Crystallographic observation of nonenzymatic RNA primer extension
Figures
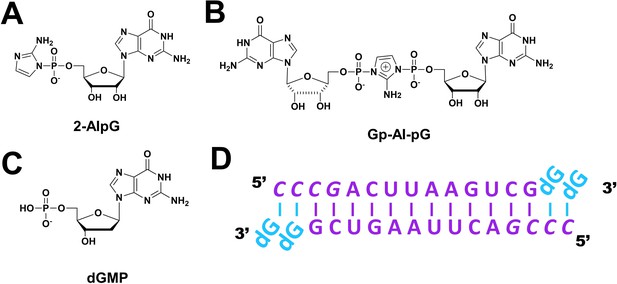
Chemical structures of mononucleotide and dinucleotide molecules in this study.
(A) guanosine-5′-phosphoro-2-aminoimidazolide (2-AIpG). (B) 2-aminoimidazolium-bridged guanosine dinucleotide intermediate (Gp-AI-pG). (C) 2'-deoxyguanosine-5'-monophosphate (dGMP). (D) Schematic of the RNA-dGMP complex used for crystallographic studies. The four italic nucleotides at the 5′-end represent locked nucleic acid.
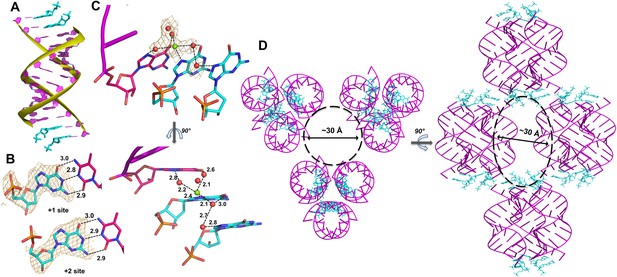
Crystal structure of RNA-dGMP complex.
(A) One dimeric RNA duplex (purple with yellow backbone) is bound by two dGMP monomers (cyan) at each end. (B) dGMP monomers in the +1 and+2 position of the template form Watson-Crick base pairs. Wheat meshes indicate the corresponding Fo-Fc omit maps contoured at 2.0 σ. (C) A Mg2+ ion with coordinating water molecules form extended contacts between the 3′ end of the primer and both template-bound monomers through hydrogen bonds with N7. Wheat meshes indicate the corresponding Fo-Fc omit maps contoured at 1.0 σ. (D) Top and side views of RNA-dGMP complex packing in crystal. Molecular packing of the RNA-dGMP complex shows a possible channel (dashed circle and oval) for diffusion of small molecules through the crystal.
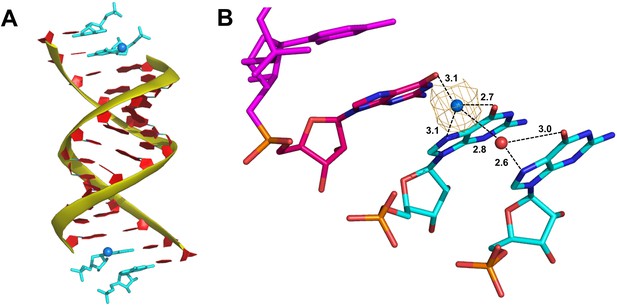
A Sr2+ ion and coordinating water molecule form an extended interaction network between the 3′ end of the primer and monomers in the +1 and+2 positions.
(A) Similar to the RNA-dGMP complex, the RNA duplex (red with yellow backbone) crystallizes with two bound GMP monomers (cyan) at each end. (B) Close up view of the template-bound GMP monomers showing a Sr2+ ion (blue sphere) with a coordinating water molecule (red sphere) interacting with the N7 of monomers in the +1 and+2 position. Wheat mesh indicates the corresponding Fo-Fc omit map contoured at 4.0 σ. Interaction distances are labeled.
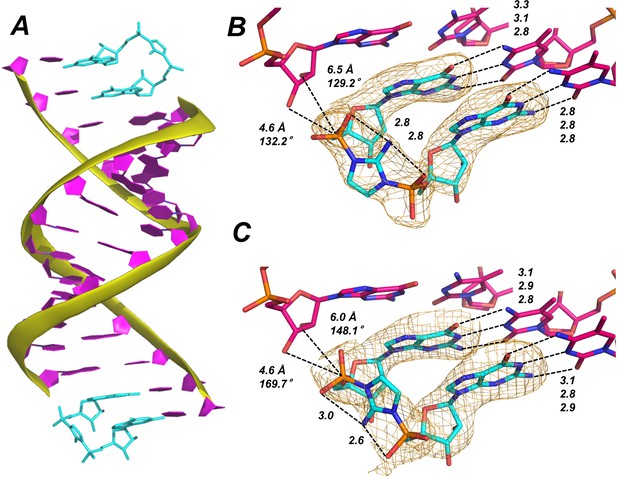
Template-bound Gp-AI-pG is pre-organized for phosphodiester bond formation.
