Structure of a TRPM2 channel in complex with Ca2+ explains unique gating regulation
Figures
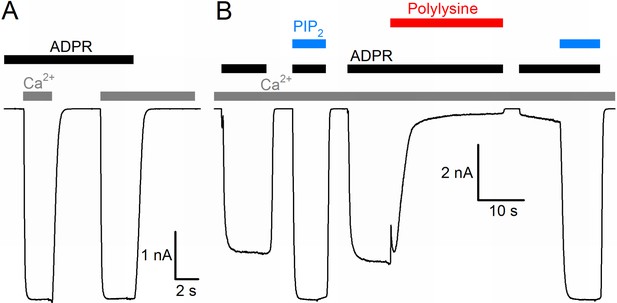
Basic functional properties of the Nematostella vectensis (nv) TRPM2 channel.
(A) Macroscopic inward Na+ currents in an inside-out patch excised from a Xenopus laevis oocyte overexpressing nvTRPM2, evoked by superfusion of the cytoplasmic patch surface with 100 μM ADPR (black bar) plus 40 μM free Ca2+ (gray bars); extracellular (pipette) [Ca2+] was ~1 nM, membrane potential was −20 mV. (B) Effects of 25 μM dioctanoyl-PIP2 (blue bars) on nvTRPM2 channel currents before and after exposure to 15 μg/ml polylysine (red bar). Conditions as in (A).
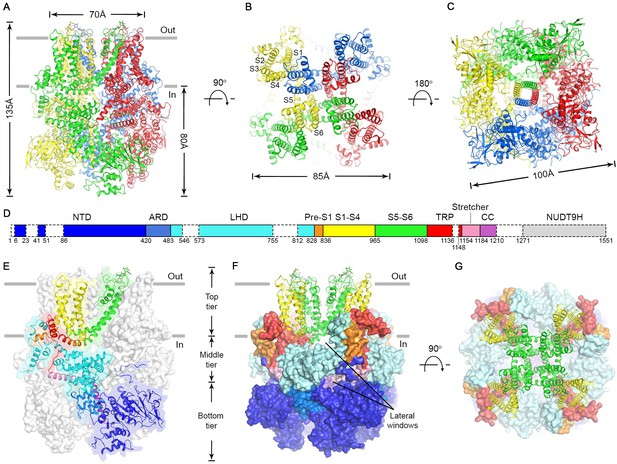
Cryo-EM structure of the nvTRPM2 channel.
(A–C) Different views of the overall structure of the nvTRPM2 tetramer. Protomers are color coded. Gray bars in (A) represent approximate membrane boundaries. Transmembrane helices S1-S6 of one subunit are labeled in (B). (D) Schematic domain structure of nvTRPM2. Regions not built in the final model are indicated with dashed boxes. (E–G) Domain organization of nvTRPM2. In (E), one subunit is shown in ribbon, the remaining subunits as gray surfaces. In (F) and (G), the transmembrane domains are represented as ribbon, the cytosolic domains as surfaces. Domain color coding in (E–G) follows that in (D). Two N-Acetylglucosamines attached to residue N1017 are shown as sticks. See also Figure 2—figure supplements 1–5.
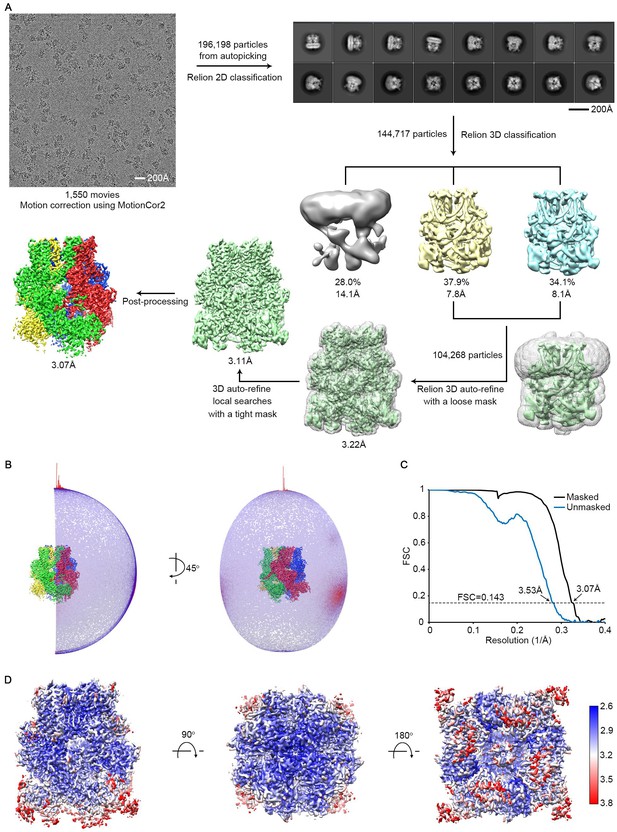
Cryo-EM structure determination and evaluation.
