High levels of histones promote whole-genome-duplications and trigger a Swe1WEE1-dependent phosphorylation of Cdc28CDK1
Figures
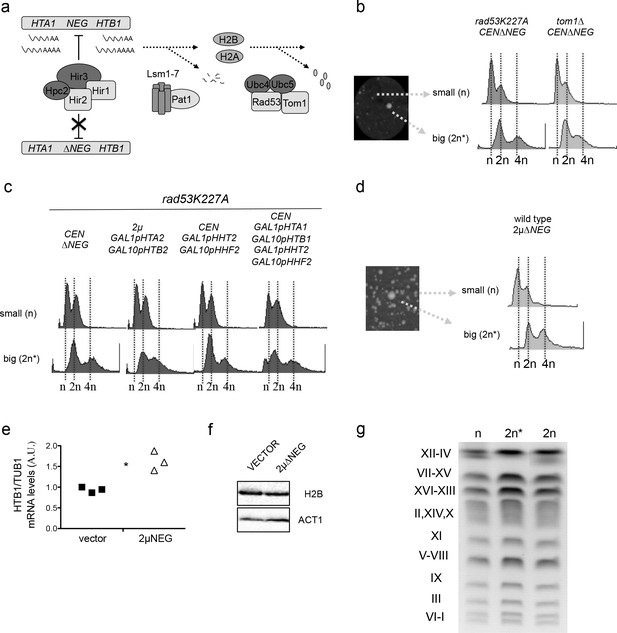
Persistent transcription of histones promotes WGDs.
(a) Schematic representation of the three pathways that negatively regulate histone levels at a transcriptional (HIR complex), post-transcriptional (Lsm1-7 Pat1 complex), or post-translational (Rad53-Tom1-Ubc4/5 complex) level. (b) Left: small and large colonies observed after transformation of rad53K227A or tom1∆ cells with the centromeric vector pHTA1-HTB1∆NEG (pCEN∆NEG). Right: corresponding FACS profiles (c) FACS profiles from large and small colonies after transformation of rad53K227A cells with the indicated vectors. Histone expression is under the control of the GAL1-GAL10 galactose-inducible divergent promoter. 2µ GAL1pHTA2 GAL10pHTB2 simultaneously expresses histones H2A and H2B, while CEN GAL1pHHT2 GAL10pHHF2 expresses H3 and H4. Simultaneous expression of the four canonical histones is driven by two centromeric vectors. Samples were grown in galactose before and after transformation. (d) FACS profiles from small and big colonies obtained after the transformation of a wild type strain with a high-copy vector that contains the HTA1-HTB1∆NEG construct (2µ∆NEG). (e) H2B (HTB1) Q-PCR mRNA levels normalised to TUB1 mRNA in wild-type cells transformed with the 2µ∆NEG or an empty vector (p=0.029; t-test paired samples). Each point represents an independent experiment (f) Representative example of H2B protein levels in wild-type cells transformed with the 2µ∆NEG or an empty vector. Act1 was used as a loading control (g) Karyotypes from wild-type haploid cells (n), diploid strains (2 n) (obtained by cross) and big colonies obtained after transformation with the 2µ∆NEG (2 n*). Karyotypes were analysed by Pulse Field Gel Electrophoresis (PFGE) in five independent clones for each strain.
-
Figure 1—source data 1
Persistent transcription of histones promotes WGDs.
- https://doi.org/10.7554/eLife.35337.004
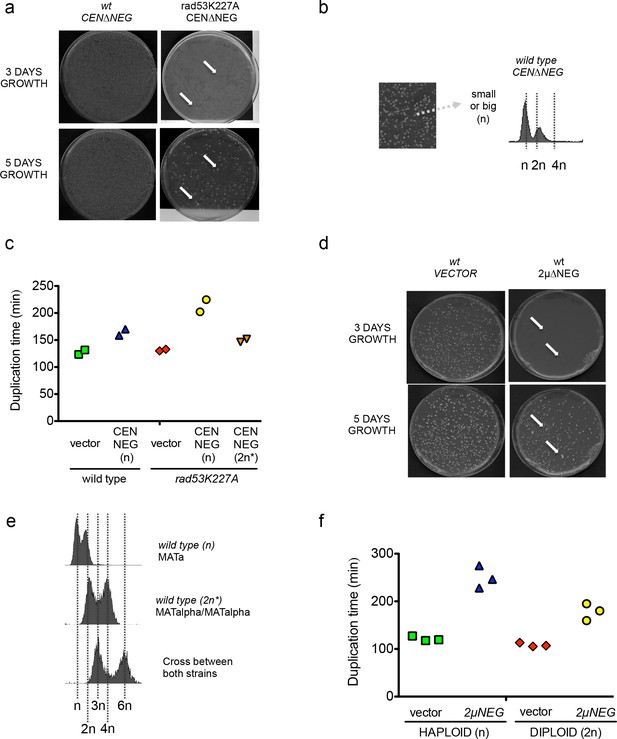
Persistent transcription of histones promotes WGDs.
