How small-molecule inhibitors of dengue-virus infection interfere with viral membrane fusion
Figures
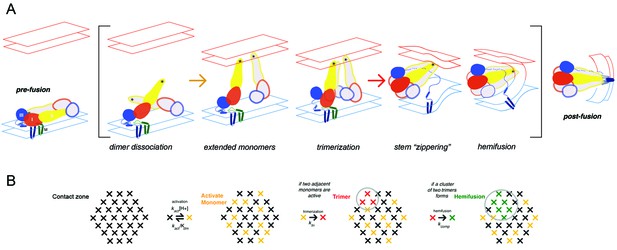
Flavivirus membrane fusion
(A) Steps in the fusion transition. Starting point and end point represent known structures; steps in brackets inferred from experimental evidence as summarized in (Chao et al., 2014). (B) Scheme for simulation of fusion reaction. Array of crosses represents a contact zone with 30 monomers.
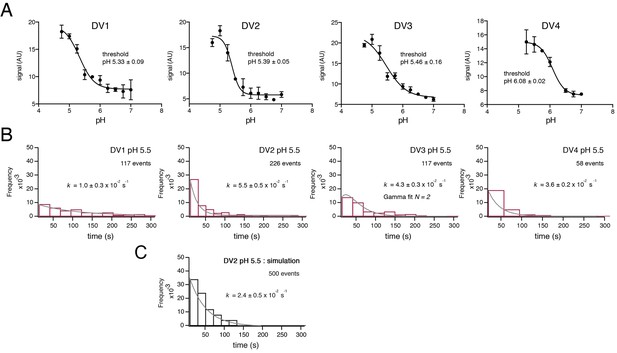
Fusion measurements.
(A) Fusion (with liposomes, in bulk solution) for VLPs of the four DV serotypes. The fluorescence from membrane-incorporated DiD is shown as a function of pH. Hemifusion (or fusion) at low pH causes dequenching of the VLP-incorporated fluorophore. (B) Histograms of single-particle fusion dwell times (between lowering of pH and observed dequenching) at pH 5.5 for each of the four DV serotypes. Curves show fit with a single exponential (DV1, DV2, and DV4) or with a gamma distribution, N = 2 (DV3). (C) Results of a simulation with parameters as described in the text. Compare with experimental data for DV2 fusion in the panel immediately above it.
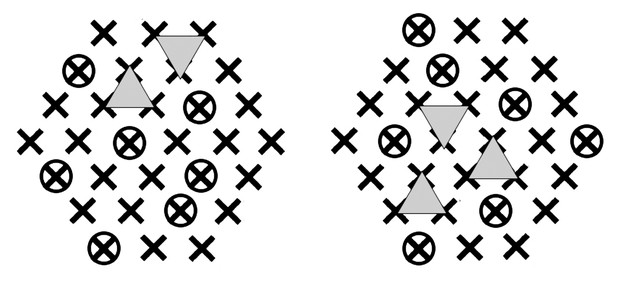
Simulation diagram.
The contact zone is represented as in Figure 1B. Circles indicate a particular choice of inihibitor-bound monomers. Two different random selections of 7 inactivated (inhibitor-bound) monomers will permit two adjacent trimers (triangles) to form, although in general later than the first possible set of adjacent trimers that would form if all monomers were potentially active. The right-hand pattern allows three possible pairs of trimers.
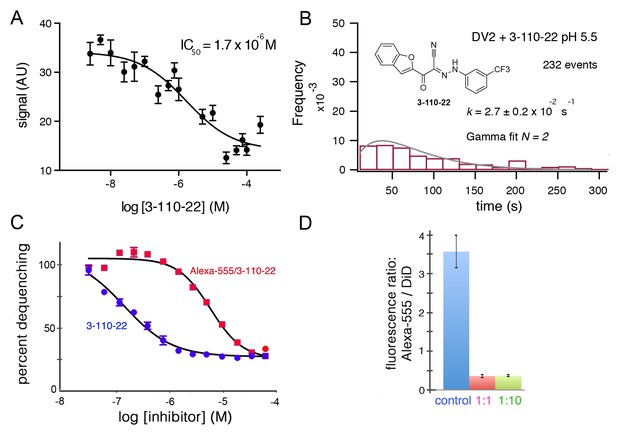
Inhibition of DV2 VLP fusion by 3-110-22.
(A) Fluorescence dequenching as a function of inhibitor concentration. (B) Single-particle dwell-time distribution at pH 5.5 in the presence of 1 μM inhibitor. (C and D) Inhibition of DV2 VLP fusion by Alexa-555/3-110-22. (C) Fluorescence dequenching as a function of inhibitor concentration. Percent dequenching calculated with 100% as DiD dequenching with no added inhibitor and 0% as dequenching with no pH drop. Error bars are SEM, n = 3. (D). Single-particle binding intensity for Alexa-555/3-110-22 in the presence of varying molar ratios of underivatized 3-110-22 (none, 1:1, 1:10). Error bars: SEM; n = 373, 370, 382 for the three sets of measurements, respectively.
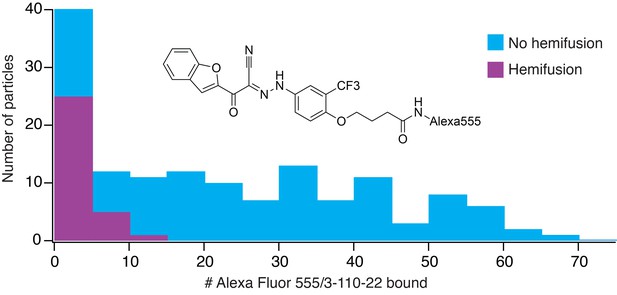
Frequency of hemifusion (measured as DiD fluorescence dequenching) as a function of number of bound Alexa-fluor-555/3-110-22 molecules.
Histogram shows the number of particles with a particular number of bound fluorescent inhibitor molecules, in bins of five. The number of particles in a bin that proceeded to hemifusion in the time of the experiment is shown as the height of the purple bar; the number of non-fusing particles is the total height of the bar (blue).
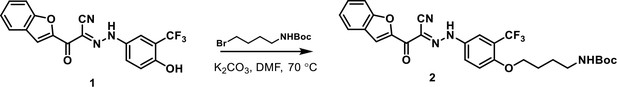
Addition of four-carbon linker with primary amine to 3-110-22
https://doi.org/10.7554/eLife.36461.007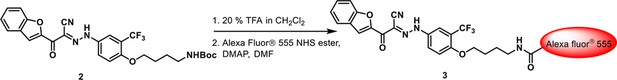
Conjugation of Alexa fluor 555 with activated 3-110-22.
https://doi.org/10.7554/eLife.36461.008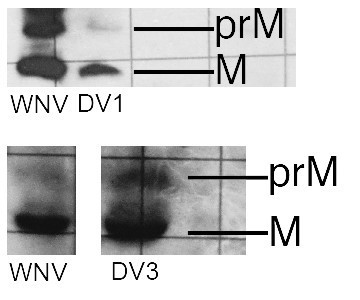
Western blots for VLPs of DV1 and DV3.
A WNV VLP control is on the left. These are photographed from a lab notebook, hence the rulings in the background.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36461.009





