to inset in (A) shows reading of individual replicate wells.
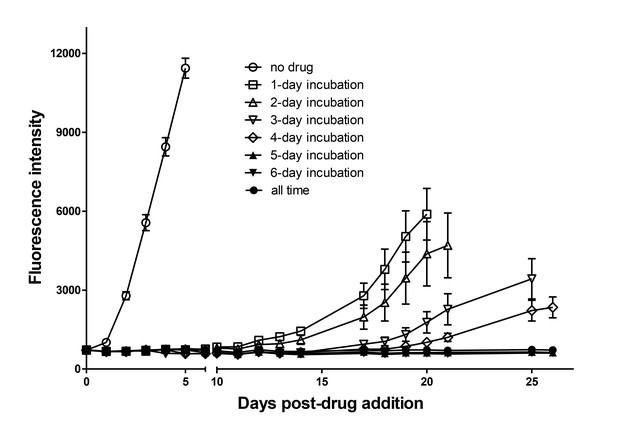
Dormant amastigotes are resistant to extended oxaborale treatment in vitro.
Vero cell cultures were infected with Tdtomato-expressing trypomastigotes (5:1 ratio parasites:cells) 24 hr prior to addition of oxyaboral AN14353 (300 nM). At various times post-treatment, AN14353 was removed from the cultures and the subsequent rebound of parasites surviving the drug treatment was determined by whole well fluorescence intensity reading. Results are representative of two independent experiments with six replicates per condition.
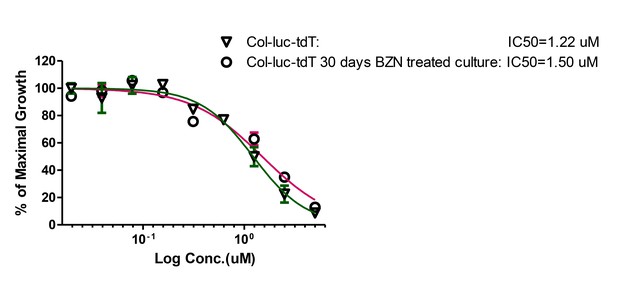
Amastigotes surviving 30 days in vitro BZN treatment have unaltered susceptibility to BZN.
Trypomastigotes harvested from infected Vero cultures pre-treatment of after treatment with BZN for 30 days were used to measure the IC50 for BZN. Assay performed once with three replicates per condition.
Videos
Time lapse video of early stage intracellular replication of T. cruzi amastigotes.
Cultures of HFF cells in 96 well glass bottom plates were infected with Tdtomato-expressing Cell Trace Violet labeled trypomastigotes in a ratio of 10:1 (parasites: host cells). After a5hrincubation, cultures were washed to remove extracellular parasites and imaged every 15 min for 48hr. Time lapse videos were generated spanning from 15:45 to 31:30hr. Note the dye-retaining amastigote (blue) remaining undivided while other amastigotes in the same cells (bright orange amastigotes) actively proliferate.
Time lapse video of early stage intracellular replication of T. cruziamastigotes.
Cultures of Vero cells in 96 well glass bottom plates were infected with Tdtomato-expressing CellTrace Violet labeled trypomastigotes in a ratio of 5:1 (parasites: host cells). After a5hrincubation, cultures were washed to remove extracellular parasites and imaged every 15 min for 48hr. Time lapse videos were generated spanning from 9:30 to 47:15hr. Note the dye-retaining amastigote (blue amastigote denoted by white asterisk at the beginning of Video 1) remaining undivided while other amastigotes in the same cell (bright orange amastigotes) actively proliferate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | Trypanosoma cruzi | RRID:NCBITaxon:5693 | ||
| Strain, strain background (Trypanosma cruzi, colombiana) | TdTomato and luciferase coexpresors; Luciferase- and Tdtomato-expressing parasites. | this paper | NA | Cotransfected with pTREX-Luciferase and pTREX-Td-tomato plasmids. |
| Strain, strain background (T. cruzi, colombiana) | Colombiana strains expresing luciferase | this paper | NA | Transfected with the pTREX-Luciferase plasmid. |
| Strain, strain background (T. cruzi, CL3) | CL-3 strain expresing luciferase | this paper | NA | Transfected with the pTREX-Luciferase plasmid. |
| Strain, strain background (T. cruzi, ARC0704) | ARC-0704 strain expresing luciferase. | this paper | NA | Transfected with the pTREX-Luciferase plasmid. |
| Strain, strain background (T. cruzi, colombiana) | TdTomato expresing parasites. | this paper | NA | Transfected with pTREX-Tdtomato plasmid. |
| Cell line | VERO 76 | American Type Culture Collection (ATCC) | CRL 1587; RRID:CVCL_0603 | |
| Cell line | Human Foreskin Fibroblasts (HFF) | other | N/A | HFF cells were a gift from Dr. D. Etheridge (University of Georgia) |
| Recombinant DNA reagent | pTREX-Luciferase (plasmid) | Canavaci et al., 2010. PMID: 20644616 | NA | Addgene 48337 |
| Recombinant DNA reagent | pTREX-Tdtomato (plasmid) | Canavaci et al., 2010. PMID: 20644616 | NA | Addgene 47975 |
| Commercial assay or kit | Click-iT EdU Imaging Kit | ThermoFisher Scientific, Waltham, MA | C10337 | Cell proliferation detection kit |
| Commercial assay or kit | Click-iT TUNEL Alexa Fluor 647 imaging assay | ThermoFisher Scientific, Waltham, MA | C10247 | Cell apoptosis detection kit |
| Commercial assay or kit | CellTrace Violet fluorescent dye | ThermoFisher Scientific, Waltham, MA | C34557 | Cell proliferation detection kit |
| Commercial assay or kit | CFSE fluorescent dye | ThermoFisher Scientific, Waltham, MA | C34554 | Cell proliferation detection kit |
| Chemical compound, drug | D-luciferin | PerkinElmer, Waltham, MA | 122799 | luciferase substract reagent |
| Chemical compound, drug | CUBIC clarifying solution | other | NA | 25 % N,N,N′,N′-Tetrakis (2-hydroxypropyl)ethylenediamine;25% urea; 15% Triton X-100 and distilled water (Susaki et al., 2015) |
| Chemical compound, drug | Benznidazole (N-benzyl-2-nitro-1-imidazolacetamida) | LAFEPE medicamentos. Brazil; Aesica Pharmaceutical, United Kingdom | BZN | |
| Chemical compound, drug | DAPI stain 4’,6-diamidino-2-phenylindole dihydrochloride | ThermoFisher Scientific, Waltham, MA | 122799 | |
| Software, algorithm | Living Image software v4.3 | Xenogen, Alameda, CA | RRID:SCR_014247 | |
| Software, algorithm | GraphPad 5.0 Prism v5.0 | GraphPad Software, La Jolla California USA, | RRID:SCR_002798 | |
| Other: Mice strains | C57BL/6NCr mice | Charles River Laboratories | C57BL/6NCrl - strain code 027 | |
| IFN-γ knockout mice | The Jackson Laboratory | B6.129S7-Ifngtm1Ts/J - stock No 002287 | ||
| SKH-1 mice | other | NA | The SKH-1 ‘hairless’ mice backcrossed to C57BL/6 were a gift from Dr. Lisa DeLouise (University of Rochester). |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34039.019