(a) Representative example of individual colonies obtained in a wild type or a rad53K227A strain after transformation with the pCEN∆NEG vector after 3 or 5 days of incubation. (b) Right: Typical FACS profile obtained in wild-type cells transformed with pCEN∆NEG. (c) Average duplication time of the indicated strains after transformation with pCEN∆NEG. Duplication times were estimated by measuring the Optical Density at 600 nm every 2 hr during 12 hr. Each point represents an independent experiment. Ploidy was measured at each time point to confirm that cells remained haploid or diploid during the time course. Cells that had already started to form diploid cells were not considered (d) Similar to (a) but in wild-type cells after transformation with the empty vector or the 2µ∆NEG vector. (e) FACS profile of a triploid strain obtained by cross between a wild-type haploid alpha strain and a 2 n* Mat a strain. (f) Similar to (c) but comparing the growth of wild-type haploid or diploid cells transformed with either an empty vector or the 2µ∆NEG. Each point represents an independent experiment.
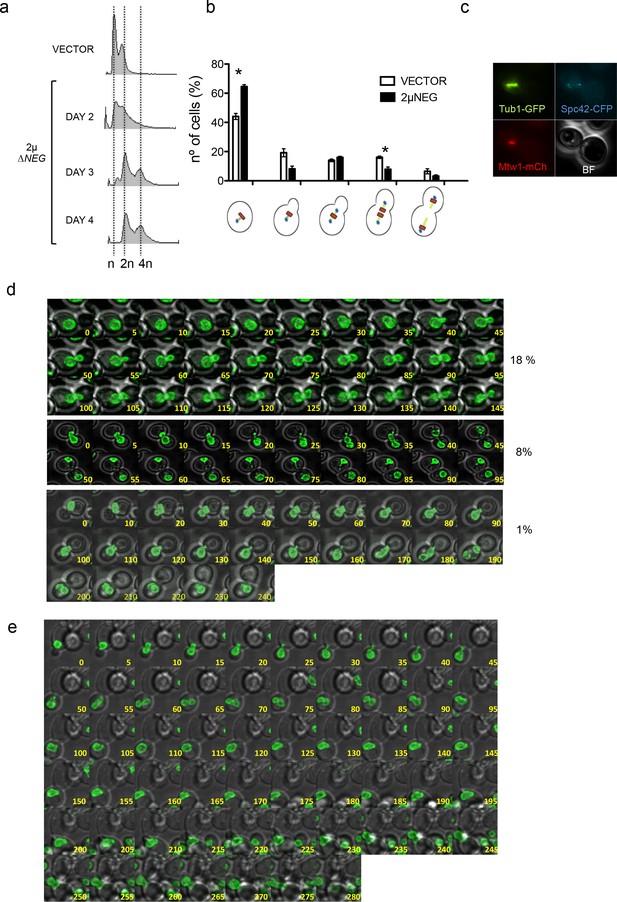
Deregulation of histone levels delays chromosome segregation and promotes aberrant cell divisions.
(a) Representative FACS profiles of haploid wild type cells after transformation with the 2µ∆NEG vector grown during several days. Samples were taken every 24 hr and analysed by FACS to estimate DNA content. (b) Quantification of the different cell cycle stages observed in exponentially growing haploid cells after transformation with the 2µ∆NEG or an empty vector (11 Z-sections, 5µ total). The strain (DVY15) carries markers for the kinetochore (Mtw1-mCherry), the spindle pole body (Spc42-CFP), and tubulin (Tub1-GFP). Three independent samples were counted for the vector (n = 986) and the 2µ∆NEG (n = 1613). Significant p-values are indicated with an asterisk (0.022 and 0.021 from left to right; t-test, paired samples, two tails). (c) Example of a cell in which the whole mitotic machinery can be observed at the daughter cell before mitosis. (d) Live microscopy (5-min interval, 11 Z-sections, 3µ total) of strain DVY12 (Nup49-GFP) after transformation with the 2µ∆NEG vector. first panel: Representative example of cells that remain with an undivided nucleus for more than 2 hr (anaphase in control cells usually takes place in less than 20 min). second panel: Representative example of a cell in which the whole nucleus migrates to the daughter before anaphase. third panel: Representative example (10-min interval) of a cell in which both nuclei remain trapped in the daughter cell. Complete movies can be found in Figure 2d-videos 1–4. (e) Representative example of a cell in which both nuclei initially trapped in the daughter cell are able to enter a new round of division, and segregate (two nuclei at time 280). Percentages are estimated from the analysis of all movies performed during the first biological replicate (n = 228). Full data from this quantification can be observed in the additional Source Data file.
-
Figure 2—source data 1
Deregulation of histone levels delays chromosome segregation and promotes aberrant cell divisions.
- https://doi.org/10.7554/eLife.35337.006
Representative example of a normal anaphase in cells transformed with an empty vector.
https://doi.org/10.7554/eLife.35337.007Representative example of cells that remain with an undivided nucleus for more than 2 hr (Figure 2d first panel).
https://doi.org/10.7554/eLife.35337.008Representative example of a cell in which the whole nucleus migrates to the daughter cell before anaphase (Figure 2d second panel).
https://doi.org/10.7554/eLife.35337.009Representative example of a cell in which both nuclei remain trapped in the daughter cell (Figure 2d third panel).
Full information about live microscopy can be found in the Materials and methods section.