(A) Flow chart of data processing. (B) Angular distributions for all the particles used in the final reconstruction. Red and higher cylinders represent more particles, blue and shorter cylinders less particles, assigned to a specific orientation. (C) Fourier shell correlation (FSC) curves between two half datasets calculated by Relion before (blue) and after (black) post-processing. (D) Local resolution estimation calculated by Blocres.
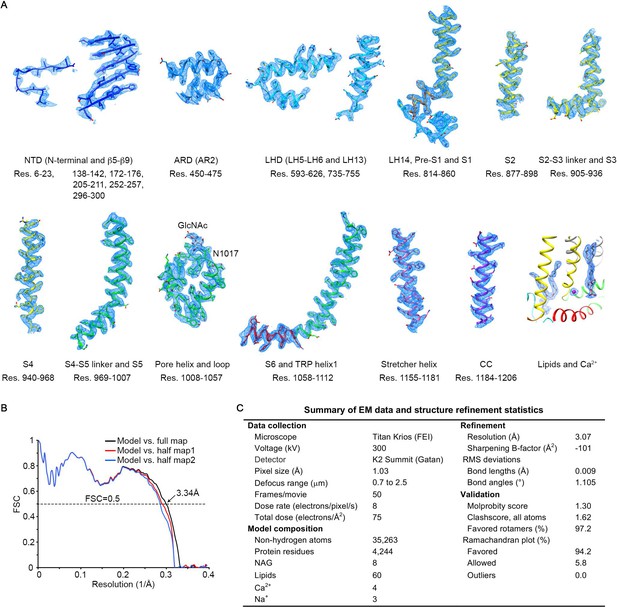
Validation of the atomic model.
(A) Local EM density maps and fitted atomic models for representative regions of the structure. (B) Fourier shell correlation (FSC) curves between the model and different maps. The model was refined against half map1. (C) Summary of data collection parameters and structure refinement statistics.
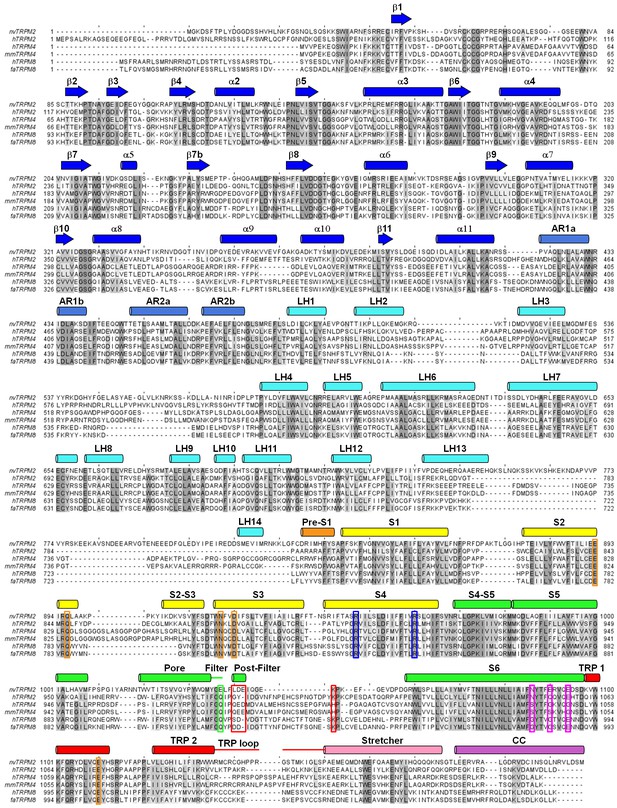
Multiple sequence alignment of TRPM proteins.
Sequence alignment of TRPM2, TRPM4, TRPM5, and TRPM8 channel orthologs (h: human, nv: Nematostella vectensis, mm: Mus musculus, fa: Ficedula albicollis) predicted by Clustal Omega (Li et al., 2015), and manually adjusted in the pore region. Conserved residues are shaded according to percentage of identity using Jalview (Waterhouse et al., 2009). Secondary structure elements are shown for nvTRPM2 above the sequences, and colored according to Figure 2. Colored boxes identify residues discussed in the text: residues involved in Ca2+ coordination (orange boxes), conserved arginines in helix S4 (blue boxes), residues that form the post-filter helix in nvTRPM2, and that corresponding to K1047 in nvTRPM2 (red boxes), negative residues in nvTRPM2 inner vestibule (pink boxes).
