BRG1 governs glucocorticoid receptor interactions with chromatin and pioneer factors across the genome
Figures
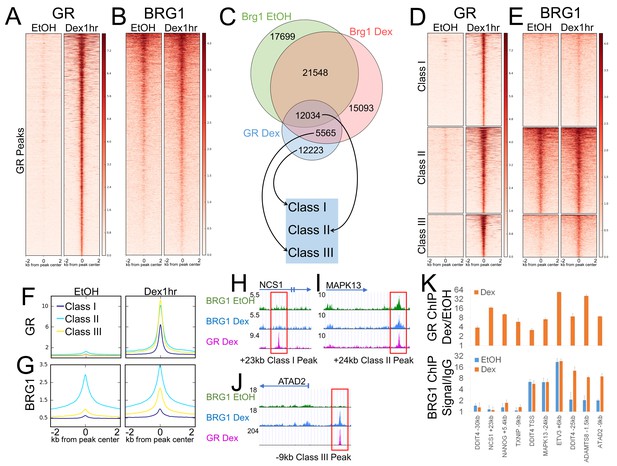
BRG1 chromatin interaction defines distinct classes of GR binding site.
(A) Heatmap demonstrating that GR is detected at 29934 binding sites following 1 hr Dex treatment. (B) Heatmaps illustrating detection of BRG1 at a subset of GR binding sites prior to hormone treatment and recruitment of BRG1 to additional sites following 1 hr of Dex treatment. (C) Venn diagram of overlap between GR, BRG1-EtOH, and BRG1-Dex peaks and the designation of three classes of GR peaks. (D) Heatmap of GR signal over GR peaks divided into three classes. (E) Heatmap of BRG1 signal over GR peaks divided into three classes. (F–G) Meta-profiles of GR and BRG1 ChIP-seq coverage over GR peak classes. (H–J) UCSC genome browser snapshots of GR, BRG1 EtOH, and BRG1 Dex ChIP-seq coverage at representative Class I, II, and III GR peaks. (K) ChIP-QPCR validation of GR and BRG1 ChIP enrichment at representative Class I, II, and III GR peaks.
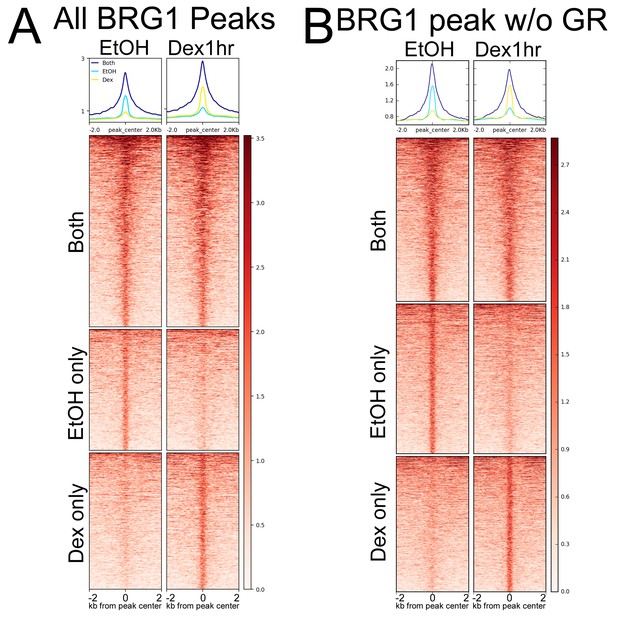
Hormone treatment reorganizes BRG1 chromatin localization.
(A) Heatmap depicting BRG1 ChIP-seq coverage at BRG1 peaks that are called in EtOH only, Dex only, or both conditions. (B) Heatmap depicting BRG1 ChIP-seq coverage at BRG1 peaks as in (A), but with peaks that overlap GR peaks removed (12034 Class II and 5565 Class III peaks removed).