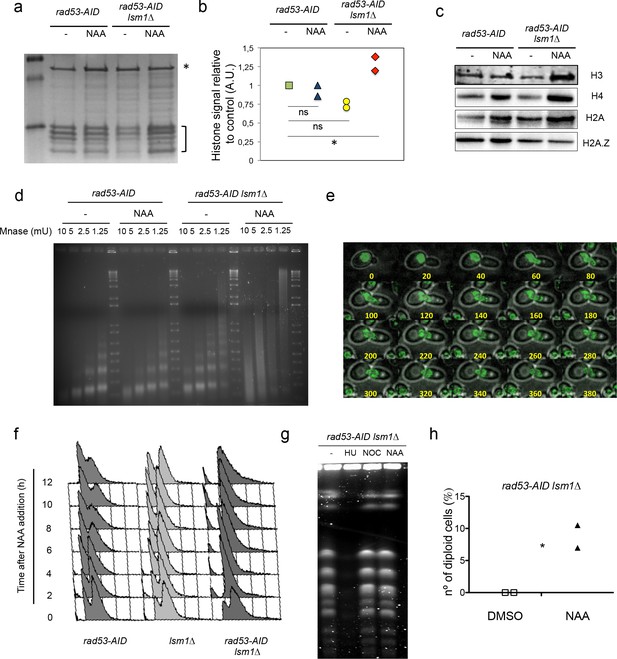
Rad53 depletion in lsm1∆ cells allows large and conditional overproduction of histones beyond S-phase.
(a) Purified core histones from rad53-AID and rad53-AID lsm1∆ cells that were treated or not 2 hr with NAA and blocked with Nocodazole 2 additional hours. The image represents a Coomassie-Blue-stained gel of histone-purified samples. (b) Quantification of histones in the samples loaded in (a). Histone signal (indicated with a bracket) was normalised using the signal of a non-specific band (indicated with an asterisk). The results were normalised to lane 1. Quantification was done using image J. Two independent experiments were performed. *p=0.026 (t-test, paired, two tails) (c) Western Blots of canonical histones H3, H4 and H2A, and the histone variant H2A.Z. performed with purified core histones samples. Ratio H2A/H4 and H2AZ/H4 are shown in Figure 3—figure supplement 3f. (d) Micrococcal nuclease (Mnase) analysis of chromatin extracted from rad53-AID and rad53-AID lsm1∆ cells treated as described in (a). (e) Live microscopy (20-min interval, 14 Z-sections, 2,89µ total) in strain DVY36 (rad53-AID lsm1∆ NUP49-GFP) after 90 min of Auxin treatment (Auxin was maintained in the media). (f) Cell cycle distribution of rad53-AID, lsm1∆ and rad53-AID lsm1∆ cells after treatment with 500 µM NAA. (g) Pulse field gel electrophoresis of rad53-AID lsm1∆ cells that were treated with 200 mM Hydroxyurea (HU), 10 µg/ml Nocodazole (NOC), or 500 µM NAA (NAA). Control asynchronous growing cells were not treated (UT). HU-treated cells are blocked in S-phase preventing chromosomes from entering the gel. Nocodazole allows full replication but blocks mitosis. (h) Percentage of rad53-AID lsm1∆ cells that have experienced or not a WGD after a 4 hr treatment with 500 µM NAA. The DNA content of all survivors was analysed by FACS. two independent experiments (n = 100 or 75) p-value=0.038 (t-test, two tails, unequal variance).
-
Figure 3—source data 1
Rad53 depletion in lsm1Δ cells allows large and conditional overproduction of histones beyond S-phase.
- https://doi.org/10.7554/eLife.35337.013
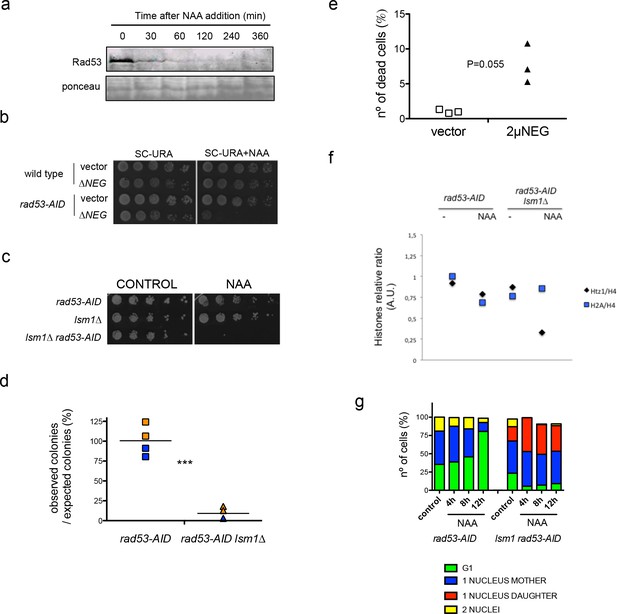
Rad53 depletion in lsm1Δ cells allows large and conditional overproduction of histones beyond S-phase.
(a) Rad53 depletion in a rad53-AID strain after the addition of 500 µM NAA (b) Plate growth assay of wild-type and rad53-AID strains transformed with an empty vector or the CEN∆NEG in the absence or presence of Auxin (200 µM). (c) Similar to (b) for rad53-AID, lsm1∆, and rad53-AID lsm1∆ strains grown in YPGAL plates with or without Auxin (500 µM). (d) Colony formation assay. Percentage of rad53-AID and rad53-AID lsm1∆ cells able to form a colony after 4 hr of Auxin treatment. Two independent experiments with two different dilutions were counted. p=0.0006 (t-test, two tails, paired samples) (e) Viability essay in wild-type cells transformed with an empty vector or the 2µ∆NEG vector using the FUN1 staining kit. Three independent experiments. p-Value p = 0.055 (t-test, one tail, paired samples). (f) Density of the bands on the western blot shown in Figure 3C were quantified with Image J. Ratio of H2AZ(Htz1)/H4 and H2A/H4 are indicated. (g) Representative example of the cell cycle distribution observed at different time points in rad53-AID and rad53-AID lsm1∆ cells after NAA is added to the media. Percentages were estimated from fixed samples stained with DAPI. 200 cells or more were counted for each condition. This experiment was performed twice and gave similar results.