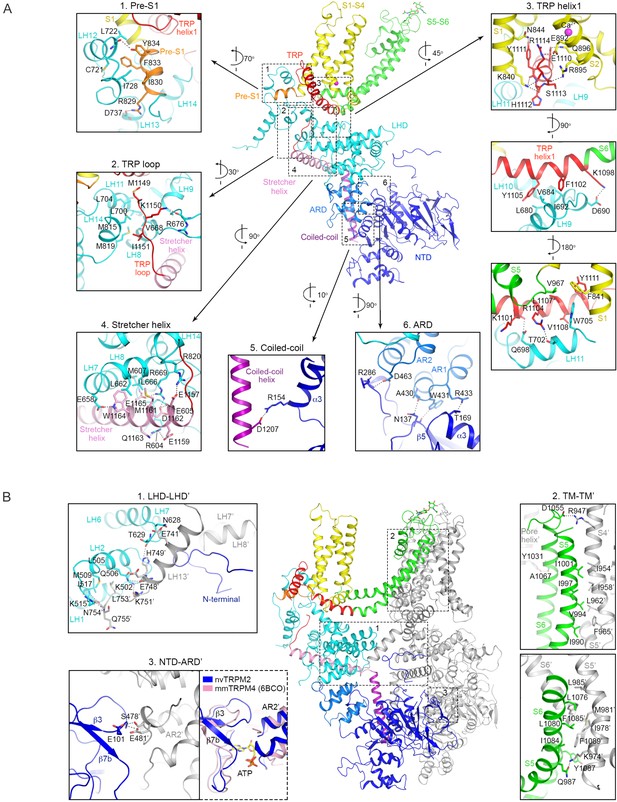
Intra- and inter-subunit interactions.
(A) Intra-subunit interactions. Interactions between different domains within one subunit shown in cartoon representation and color-coded by domains (see Figure 2D). (B) Inter-subunit interactions. Interactions between two adjacent subunits shown in cartoon representation. One subunit is color-coded by domains (see Figure 2D), the other subunit is gray. Numbered insets in (A) and (B) magnify regions marked by dashed boxes, and display side chains involved in hydrophobic (<4.0 Å) and electrostatic interactions (<3.5 Å) as sticks. Electrostatic interactions are further indicated with dotted lines. A comparison of the ATP binding site of mmTRPM4 (PDB: 6BCO) with the corresponding region of nvTRPM2 is attached to inset three with a dotted box.
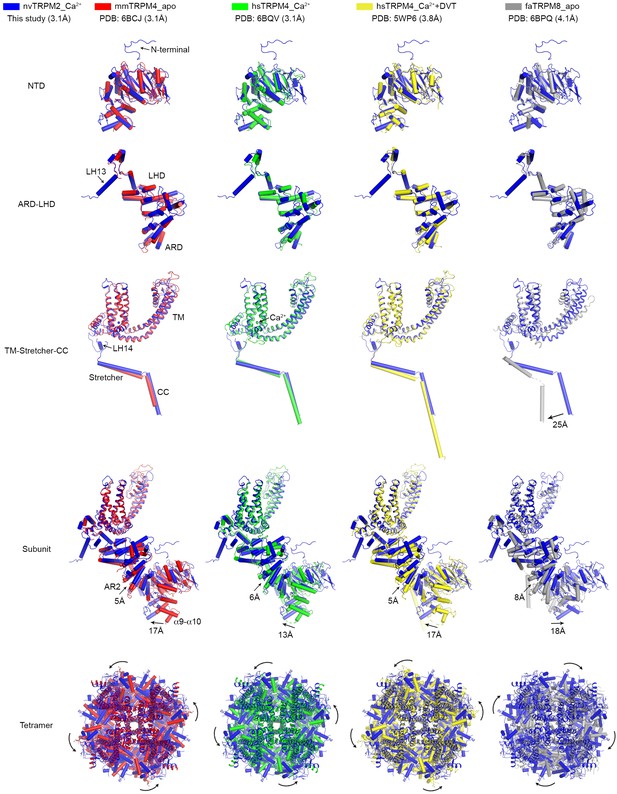
Structural comparisons between individual domains, one subunit, or the entire tetramer of nvTRPM2 and other TRPM family channels.
The superposition was performed for individual domains (NTD, ARD-LHD, and TM-Stretcher-CC), the integral subunit, as well as the entire tetramer. The root-mean-square deviations (RMSDs) between nvTRPM2 (blue) and mmTRPM4_apo (red, PDB: 6BCJ), hsTRPM4_Ca2+ (green, PDB: 6BQV), hsTRPM4_Ca2++DVT (yellow, PDB: 5WP6), and faTRPM8_apo (gray, PDB: 6BPQ) are 1.9 Å (299 Cα atoms), 1.8 Å (269 Cα atoms), 2.0 Å (293 Cα atoms), and 2.2 Å (230 Cα atoms), respectively, for the NTD, 1.7 Å (248 Cα atoms), 1.6 Å (232 Cα atoms), 1.8 Å (227 Cα atoms), and 1.8 Å (222 Cα atoms), respectively, for the ARD-LHD, and 2.4 Å (318 Cα atoms), 2.4 Å (333 Cα atoms), 2.9 Å (316 Cα atoms), and 3.2 Å (207 Cα atoms), respectively, for the TM-Stretcher-CC.