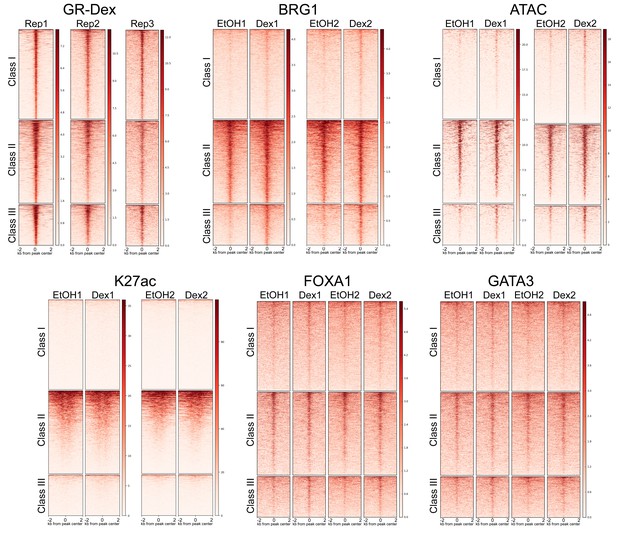
Biological replication of Next Generation Sequencing data sets.
Heatmaps depicting consistency between biological replicate ChIP-seq and ATAC-seq experiments over GR peak classes
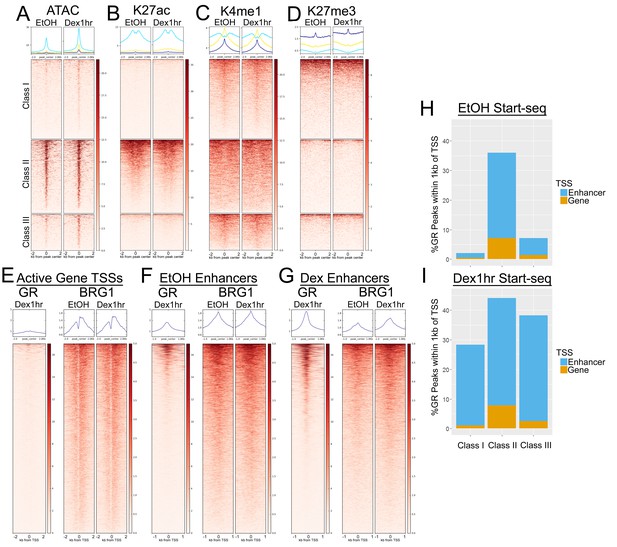
Class II GR peaks are associated with open and transcriptionally active chromatin.
(A) ATAC-seq accessibility/short reads are enriched over Class II GR peaks independently of hormone treatment. (B) H3K27ac ChIP-seq coverage is specifically enriched over Class II GR peaks independently of hormone treatment. (C) H3K4me1 ChIP-seq is enriched over Class I and III peaks, whereas Class II peaks have a broader enrichment pattern with a trough directly over the GR peak. (D) H3K27me3 is not strongly enriched at any of the GR peak classes. (E–G) Heatmaps depicting GR and BRG1 ChIP-seq signal around active gene TSSs (E), enhancer TSSs from EtOH/untreated cells (F), and enhancer TSSs from Dex 1 hr cells (G). (H–I) Stacked barplots showing the percentage of GR peaks that are within 1 kb of an active gene TSSs (orange) or enhancer TSSs (blue) in using Start-seq TSS calls from untreated cells (H) and Dex 1 hr cells (I).
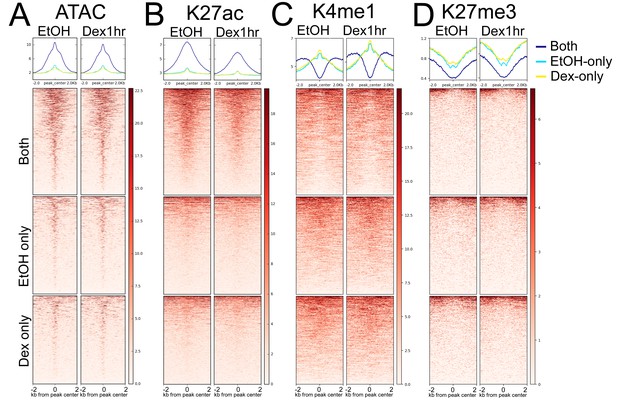
BRG1 peaks without GR are accessible and have active chromatin marks.