Videos depicting several examples of the normal behavior of rad53-aid lsm1∆ NUP49-GFP cells after a pre-treatment of 90 min with NAA.
NAA was maintained in the media during the time course.
Videos depicting several examples of the normal behavior of rad53-aid lsm1∆ NUP49-GFP cells after a pre-treatment of 90 min with NAA.
NAA was maintained in the media during the time course.
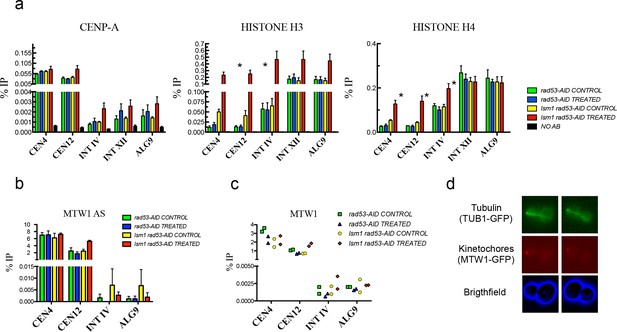
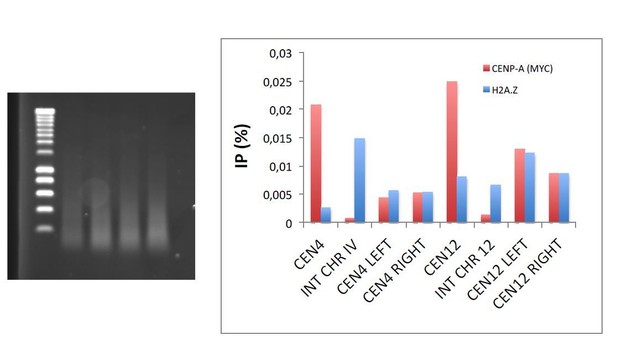
High levels of histones do not affect CENP-A recruitment to centromeres or the attachment of chromosomes to the spindle axis.
(a) Relative levels of histones H3, H4, Cse4 (CENP-A) measured by ChIP-Q-PCR at the indicated loci in rad53-AID (DVY22) and rad53-AID lsm1∆ (DVY23) cells. Samples were obtained from cultures treated (or not) with Auxin for 2 hr and incubated 2 additional hours with Nocodazole. CEN4 and CEN12 correspond to the centromeric region of Chromosomes IV and XII. INT IV and INT XII to intergenic regions in the same chromosomes. ALG9 corresponds to a small amplicon in the coding region of ALG9. p-Values were obtained from a Student T-test (paired samples, two tails) that compares rad53-AID-untreated cells versus rad53-AID-lsm1∆-treated cells *p<0.05. (b) MTW1 chromatin association (ChIP-qPCR) obtained in two independent experiments in which asynchronous growing rad53-AID and rad53-AID lsm1∆ cells were treated or not with NAA for 4 hr (c) Same as in b but using samples that were synchronised with NAA and Nocodazole as in (a). (d) Two representative examples of the normal distribution of kinetochores (MTW1-mCherry) along the spindle axis (Tub1-GFP) in rad53-AID lsm1∆ cells after 4 hr of Auxin treatment.
-
Figure 4—source data 1
High levels of histones do not affect CENP-A recruitment to centromeres or the attachment of chromosomes to the spindle axis.
- https://doi.org/10.7554/eLife.35337.017
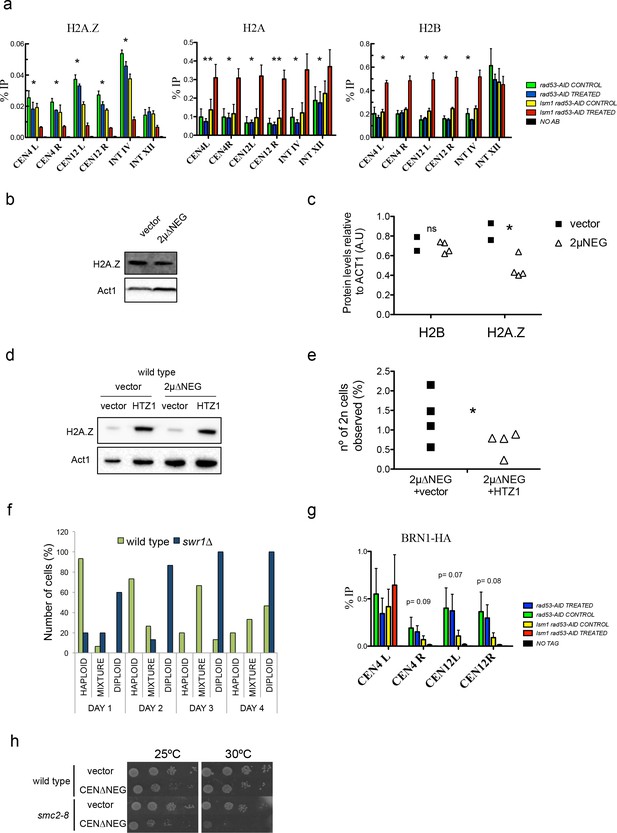
High levels of histones decrease Htz1H2AZ and condensin incorporation into pericentromeric chromatin.