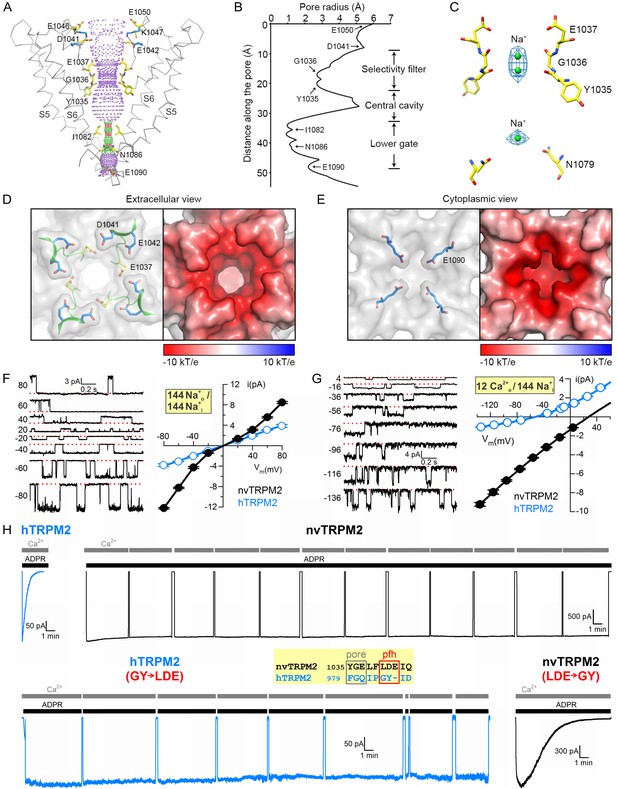
Pore of the nvTRPM2 channel.
(A) Ribbon representation of the ion pore, front and rear subunits are removed for clarity. The dotted mesh distinguishes regions that are too tight (red, radius <1.15 Å) or just spatious enough (green, 1.15 - 2.30 Å) for a single water molecule to pass, as well as regions with a radius larger than 2.30 Å (purple). Residues facing the pore are shown as sticks. The E1042 and K1047 side chains (blue sticks) form a salt bridge (blue dotted line). (B) Van der Waals radius of the pore along the central axis. (C) Electron densities in the selectivity filter (blue meshes), and nearby residues (sticks). The Na+ ions within the pore are represented as green spheres. (D) Extracellular view of the pore. Left panel: Surface representation; the selectivity filter and post-filter helix are shown in ribbon, the side chains of some important acidic residues as sticks. Right panel: Electrostatic property of the surface calculated at pH 7 and 0.15 M concentrations of monovalent cations and anions. (E) Cytoplasmic view of the pore. A constricting acidic residue (E1090) is shown as sticks in the left panel. (F–G) Unitary currents (left) and unitary current-voltage (i–V) relationships (right; solid black symbols (mean ± SEM)) of nvTRPM2 channels in symmetrical 144 mM Na+ as the cation (F), or with 12 mM Ca2+ in the extracellular (pipette) and 144 mM Na+ in the intracellular (bath) solution (G). Open blue symbols (mean ± SEM) in the i-V graphs, replotted from (Tóth and Csanády, 2012), represent hTRPM2. Smooth fitted curves in (G) were used to estimate reversal potentials under bi-ionic conditions (see Materials and Methods). (H) Inactivation of WT hTRPM2 (top left) and of nvTRPM2 with residues 1040–1042 (LDE) replaced by a GY doublet (bottom right), and lack of inactivation of WT nvTRPM2 (top right) and of hTRPM2 with residues 984–985 (GY) replaced by an LDE triplet (bottom left). Membrane potential was −20 mV, currents were activated by cytosolic exposures to 100 μM ADPR (black bars; 32 μM for hTRPM2) plus 125 μM Ca2+ (gray bars). Inset: Sequence alignment highlighting target residues (red box) swapped between nv and hTRPM2; pfh, post-filter helix. See also Figure 3—figure supplements 1–3.
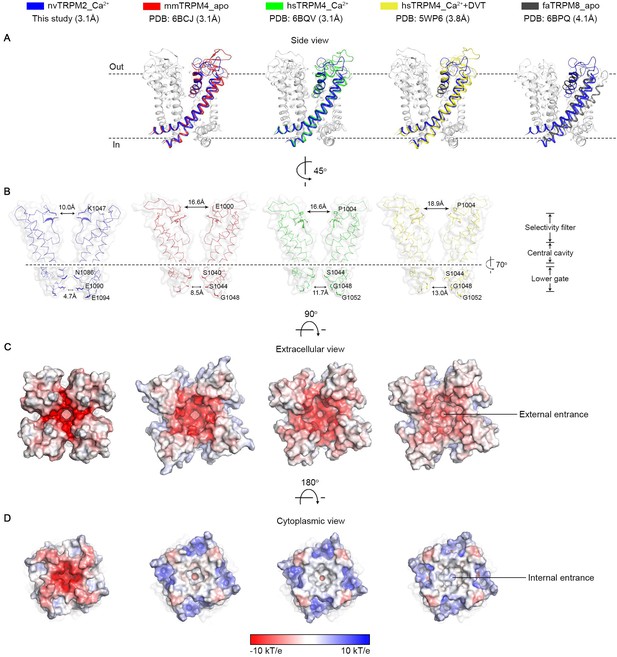
Structural comparisons between the ion channel pore of nvTRPM2 and of other TRPM family channels.