BRG1 peaks without GR that are hormone-independent (labeled ‘Both’) have enrichment patterns for ATAC accessibility, H3K27ac, and H3K4me1 that are similar to Class II peaks. EtOH-specific and Dex-specific BRG1 peaks have patterns more similar to Class I or III peaks.
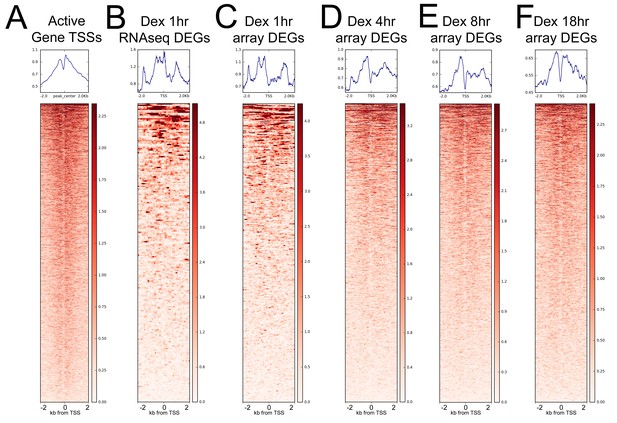
GR is weakly enriched around TSSs.
Heatmaps showing GR ChIP-seq coverage at (A) active gene TSSs (same plot as Figure 2e with adjusted y and z scales), (B) TSSs of RNA-seq identified DEGs following 1 hr Dex treatment, (C) TSSs of microarray-identified DEGs following 1 hr Dex treatment, (D) TSSs of microarray-identified DEGs following 4 hr Dex treatment, (E) TSSs of microarray-identified DEGs following 8 hr Dex treatment, and (F) TSSs of microarray-identified DEGs following 18 hr Dex treatment.
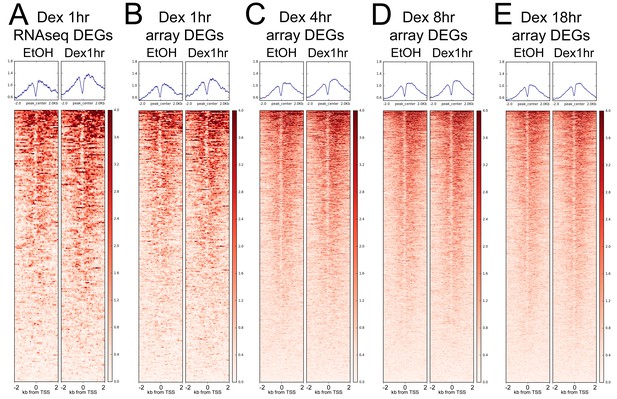
BRG1 is enriched around TSSs.
Heatmaps showing BRG1 +/- Dex at ChIP-seq coverage at (A) TSSs of RNA-seq identified DEGs following 1 hr Dex treatment, (B) TSSs of microarray-identified DEGs following 1 hr Dex treatment, (C) TSSs of microarray-identified DEGs following 4 hr Dex treatment, (D) TSSs of microarray-identified DEGs following 8 hr Dex treatment, and (E) TSSs of microarray-identified DEGs following 18hr Dex treatment.
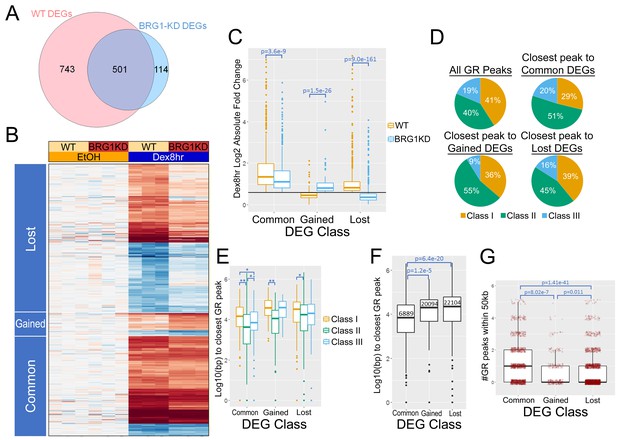
BRG1 is required for the Dex-induced transcriptional response.