(A) Structure of RNA duplex (purple with yellow backbone) bound by two Gp-AI-pG molecules (cyan) obtained after 4 hr of soaking the RNA-dGMP complex in buffer containing Gp-AI-pG. (B) Close-up of template-bound Gp-AI-pG modeled with the 2-amino group of the imidazolium bridge pointing toward the major groove and (C) minor groove. Distances for hydrogen bonds and in-line attack by the primer 3′-hydroxyl are indicated by dashed lines. Wheat meshes indicate the corresponding Fo-Fc omit maps contoured at 1.5 σ.
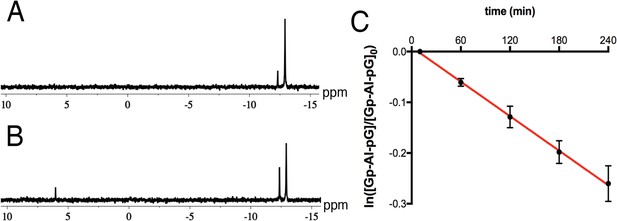
Decay of the Gp-AI-pG intermediate in crystal soaking buffer.
31P NMR spectra of 3.5 mM Gp-AI-pG after 10 min (A) or 4 hr (B) of incubation in crystal soaking buffer (5% v/v (+/-)−2-methyl-2, 4-pentanediol, 20 mM sodium cacodylate pH 7.0, 6 mM spermine tetrahydrocholoride, 40 mM sodium chloride). The peak at −12.91 ppm corresponds to Gp-AI-pG, and −12.26 ppm to 2-AIpG. The peak at 6.01 ppm may correspond to reaction with the buffer. The peak for GMP is too broad to be observed at these concentrations. (C) Quantification of the decay of Gp-AI-pG in a pseudo-first order rate plot. The slope is equal to the negative of the first order rate constant k = (6.8 ± 0.1) x 10−2 h−1. This corresponds to a half-life of 10.2 ± 0.2 hr under these conditions. The experiment was performed in triplicate.
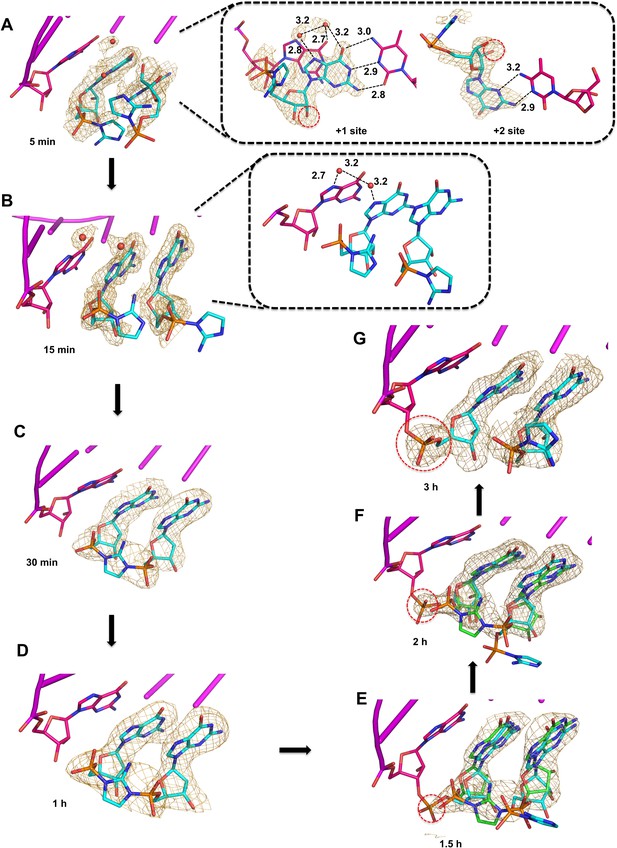
Observation of imidazolium-bridge and phosphodiester bond formation within crystals soaked with 2-AIpG.