(A) Superposition of the pore region (S5–S6) between nvTRPM2 and other TRPM family structures. The same structures as those in Figure 2—figure supplement 5 were used for comparison. Only one subunit is colored, the other three are gray. The root-mean-square deviations (RMSDs) between nvTRPM2 (blue) and mmTRPM4_apo (red), hsTRPM4_Ca2+ (green), hsTRPM4_Ca2++DVT (yellow), and faTRPM8_apo (gray) are 1.5 Å (457 Cα atoms), 1.6 Å (448 Cα atoms), 1.6 Å (444 Cα atoms), and 3.4 Å (267 Cα atoms), respectively. (B) Comparison of diameters of the external and internal vestibule of nvTRPM2 and of various TRPM4 structures. Side chains important for shaping both vestibules are highlighted as sticks. (C–D) Electrostatic surface representations showing the external (C) and internal (D) entrances of the pore for nvTRPM2 and for various TRPM4 structures.
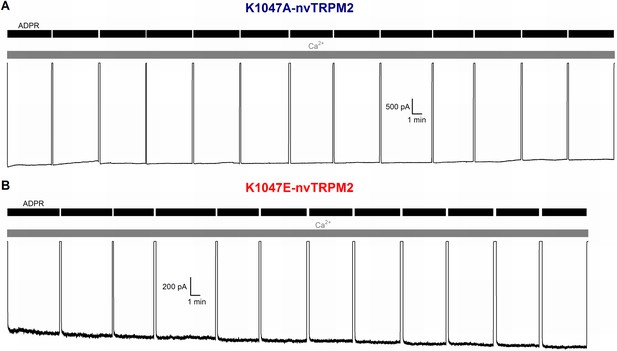
Disruption of post-filter salt bridge does not cause inactivation.
(A–B) Lack of inactivation of nvTRPM2 channels in which the E1042-K1047 salt bridge has been disrupted by mutation K1047A (A) or K1047E (B). Membrane potential was −20 mV, currents were activated by cytosolic exposures to 100 μM ADPR (black bars) plus 125 μM Ca2+ (gray bars).
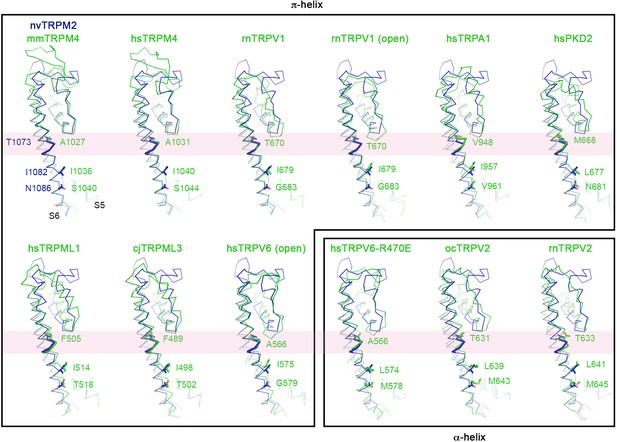
Alpha-pi-alpha helical transition in S6 of TRP family channels.
Pairwise structural superposition of the pore domain of one nvTRPM2 subunit (blue) with those of other TRP channels (green), including mm (Mus musculus) and hs (Homo sapiens) TRPM4 (PDB: 6BCJ and 6BQV), rn (Rattus norvegicus) TRPV1 (PDB: 5IRZ and 5IRX), hsTRPA1 (PDB: 3J9P), hsPKD2 (PDB: 5T4D), hsTRPML1 (PDB: 5WJ5), cj (Callithrix jacchus) TRPML3 (PDB: 5W3S), hsTRPV6 (PDB: 6BO8 and 6BOA), oc (Oryctolagus cuniculus) TRPV2 (PDB: 5AN8), and rnTRPV2 (PDB: 5HI9). The two structures that are in the open conformation are indicated, all other structures represent the closed conformation. The α- to π-helix transition regions in S6 are shown as thicker ribbon and highlighted with pink bands. Structures that are π- or α-helical in this region are boxed separately. In all structures the residue corresponding to the alanine gating hinge of TRPV6, and the two residues that form the lower gate, are labeled and shown with side chains.