(A) Venn diagram showing overlap between DEGs in 8 hr Dex treatment vs EtOH in WT and BRG1-KD cells. (B) Heatmap depicting log2 fold change of DEGs in 8 hr Dex treatment vs EtOH in WT and BRG1-KD cells. (C) Box plots of log two absolute fold changes of 8 hr Dex DEGs. Black line depicts 1.5 fold change. (D) Pie charts depicting the class of the closest GR peak to each DEG TSS. (E) Box plots depicting the log 10 distance from the DEG TSSs to their closest GR peaks divided into GR peak classes. *=p value<0.05, **=p value<0.001 (F) Box plots showing the distance from DEG TSSs to their closest GR peaks. Median distances are labeled in base pairs. (G) Box and scatter plots depicting the number of GR peaks within 50 kb of the TSS of 8 hr Dex DEGs. Outlier peaks with more than 5 GR peaks within 50 kb of the TSS were omitted from the graph for display purposes. All p-values were calculated using Wilcoxon rank sum tests.
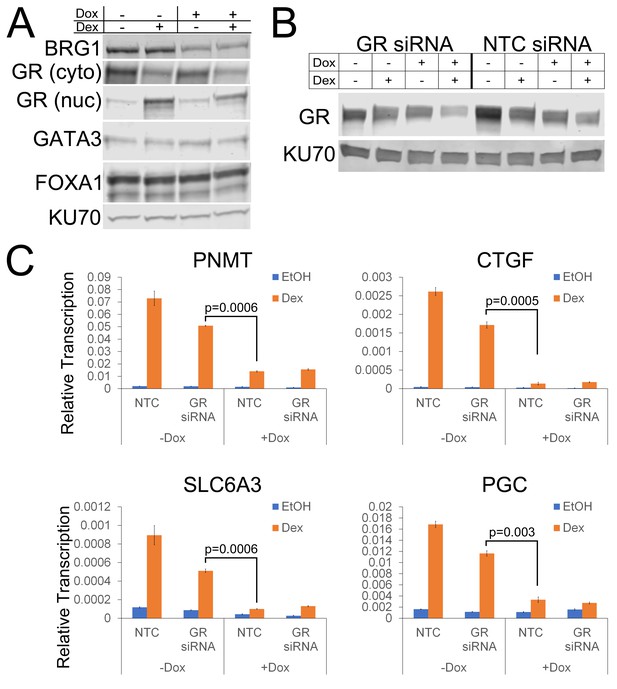
BRG1 and GR knockdown in A1A3 cells.
(A) Western blots depicting protein levels of BRG1, cytoplasmic GR, nuclear GR, GATA3, FOXA1, and KU70 in A1A3 cells following BRG1 knockdown by doxycycline treatment. (B) Western blots depicting protein levels of GR and KU70 following transfection with GR siRNA and/or BRG1 knockdown by doxycycline treatment. (C) QPCR demonstrating the effects of GR siRNA and BRG1 knockdown on ‘lost’ target genes. Note that while GR siRNA partially reduces dex-dependent induction of transcription, BRG1 knockdown results in a greater loss of dex-dependent induction.
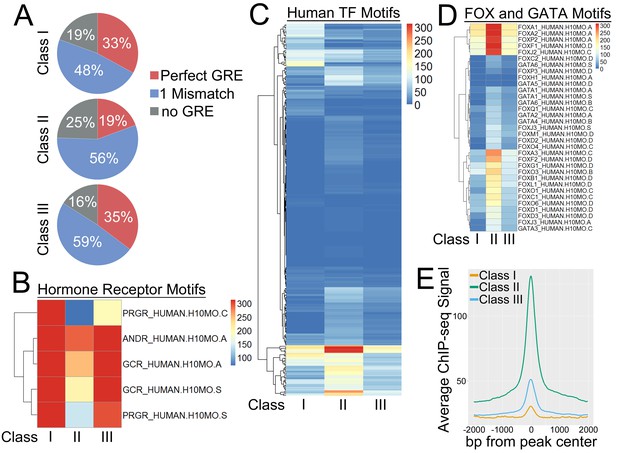
GR peak classes have distinct underlying DNA sequences and transcription factor motifs.
