BHLHE40, a third transcription factor required for insulin induction of SREBP-1c mRNA in rodent liver
Figures
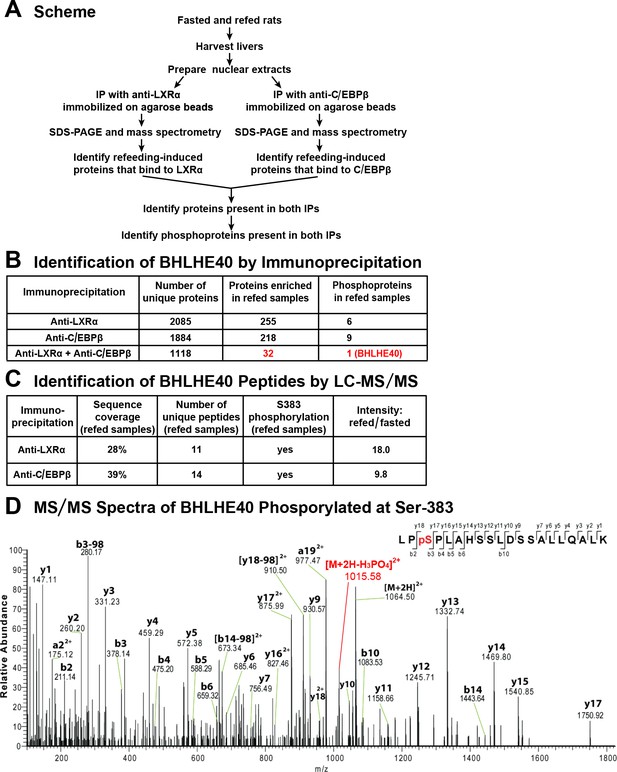
Co-immunoprecipitation of BHLHE40 phosphorylated at serine-383 (S383).
(A) Protocol for identifying the phosphoproteins that are induced by feeding and interact with both LXRα and C/EBPβ. Rats were either fasted for 48 hr, or fasted for 48 hr and then refed with a high-carbohydrate diet for 6 hr. Nuclear extracts were prepared from livers and subjected to immunoprecipitation with anti-LXRα or with anti-C/EBPβ, after which the immunoprecipitates were subjected to 10% SDS-PAGE and analyzed by mass spectrometry. Phosphoproteins that were enriched (threefold or greater) in refed samples and co-immunoprecipitated with both anti-LXRα and anti-C/EBPβ were identified. (B) LC-MS/MS analysis of immunoprecipitated proteins. (C) Identification of BHLHE40 peptides. The intensity ratio (refed/fasted) was calculated using the summed intensity of all peptides in the protein. (D) MS/MS spectra data of BHLHE40 phosphorylated at Ser-383. LC-MS/MS sample preparation and analysis are described in Materials and methods.
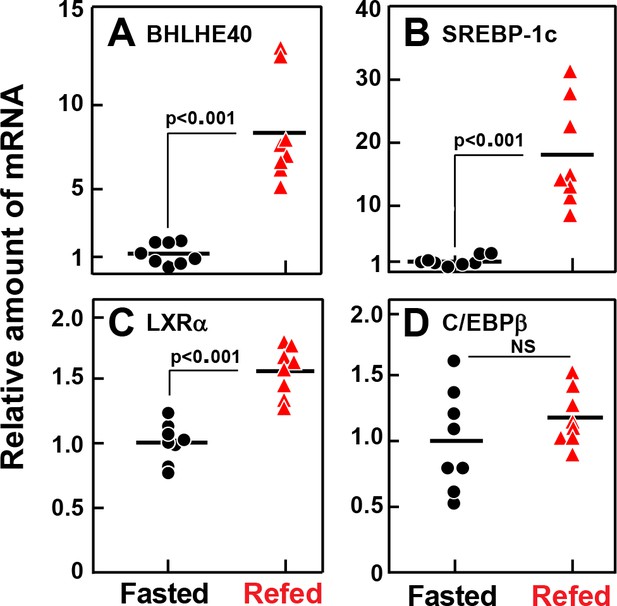
Relative amounts of mRNA for BHLHE40 (A), SREBP-1c (B), LXRα (C), and C/EBPβ (D), in livers of rats subjected to fasting and refeeding.
Male rats (age 2–3 months) were either fasted for 48 hr, or fasted for 48 hr and then refed with a high-carbohydrate diet for 6 hr. Total liver RNA from eight rats in each group was subjected to quantitative RT-PCR. Each black circle (fasted rats) or red triangle (refed rats) represents an individual animal. Each value in the refed group represents the amount of mRNA relative to the mean amount in the fasted group, which is arbitrarily defined as 1.0. Mean Ct values for BHLHE40, SREBP-1c, LXRα, and C/EBPβ in the fasted groups were 24.9, 23.9, 24.9, and 24.4, respectively. NS, not significant. p-Values calculated using Student’s t-test.