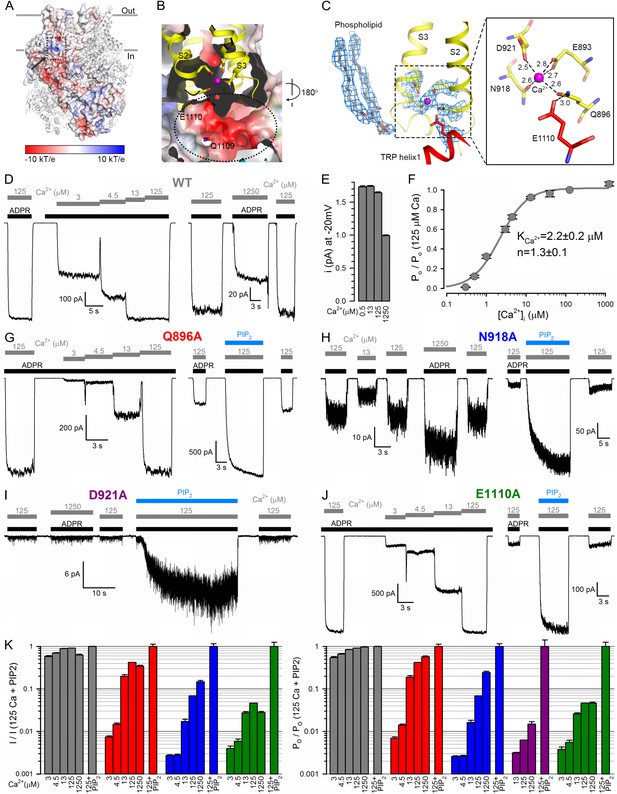
The nvTRPM2 Ca2+binding site.
(A) The Ca2+ binding site is located close to the inner leaflet of the membrane. One subunit is represented as electrostatic surface, the remaining three are shown as ribbon. The position of the Ca2+ binding site (hidden behind surface) is indicated with a dashed box and the vestibule of the peripheral tunnel is marked with an arrow. (B) A cross section of the Ca2+ binding site. Ca2+ is shown as a magenta sphere. Side chains of residues from S2 and S3 involved in Ca2+ coordination, as well as nearby side chains of residues in TRP helix 1, are shown as sticks. The peripheral tunnel located between the transmembrane and the TRP domain is indicated with a dashed ellipse. (C) Local EM densities and geometry of the Ca2+ binding site. The nearby phospholipid with a poorly resolved head group was modeled as a phosphatidic acid. (D, G–J) Macroscopic inward Na+ currents through WT (D), Q896A (G), N918A (H), D921A (I), and E1110A (J) nvTRPM2, evoked by cytosolic exposures to 100 μM ADPR (black bars) and various concentrations (in μM) of free Ca2+ (gray bars), with or without 25 μM dioctanoyl-PIP2 (blue bars). Extracellular (pipette) [Ca2+] was ~1 nM, membrane potential was −20 mV. (E) Absolute values of WT nvTRPM2 unitary current amplitudes (mean ± SEM) under conditions similar to those in panel D: at −20 mV membrane potential in the presence of symmetrical 144 mM Na+, but various cytosolic [Ca2+] (in μM). (F) Dependence on cytosolic [Ca2+] of nvTRPM2 open probability (Po; mean ±SEM), normalized to that in 125 μM Ca2+ (Po;125), calculated as Po/Po;125=(I/I125)/(i/i125) (I, macroscopic current; i, unitary current). Gray curve is a fit to the Hill equation with parameters plotted. (K) Dependence on cytosolic [Ca2+] of macroscopic current (left; mean ± SEM) and of channel open probability (right; mean ± SEM), normalized to the values observed in the presence of 125 μM Ca2+ + 25 μM dioctanoyl-PIP2 (see Materials and Methods), for WT (gray), Q896A (red), N918A (blue), D921A (purple), and E1110A (green) nvTRPM2. Fractional Po was calculated as in (F), except for D921A for which it was estimated using dwell-time analysis (see Materials and methods) as currents in the absence of PIP2 were too small for reliable cursor measurement. See also Figure 4—figure supplement 1.
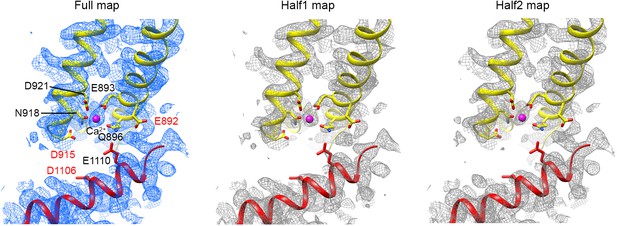
Density maps around the Ca2+binding site.
The full map and both half maps are presented. Residues that participate in Ca2+ binding are labeled with black font. Some adjacent acid amino acids not involved in hydrogen bonds or ion bindings are labeled with red font. Note strong side-chain densities for the acidic amino acids that participate in Ca2+ binding (D921 and E893), but poor side-chain densities for those that are not involved in hydrogen bonding or Ca2+ coordination (E892, D915, and D1106).