(A) Class II GR peaks have fewer perfect GRE motifs than Class I or Class III peaks. (B) Heatmap of -log10 p-values for enrichment of hormone receptor motifs under GR peak classes. (C) Heatmap of -log10 p-values for enrichment of human transcription factor motifs under GR peak classes. (D) Heatmap of -log10 p-values for enrichment of FOX and GATA motifs under GR peak classes. (E) Meta-profile of average coverage of 25 ENCODE transcription factor ChIP-seq MCF7 datasets over GR peak classes.
-
Figure 4—source data 1
Table of MCF7 ENCODE ChIP-seq data sets.
Table of ENCODE ChIP-seq data sets from MCF7 cells used to calculated average transcription factor ChIP-seq meta-plot in Figure 4E.
- https://doi.org/10.7554/eLife.35073.014
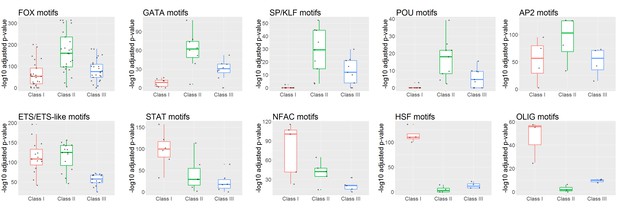
Transcription factor motifs enriched under distinct GR peak classes.
Box plots of -log10 p-values of families of transcription factor motifs under Class I, II, and III GR peaks. Note that Fox, Gata, SP/KLF, and Pou family motifs are most strongly enriched under Class II peaks, while Stat, NFAC, Olig, and HSF motifs are most strongly enriched under Class I peaks.
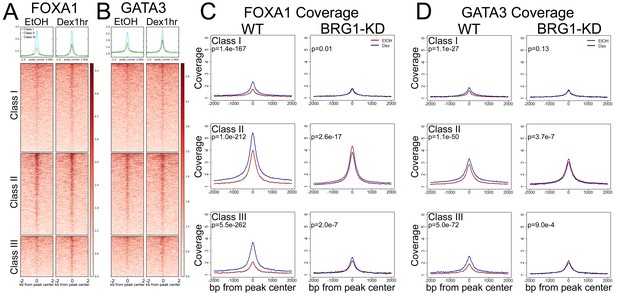
BRG1 is required for Dex-induced recruitment of pioneer factors.
(A–B) FOXA1 and GATA3 ChIP-seq coverage is enriched across all GR peak classes, but shows strongest enrichment over Class II peaks. (C–D) Meta-profiles of FOXA1 and GATA3 ChIP-seq signal over GR peak classes in A1A3 cells. In wild-type cells, Dex treatment induced recruitment of additional FOXA1 and GATA3 to all three GR peak classes. This recruitment is lost following BRG1-KD. To calculate P-values in (C) and (D), we used the average signal in the 500 bp window centered on the center of the GR peak, and performed unpaired Wilcoxon Rank Sum/Mann-Whitney test.
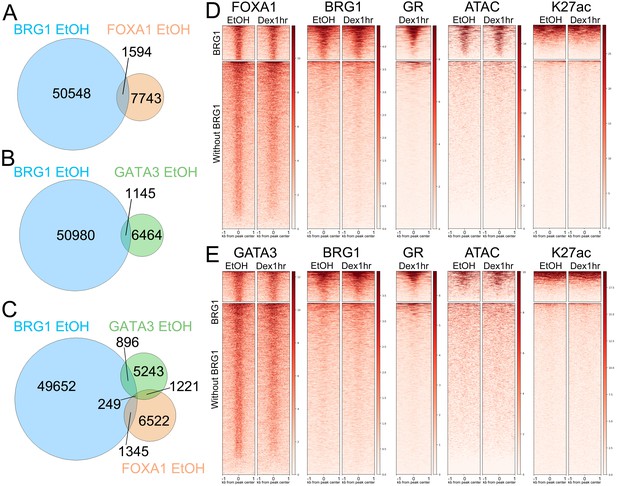
BRG1 binding to pioneer factor binding sites is predictive of GR binding upon Dex treatment.