(a) Relative levels of histones H2B, H2A and H2A.Z at the indicated loci measured by ChIP-Q-PCR in rad53-AID (DVY22) and rad53-AID lsm1∆ (DVY23) cells. Samples were obtained from cultures treated (or not) with Auxin for 2 hr and incubated 2 additional hours with Nocodazole. This synchronisation was performed to avoid cell-cycle-related differences. CEN4 (or 12) L (left) and R (right) correspond to the pericentromeric regions of centromeres 4 and 12. p-Values were obtained from a Student T-test (paired samples, two tails) that compares rad53-AID-untreated cells versus rad53-AID-lsm1∆-treated cells *p<0.05; **p<0.01 (b) Representative example of H2A.Z protein levels obtained in wild-type cells transformed with an empty vector or the 2µ∆NEG vector (c) Quantification of protein levels of H2B and H2A.Z relative to Act1. p-Value for H2B differences (t-test, paired, one tail) 0.36; p-value for H2A.Z=0.033 (d) Representative example of H2A.Z protein levels obtained in wild-type cells transformed with either an empty vector or the 2µ∆NEG vector. These cells carry a second empty vector (prs425) or the multicopy vector prs425-HTZ1 driving the overexpression of HTZ1 (H2A.Z) (e) WGD events observed in cells transformed with the indicated plasmids. WGDs were estimated as in Figure 1 (described also in the Supplemental Experimental Procedures). p=0.045 (t-test, paired, one tail) four independent experiments (f) Percentage of cells with a haploid, mixed, or diploid DNA content during a 4 days (D1–D4) time-course in wild type or swr1∆ cells after transformation with the 2µ∆NEG vector. Percentages were obtained from the analysis of 15 clones for each background. (g) Relative levels of Brn1 associated to chromatin measured by ChIP-Q-PCR. Experiments and statistical tests were carried out as in (a) in strains DVY32 and DVY33 expressing HA-tagged Brn1. This experiment was performed five times with five independent biological replicates. (h) Plate growth assay of a wild-type strain and a condensin thermosensitive mutant (smc2-8) transformed with an empty vector or the centromeric version of the ∆NEG vector (CEN∆NEG).
-
Figure 5—source data 1
High levels of histones decrease Htz1H2A.Z and condensin incorporation into pericentromeric chromatin.
- https://doi.org/10.7554/eLife.35337.020
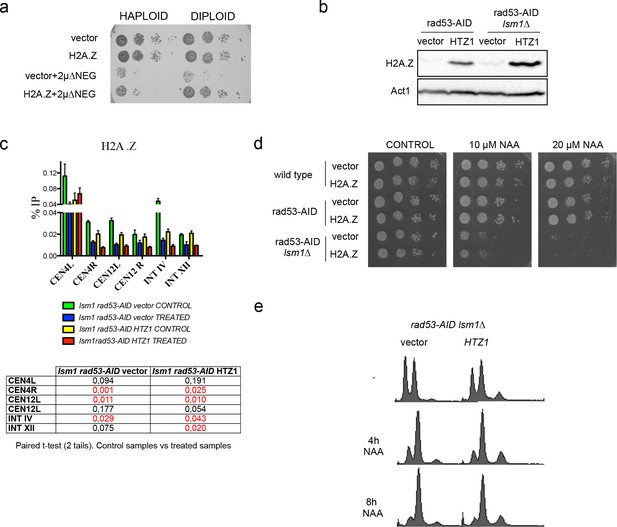
High levels of histones decrease Htz1H2A.Z and condensin incorporation into pericentromeric chromatin.
(a) Plate growth assay that compares the growth rate of haploid and diploid cells transformed with an empty LEU2 vector (prs425) or the vector prs425-HTZ1 in combination with an empty vector or the 2µ∆NEG vector (b) Protein H2A. Z levels in rad53-AID and rad53-AID lsm1∆ cells transformed with the prs425-HTZ1. Cells were treated with Auxin and Nocodazole as previously described (c) H2A.Z ChIP Q-PCR experiment similar to the one shown in Figure 5a in which H2A.Z incorporation was measured in rad53-AID and rad53-AID lsm1∆ cells transformed with an empty vector or the prs425-HTZ1. p-values (t-test; two tails; paired) are shown below. (d) Plate growth assay of rad53-AID and rad53-AID lsm1∆ overexpressing (or not) H2A.Z in the presence of different sublethal concentrations of NAA (e) FACS profiles of rad53-AID lsm1∆ cells transformed with an empty vector or the prs425-HTZ1 vector.