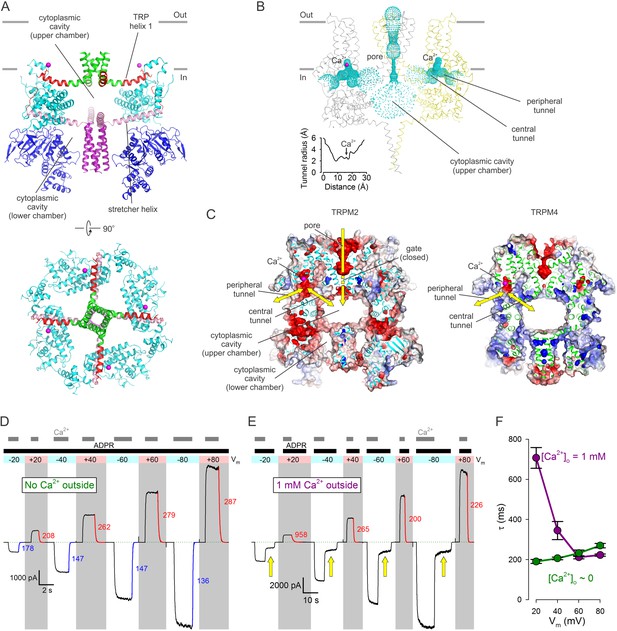
Cytoplasmic cavities and tunnels.
(A) Architecture of the cytoplasmic cavity viewed from an angle parallel (top) or perpendicular (bottom) to the membrane plane. The NTD (blue), the LHD (cyan), the cytoplasmic ends of transmembrane helix S6 (green), TRP helix 1 and the TRP loop (red), the stretcher helix (pink), and the CC (purple) are shown as cartoon, Ca2+ ions as magenta spheres, and the E1110 side chain as sticks. In the top panel two diagonally opposing subunits are shown, in the bottom panel the NTD is removed for clarity. (B) Ribbon representation of the top and middle tiers and the CC helices, front and rear subunits omitted for clarity. Dotted mesh represents the contiguous surface that lines the pore, the upper chamber, the central and peripheral tunnels, and the Ca2+ binding sites. Inset plots the van der Waals radius of the tunnel along its central axis. (C) Longitudinal cross sections through nvTRPM2 (left) and hTRPM4 (right; PDBID:6BQV), showing connectivities and surface electrostatics for the upper chamber, a central and peripheral tunnel, and the corresponding Ca2+ binding site. Yellow arrows highlight possible pathways for Ca2+ flux. (D–E) Macroscopic nvTRPM2 currents evoked at various membrane potentials (colored bars and shading) by cytosolic exposures to 100 μM ADPR +125 μM Ca2+, in the presence of either ~1 nM (D) or 1 mM (E) free Ca2+ in the extracellular (pipette) solution. Colored curves are single exponentials fitted to the current decay time courses that follow cytosolic Ca2+ removal, colored numbers are time constants (in ms). Green dotted line marks zero-current level. Yellow arrows in (E) highlight current fractions that survive removal of cytosolic Ca2+. (F) Voltage dependence of closing time constants (mean ± SEM) upon cytosolic Ca2+ removal, in the presence (purple symbols) or absence (green symbols) of extracellular Ca2+.
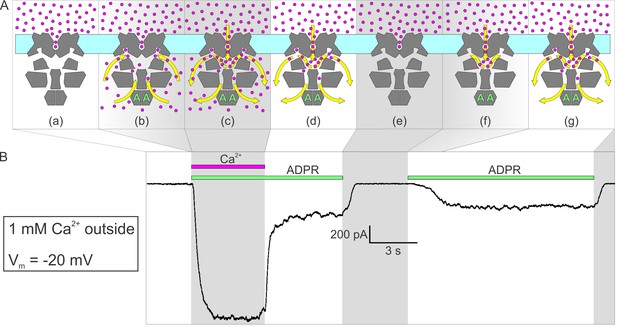
Activation by ADPR of nvTRPM2 current in the presence of external but absence of cytosolic Ca2+.
(A) Cartoon interpretation of the molecular events that occur during consecutive time intervals (sections a-g, also identified by intermittent gray shading in (A)-(B)) of the current recording in (B). Membrane, light cyan; nvTRPM2 protein, dark gray; Ca2+ ions, magenta spheres; ADPR, green letters ‘A’; direction of Ca2+ flow, yellow arrows. (B) In the presence of 1 mM external Ca2+, at −20 mV membrane potential, a fraction of the macroscopic nvTRPM2 current evoked by cytosolic exposure to 100 μM ADPR +125 μM free Ca2+ survives cytosolic Ca2+ removal, and subsides only upon removal of ADPR (see Figure 5E). A second application of 100 μM ADPR, without cytosolic Ca2+, activates a current comparable to that which survived prior Ca2+ removal. Note delayed current activation following exposure to ADPR alone.
Videos
Cytosolic chambers and tunnels in nvTRPM2.