Wheat meshes in all structures indicate the corresponding Fo-Fc omit maps contoured at 1.5 σ. (A) Structure after 5 min of soaking shows 2-AIpG monomer binding template through Watson-Crick and non-canonical base pairing. Inset also shows electron density belonging to 2′-hydroxyl (red dashed circle) and water molecules (red dots) that interact with N7 guanosine bases. (B) Structure after 15 min soaking shows both 2-AIpG monomers bound through Watson-Crick base pairing. Inset shows two water molecules that bridge the first monomer to the primer. (C) Structures after 30 min and (D) 1 hr of soaking shows the imidazolium-bridged Gp-AI-pG intermediate bound to the template. (E) Structures after 1.5 hr, and (F) 2 hr show a mix of template-bound Gp-AI-pG and extended primer (red dashed circle) (G) After 3 hr soaking, the electron density predominantly corresponds to the extension product.
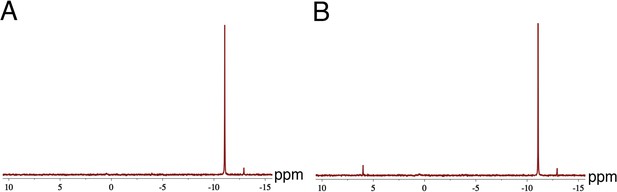
Decay of the 2-AIpG monomer in crystal soaking buffer.
31P NMR spectra of 20 mM 2-AIpG after 3 min (A) or 3 hr (B) of incubation in crystal soaking buffer (5% v/v (+/-) −2-methyl-2, 4-pentanediol, 20 mM sodium cacodylate pH 7.0, 6 mM spermine tetrahydrocholoride, 40 mM sodium chloride) with 20 mM MgCl2. The large peak at −11.11 ppm corresponds to 2-AIpG. During the three hours of incubation, only 7.8% of the monomer decays, mostly to an unknown product corresponding to the peak at 5.99 ppm that may represent reaction with a buffer component. The small peak at −12.92 ppm corresponds to Gp-AI-pG. The experiment was performed in triplicate.
Tables
Crystallographic and structural features of RNA-ligand complexes.
https://doi.org/10.7554/eLife.36422.008| RNA- dGMP | RNA- Gp-AI-pG | Time-resolved structures (RNA-2-AIpG) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Stage 1 (5 min) | Stage 2 (15 min) | Stage 3 (30 min) | Stage 4 (1 h) | Stage 5 (1.5 h) | Stage 6 (2 h) | Stage 7 (3 h) | |||
| PDB code | 6C8D | 6C8E | 6C8I | 6C8J | 6C8K | 6C8L | 6C8M | 6C8N | 6C8O |
| Ligand binding modes | Watson-Crick | Watson-Crick | mixed | Watson-Crick | Watson-Crick | Watson-Crick | Watson-Crick | Watson-Crick | Watson-Crick |
| N7 interaction | Mg2+ | N.D. | H2O | H2O | N.D. | N.D. | N.D. | N.D. | N.D. |
| 3ʹ-O-P distance, Å | ~6.1 | 4.6 | 3.7–4.1 | ~5.0 | ~4.3 | 4.1–5.0 | 1.6–4.2 | 1.6–4.1 | 1.6 |
| P-P distance between monomers, Å | N.D. | ~5.1 | N.D. | 6.2 | 5.2 | 5.2–5.4 | 5.5–7.1 | 5.0–7.3 | ~6.2 |
| N7-N7 distance between primer and + 1 monomer, Å | 4.2 | 3.7–4.0 | 3.9–4.1 | 4.0 | 3.7–4.0 | 3.7–4.2 | 3.7–4.3 | 3.7–4.1 | 4.1 |
-
N.D.: not detectable
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Sequence-based reagent | synthetic RNA | Exiqon Inc. | ||
| Commercial kit | Crystallization Screening kits | Hampton research, Inc. | ||
| Software | Refmac5 | University of Cambridge DOI: 10.1107/S0907444996012255 | RRID:SCR_014225 | |
| Software | HKL2000 | HKL Research Inc. DOI: 10.1016/S0076-6879(97)76066-X | RRID:SCR_015547 | |
| Software | Phaser 2.7 | University of Cambridge DOI: 10.1107/S0021889807021206 | RRID:SCR_014219 | |
| Software | MestReNova | Mestrelab Research, Inc. | ||
| Software | Pymol2 | Schrödinger, Inc. | RRID:SCR_000305 |
Additional files
-
Supplementary file 1
Table S1: Data collection statistics.
Table S2: Structure refinement statistics.
- https://doi.org/10.7554/eLife.36422.011
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36422.012