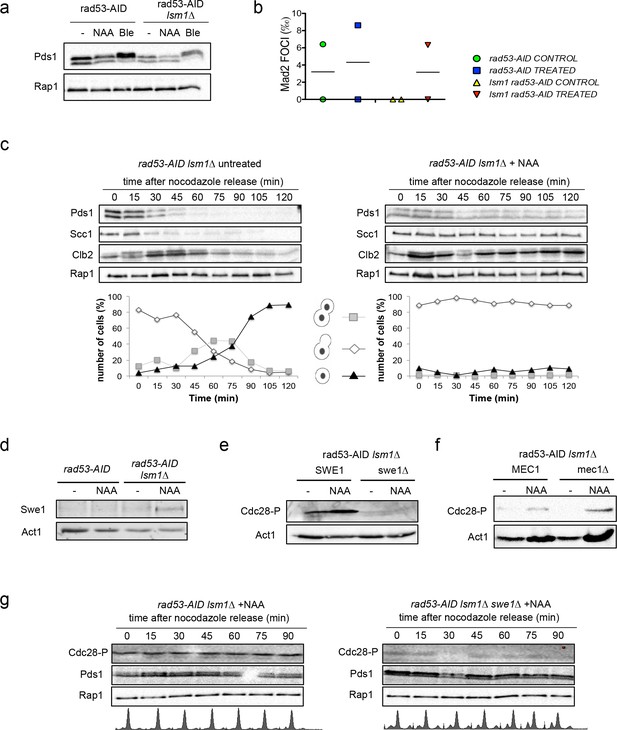
Histone accumulation triggers a Swe1-dependent Cdc28CDK1 phosphorylation.
(a) Pds1 phosphorylation (Pds1-HA) in rad53-AID and rad53-AID lsm1∆ cells either after 4 hr treatment with NAA or bleomycin (Ble, 15 µg/ml). Pds1 appears as a doublet. Its phosphorylation can be visualised as a shifted band after bleomycin treatment. (b) Average number of Mad2 foci (Mad2-GFP) that co-localise with kinetochores (Mtw1-mCHERRY) in rad53-AID and rad53-AID lsm1∆ cells after 4 hr of treatment with either DMSO or Auxin. Two independent experiments were performed in which at least 100 cells were counted. (c) Cell-cycle progression from G2/M to the next G1 in rad53-AID lsm1∆ cells after treatment or not with Auxin. Exponentially growing cells were treated (or not) with Auxin for 2 hr and incubated for 2 additional hours with Nocodazole. Cells were then washed to eliminate Nocodazole and released into media containing Auxin and alpha-factor to arrest cells able to enter mitosis the next G1. This experiment was performed in cells carrying either a PDS1-HA tag or a SCC1-MYC tag. The presence of Clb2 and Rap1 was monitored with anti-Clb2 and anti-Rap1 antibodies. Rap1 was used in both as a loading control. A representative full example of the experiment for each strain is shown in the Figure 6—figure supplement 1a and b. This data file also includes the results obtained with the single mutant that are not shown in the this figure. Graphs below each panel represent the progression of cells to the next G1. These percentages were estimated from fixed cells that were analysed using DAPI staining. (d) Swe1 protein levels in rad53-AID and rad53-AID lsm1∆ cells treated as indicated. (e and f) Cdc28CDK1 phosphorylation levels in the indicated strains. Cdc28CDK1 phosphorylation is monitored with phospho-cdc2 (Tyr15) antibodies. Cells were treated as indicated (g) Cell-cycle progression from G2/M to the next G1 in rad53-AID lsm1∆ and rad53-AID lsm1∆ swe1∆ similar to the one described in (6d), using the strain that expresses Pds1-HA. The results obtained with the single mutant rad53-AID are shown in the Figure 6—figure supplement 1c. All the experiments observed in Figure 6 were performed twice and gave similar results.
-
Figure 6—source data 1
Histone accumulation triggers a Swe1-996 dependent Cdc28CDK1 phosphorylation.
- https://doi.org/10.7554/eLife.35337.023
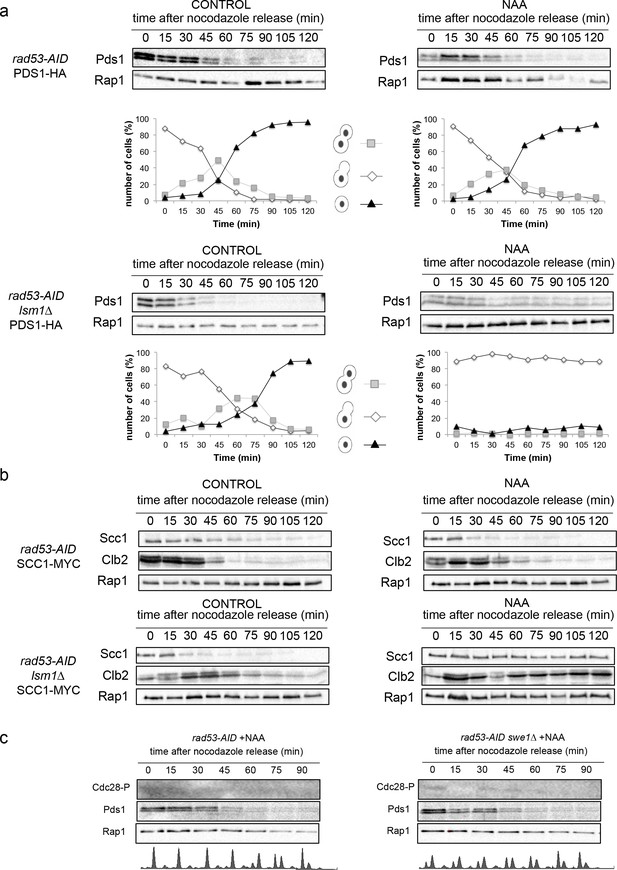
(a and b) Representative full examples of the experiments described in Figure 6d.
Kinetics of rad53-AID and rad53-AID lsm1∆ cells expressing either (a) the PDS1-HA tagged protein, or (b) the SCC1-MYC tag. (c) Kinetics of the single mutant rad53-AID corresponding of the experiment (6 hr) is shown.