(A) Overlap between BRG1 EtOH and FOXA1 EtOH peaks. (B) Overlap between BRG1 EtOH and GATA3 EtOH peaks. (C) Three-way Venn diagram showing overlap of all three factors (D) Heatmaps depicting FOXA1, BRG1, GR, ATAC nucleosome free reads, and K27ac over FOXA1 EtOH peaks divided into ‘with BRG1’ and ‘without BRG1’ subsets. (E) Heatmaps depicting GATA3, BRG1, GR, ATAC nucleosome free reads, and K27ac over GATA3 EtOH peaks divided into ‘with BRG1’ and ‘without BRG1’ subsets. Note that in both (D) and (E) GR binding to FOXA1 and GATA3 peaks is largely restricted to ‘with BRG1’ peaks.
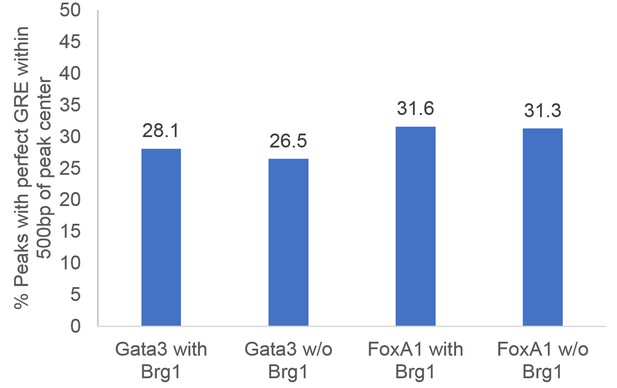
Proportion of GREs under pioneer factor peaks.
Percentage of GATA3 and FOXA1 peaks that have perfect GRE motifs within 500 bp of the center of the peak. The percentage of peaks with perfect GRE motifs is similar between peaks ‘with BRG1’ and peaks ‘without BRG1.’.
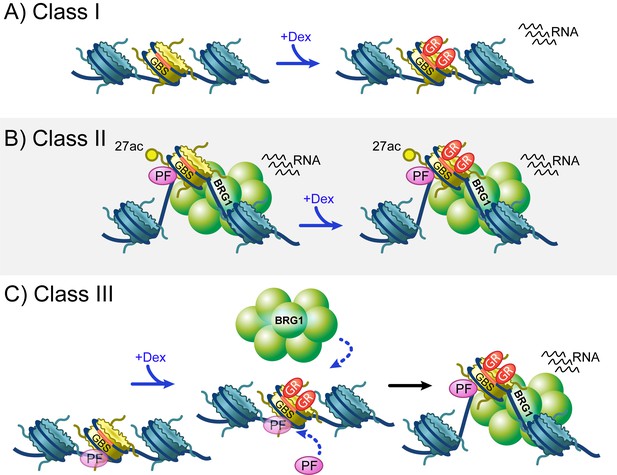
Overview of three classes of GR binding site.