(Right) Vertical cross section along nvTRPM2 pore axis: 4 Å slab represented in spacefill with surface rendering. Domain color coding as in Figure 1E. Sodium ions in the pore and bound Ca2+ ions are shown as red and magenta spheres, respectively. (Left) Sequential horizontal cross sections perpendicular to the nvTRPM2 pore axis: 4 Å slabs represented in spacefill with surface rendering, moving through the structure in 1 Å steps. The actual position of the slab on the left is illustrated by the moving black box on the right. Apertures that connect the upper chamber with the cytosol (lower and upper lateral windows, tunnels) are labeled.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| cell line (Spodoptera frugiperda) | Sf9 | ATCC | CRL-1711 | |
| cell line (Homo sapiens) | HEK293S GnTI- | ATCC | CRL-3022 | |
| biological sample (Xenopus laevis) | Xenopus laevis oocytes | African Reptile Park < mandyvorster@xsinet.co.za> | RRID:NXR_0.0080 | |
| commercial assay or kit | CNBR-activated sepharose beads | GE Healthcare | 17043001 | |
| commercial assay or kit | Superose 6, 10/300 GL | GE Healthcare | 17517201 | |
| commercial assay or kit | HiSpeed Plasmid Midi Kit | Qiagen | Catalog #12643 | |
| commercial assay or kit | QuikChange XL Site- Directed Mutagenesis Kit | Agilent Technologies | Catalog #200521 | |
| commercial assay or kit | mMESSAGE mMACHINE T7 Transcription Kit | ThermoFisher | Catalog #AM1344 | |
| chemical compound, drug | 2,2-didecylpropane-1,3- bis-β-D-maltopyranoside (LMNG) | Anatrace | NG310 | |
| chemical compound, drug | Cholesteryl hemisuccinate (CHS) | Anatrace | CH210 | |
| chemical compound, drug | Digitonin | Sigma-Aldrich | D141 | |
| chemical compound, drug | sf-900 II SFM medium | Gibco | Cat#10902088 | |
| chemical compound, drug | Cellfectin II reagents | Invitrogen | Cat#10362100 | |
| chemical compound, drug | Freestyle 293 medium | Gibco | Cat#12338018 | |
| chemical compound, drug | HI FBS | Gibco | Cat#16140071 | |
| chemical compound, drug | Antibiotic-Antimycotic (100X) | Gibco | Cat#15240062 | |
| chemical compound, drug | Gentamicin sulphate | Sigma-Aldrich | G1397-10mL | |
| chemical compound, drug | Collegenase type II | Gibco by life technologies | 17107–0125 | |
| chemical compound, drug | Adenosine 5′- diphosphoribose sodium salt | Sigma-Aldrich | A0752 | |
| chemical compound, drug | PtdIns-(4,5)-P2 (1,2- dioctanoyl) (sodium salt) | Cayman Chemical | 64910 | |
| software, algorithm | Seriel EM | DOI: 10.1016/j.jsb.2005.07.007 | http://bio3d.colorado.edu/SerialEM | |
| software, algorithm | MotionCor2 | DOI: 10.1038/nmeth.4193 | http://msg.ucsf.edu/em/software/motioncor2.html | |
| software, algorithm | Gctf | DOI: 10.1016/j.jsb.2015.11.003 | https://www.mrc-lmb.cam.ac.uk /kzhang/ | |
| software, algorithm | Gautomatch | other | https://www.mrc-lmb.cam.ac.uk /kzhang/ | Downloaded from a personal URL |
| software, algorithm | RELION 2.1 | DOI: 10.7554/eLife.18722 | http://www2.mrc-lmb.cam.ac.uk /relion | |
| software, algorithm | SWISS-MODEL | DOI: 10.1093/nar/gku340 | https://swissmodel.expasy.org | |
| software, algorithm | COOT | DOI: 10.1107/S0907444910007493 | https://www2.mrc- lmb.cam.ac.uk/personal/ pemsley/coot | |
| software, algorithm | PHENIX | DOI: DOI: 10.1107/S0907444909052925 | https://www.phenix-online.org | |
| software, algorithm | Blocres | DOI: 10.1016/j.jsb.2006.06.006 | https://lsbr.niams.nih.gov/bsoft /programs/blocres.html | |
| software, algorithm | MolProbity | DOI: 10.1107/S0907444909042073; 10.1093/nar/gkm216 | http://molprobity.biochem.duke .edu | |
| software, algorithm | Chimera | DOI: 10.1002/jcc.20084 | https://www.cgl.ucsf.edu/chimera | |
| software, algorithm | Pymol | PyMOL | http://www.pymol.org | |
| software, algorithm | HOLE | PMID: 9195488 | http://www.holeprogram.org | |
| software, algorithm | APBS | DOI: 10.1093/nar/gkm276; 10.1073/pnas.181342398 | http://www.poissonboltzmann.org | |
| software, algorithm | Pclamp9 | Molecular Devices | RRID:SCR_011323 | |
| other | R1.2/1.3 400 mesh Au holey carbon grids | Quantifoil | 1210627 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36409.018