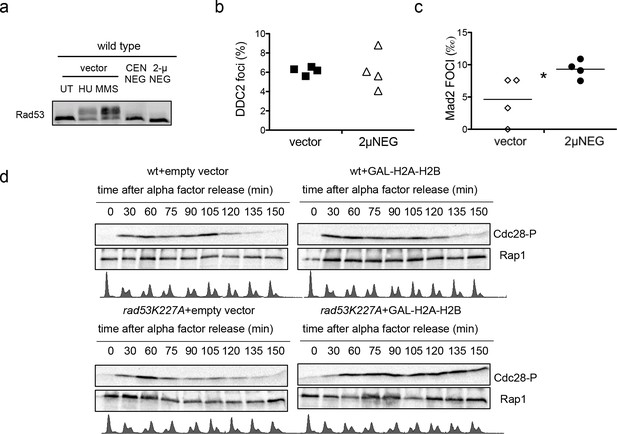
Plasmid-driven overexpresson of histones promotes Cdc28CDK1 phosphorylation.
(a) Rad53 phosphorylation in wild type cells transformed with an empty vector (UT), the centromeric, or the 2µ version of HTA1-HTB1∆NEG (CEN∆NEG and 2µ∆NEG respectively). Phosphorylated Rad53 appears as a shifted band clearly observed after treatment with 200 µM Hydroxyurea (HU) or 0.03% MMS (MMS). (b) Average and SEM (four independent experiments, 200 cells counted for each) of wild-type cells exhibiting Ddc2-Foci after transformation with an empty vector or the 2µ∆NEG vector. DNA content was measured for each sample to confirm that cells still remain haploid (not shown). (c) Average number of Mad2 foci (Mad2-GFP) that co-localise to kinetochores (Mtw1-mCHERRY) in wild-type cells after transformation with an empty vector or the 2µ∆NEG vector. DNA content was measured for each sample to confirm that cells still remain haploid (not shown). p-value=0.031 (t-test, paired, two tails) (d) Phosphorylation of Cdc28 upon H2A-H2B overexpression. Wild type and rad53K227A cells were transformed with either a control vector or a high-copy vector that expresses histones H2A and H2B from a galactose-inducible promoter. Cells were grown in raffinose, arrested in G1 with alpha-factor, incubated 2.5 hr with galactose to induce H2A-H2B expression, and released from alpha-factor block to follow cell cycle progression (FACS) and Cdc28 phosphorylation. Alpha factor was re-added 75 min after to re-arrest cells in G1. These experiment was performed twice and gave similar results.
-
Figure 7—source data 1
Plasmid-driven overexpression of histones promotes Cdc28CDK1 phosphorylation
- https://doi.org/10.7554/eLife.35337.025
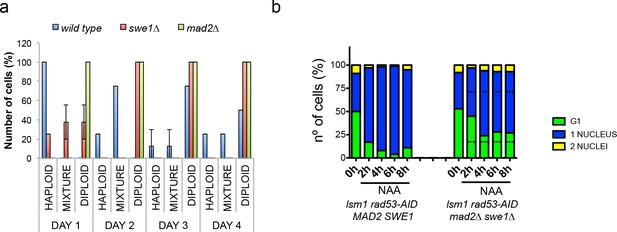
Deletion of SWE1 or MAD2 enhances WGDs.
(a) Percentage of cells with a haploid, mixed, or diploid DNA content during a 4 days (D1–D4) time-course in wild type, swe1∆, or mad2∆ cells after transformation with the 2µ∆NEG vector. These kinetics were performed as the ones shown in Figure 2a. Eight clones were analysed for each genetic background. DNA content of the indicated mutant cells is measured after transformation with 2µ∆NEG. Samples were taken every 24 hr and analysed by FACS to estimate DNA content. Eight independent clones were analysed for each genetic background. (b) Number of unbudded and budded cells with one or two well-differentiated nuclei observed in rad53-AID lsm1∆ and rad53-AID lsm1∆ mad2∆ swe1∆ cells after NAA treatment. Percentages were estimated by DAPI in fixed samples. At least 100 cells were counted. p-Values were obtained for each cell type with a one-way ANOVA test (**p<0.01 ***p<0.001) in which two independent experiments were compared.
-
Figure 8—source data 1
Deletion of SWE1 or MAD2 enhances WGDs.
- https://doi.org/10.7554/eLife.35337.028
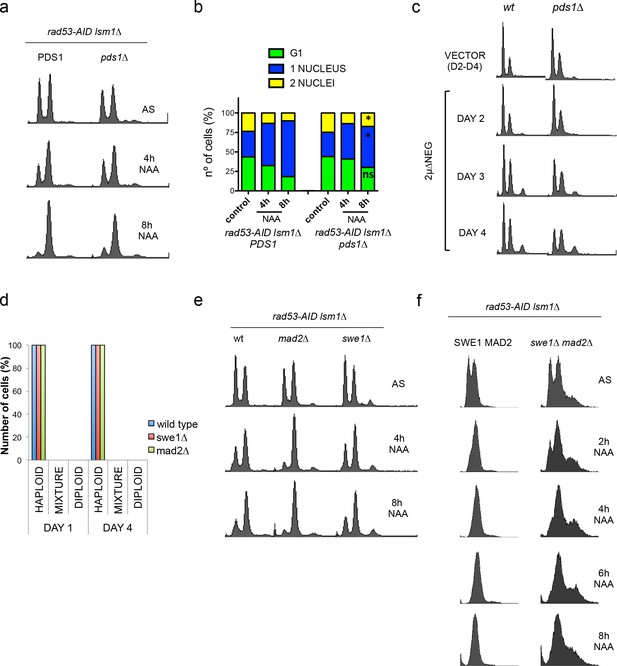
Deletion of SWE1 or MAD2 enhances WGDs.