(A) Class I GR binding sites (GBSs) reside within relatively closed regions of chromatin and have little dex-dependent chromatin remodeling or recruitment of BRG1 and pioneer factors. Despite this, greater than a quarter of Class I GBSs exhibit dex-dependent transcription. (B) Class II GBSs represent GR binding to active and occupied regions of chromatin. These GBSs are bound by BRG1 prior to hormone treatment, and also exhibit hormone independent H3K27 acetylation, chromatin accessibility, pioneer factor binding, and transcriptional activity. (C) Class III GBSs behave the most like the model described at the MMTV promoter. Upon hormone treatment, GR binds to regions of relatively inaccessible chromatin that may be pre-occupied by low levels of pioneer factors. Upon GR binding, BRG1 and additional pioneer factors are recruited, chromatin remodeling yields increased accessibility, and more than a third of these GBSs gain transcriptional activity.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H.sapiens) | A1-2 | PMID: 7838148 | RRID:CVCL_0I95 | T47D derivative with incorporated rat GR |
| Cell line (H.sapiens) | A1-A3 | PMID: 22451486 | A1-2 derivative with incorporated BRG1 shRNA | |
| Antibody | anti-BRG1 | PMID: 26055322 | lab-made, ChIP = 1 ug/100 ug chromatin, Western Blot = 0.1 ug/ml | |
| Antibody | anti-GR | Santa Cruz | M-20, sc-1004, RRID:AB_2155786 | ChIP = 1 ug/100 ug chromatin, Western Blot = 0.1 ug/ml |
| Antibody | anti-FOXA1 | Abcam | ab23738, RRID:AB_2104842 | ChIP = 1 ug/100 ug chromatin, Western Blot = 0.1 ug/ml |
| Antibody | anti-GATA3 | Cell Signaling | D13C9, RRID:AB_10834528 | ChIP = 1 ug/100 ug chromatin, Western Blot = 0.1 ug/ml |
| Antibody | anti-H3K27ac | Abcam | ab4729, RRID:AB_2118291 | ChIP = 1 ug/100 ug chromatin |
| Antibody | anti-H3K27me3 | Active Motif | 39155, RRID:AB_2561020 | ChIP = 1 ug/100 ug chromatin |
| Antibody | anti-H3K4me1 | Abcam | ab8895, RRID:AB_306847 | ChIP = 1 ug/100 ug chromatin |
| Antibody | anti-KU70 | Santa Cruz | H-308, sc-9033, RRID:AB_650476 | Western Blot = 0.1 ug/ml |
| Sequence-based reagent | GR siRNA | Dharmacon | ON-TARGETplus J-089504–07 | UUACAAAGAUUGCAGGUAU |
| Sequence-based reagent | Non-targeting Control siRNA | Dharmacon | ON-TARGETplus Not-targeting Pool D-001810-10-20 | |
| Commercial assay or kit | Nextera XT library generation kit | Illumina | 15032350 | |
| Commercial assay or kit | SuperScript III First-Strand kit | Invitrogen | 18080–051 | |
| Commercial assay or kit | iScript cDNA Sythesis kit | Bio-Rad | 170–8891 | |
| Commercial assay or kit | ssoAdvanced Universal SYBR Green Supermix | Bio-Rad | 172–5274 | |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | 74104 | |
| Commercial assay or kit | RNA 6000 RNA Pico Kit | Agilent Technologies | 5067–1513 | |
| Commercial assay or kit | QiaQuick PCR purification kit | Qiagen | 28104 | |
| Commercial assay or kit | HALT protease inhibitors | ThermoFisher | 78430 | |
| Chemical compound, drug | Dexamethasone | Sigma | D4902 | 100 nM |
| Chemical compound, drug | Doxycycline | Sigma | D9891 | 10 ug/ml |
| Software, algorithm | Cutadapt | DOI: http://dx.doi.org/10.14806/ej.17.1.200 | RRID:SCR_011841 | |
| Software, algorithm | Sickle | https://github.com/najoshi/sickle | RRID:SCR_006800 | |
| Software, algorithm | Bowtie2 | PMID: 22388286 | RRID:SCR_005476 | |
| Software, algorithm | Samtools | PMID: 19505943 | RRID:SCR_002105 | |
| Software, algorithm | MACS2 | PMID: 18798982 | RRID:SCR_013291 | |
| Software, algorithm | Homer | PMID: 20513432 | RRID:SCR_010881 | |
| Software, algorithm | Bedtools | PMID: 20110278 | RRID:SCR_006646 | |
| Software, algorithm | Deeptools | PMID: 27079975 | ||
| Software, algorithm | AME | DOI: https://doi.org/10.1186/1471-2105-11-165 | RRID:SCR_001783 | http://meme-suite.org/tools/ame |
| Software, algorithm | STAR | PMID: 23104886 | RRID:SCR_015899 | |
| Software, algorithm | Salmon | PMID: 28263959 | ||
| Software, algorithm | limma-voom | PMID: 24485249 | RRID:SCR_010943 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35073.019