(a) DNA content of rad53-AID lsm1∆ and rad53-AID lsm1∆ pds1∆ cells after 4 and 8 hr of Auxin treatment. (b) Number of unbudded and budded cells with one or two well-differentiated nuclei in the cell populations shown in (a). Percentages were estimated by DAPI in fixed sample. At least 100 cells were counted. p-Values were obtained from a Student T-test (two tails, paired samples) that compares rad53-AID lsm1∆ untreated versus treated cells. *p<0.05. (c) Representative FACS profiles (four independent colonies tested for each) of haploid wild-type cells or pds1∆ cells after transformation with the 2µ∆NEG vector grown during several days. Samples were taken every 24 hr and analysed by FACS to estimate DNA content. All the experiments that involve PDS1 deletion were carried out at 20°C. This deletion is lethal at temperatures higher than 23°C. (d) Percentage of cells with haploid, mixed, or diploid DNA content during a 4 days (D1–D4) time-course in wild type, swe1∆ or mad2∆ cells after transformation with an empty vector. These kinetics were performed as those shown in Figure 8a. Four independent clones (three for mad2∆) were analysed for each genetic background. Samples were taken at day 1 and day 4 and analysed by FACS to estimate DNA content. (e and f) Cell cycle distribution of rad53-AID lsm1∆ cells and isogenic strains that carry an additional deletion of SWE1, MAD2 (d) or both (e) after treatment with 500 µM NAA.
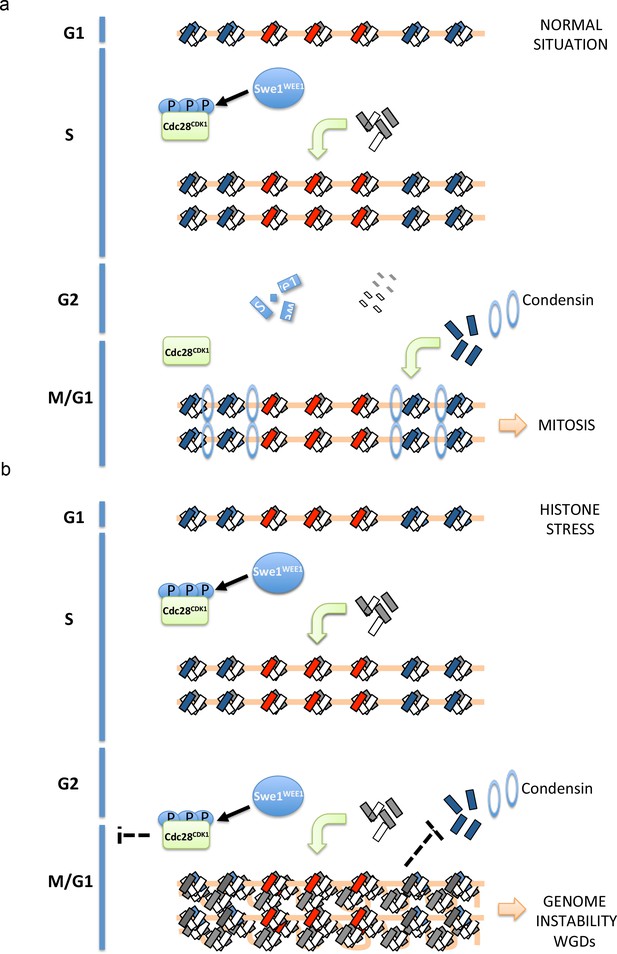
Proposed model to explain how histone-stress impacts mitosis.
Histone dimers are depicted as small rectangles (H3-H4 in white, H2A-H2B in grey and H2A.Z-H2B in blue). During an unperturbed cell cycle (a, upper scheme), canonical histones and Swe1WEE1 increase during replication. Swe1WEE1 will phosphorylate Cdc28CDK1 and maintain it inactive. During G2, histone synthesis will be repressed and all histones (mRNAs and proteins) that are not incorporated to chromatin as well as Swe1WEE1 will be degraded. During mitosis, Htz1H2A.Z will stabilise condensin recruitment at pericentromeric regions allowing the proper function of this complex in chromosome segregation. When histone degradation is compromised (b, lower scheme), cells reach G2 with high levels of histones (histone-stress). This accumulation of histones will promote Cdc28CDK1 phosphorylation and inactivation. This inhibition will delay the entry into mitosis and presumably give time to the cell to lower histone levels. Since this phosphorylation depends on Swe1WEE1, we propose that histone-stress promotes Cdc28CDK1 phosphorylation through a stabilisation of Swe1WEE1. Cells unable to efficiently lower histone levels after replication will increase the amount of canonical nucleosomes incorporated at centromeres and pericentromeres. We propose that this increase in nucleosome density will decrease the efficient exchange of histone H2A by histone Htz1H2A.Z, and reduce Htz1H2A.Z incorporation. This defect in incorporation would consequently lead to a less stable association of condensin to pericentromeres and trigger chromosome segregation defects.
Additional files
-
Supplementary file 1
Supplementary files 1a-c.
- https://doi.org/10.7554/eLife.35337.030
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35337.031