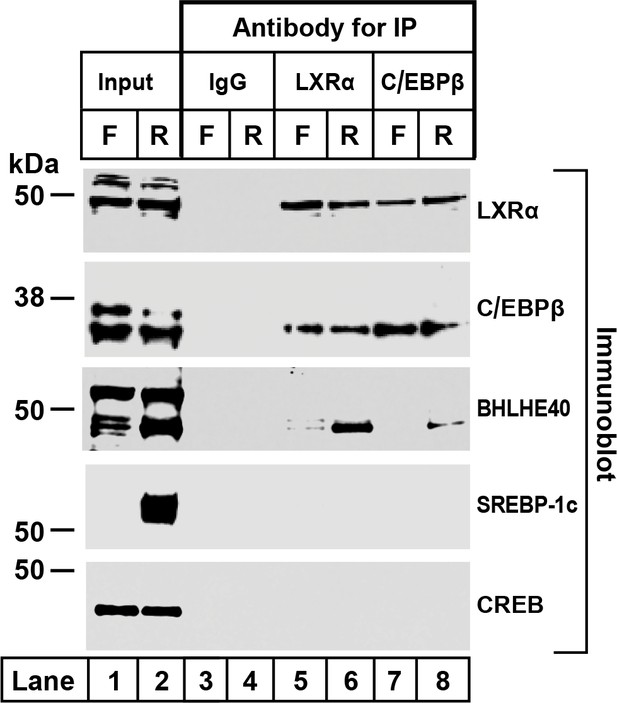
Co-immunoprecipitation of LXRα, C/EBPβ, and BHLHE40 in livers from fasted and refed rats.
Nuclear extracts were prepared from livers of rats subjected to 48 hr fasting (F) or 48 hr fasting followed by 6 hr refeeding (R). Immunoprecipitations were carried out with agarose beads conjugated with the indicated antibody as described in Materials and methods. After centrifugation, the beads were washed and boiled in SDS loading buffer to elute the proteins. Aliquots of the input (30% of total) and eluates from the immunoprecipitations (100% of eluated fraction) were subjected to SDS-PAGE and immunoblotted with the indicated antibodies as follows: 2 µg/ml of polyclonal anti-LXRα, a 1:1000 dilution of polyclonal anti-C/EBPβ, 1 µg/ml of polyclonal anti-BHLHE40, 1 µg/ml of monoclonal anti-SREBP-1, or a 1:1000 dilution of monoclonal anti-CREB. Proteins were detected with the LI-COR Odessy Infrared Imaging System using a 1:5000 dilution of anti-rabbit IgG conjugated to horseradish peroxidase.
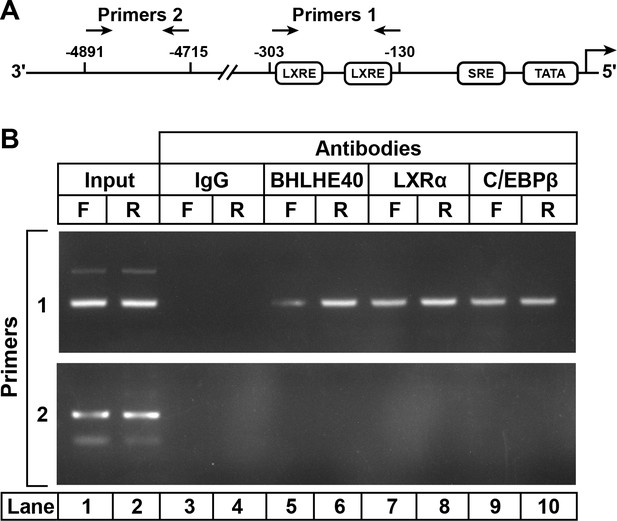
ChIP assay to demonstrate BHLHE40 binding to the enhancer/promoter region of rat SREBP-1c gene.
(A) Schematic diagram of rat SREBP-1c enhancer/promoter region, showing the location of the two LXREs. Arrows denote the location of the primers used in the ChIP assays. (B) ChIP assay was performed with livers from rats that had been subjected to either 48 hr fasting (F) or 48 hr fasting followed by 6 hr refeeding (R), as described in Materials and methods. After crosslinking, the sheared chromatin was incubated with 5 μg/mL of one of the following antibodies: anti-IgG (control), anti-BHLHE40, anti-LXRα, or anti-C/EBPβ, after which the DNA was purified from each immunoprecipitate and subjected to PCR with primer pairs 1 or 2, and the products visualized on an agarose gel. The DNA sequences for primers 1 and 2 are described in Table 2 in the paper by Tian et al. (2016).
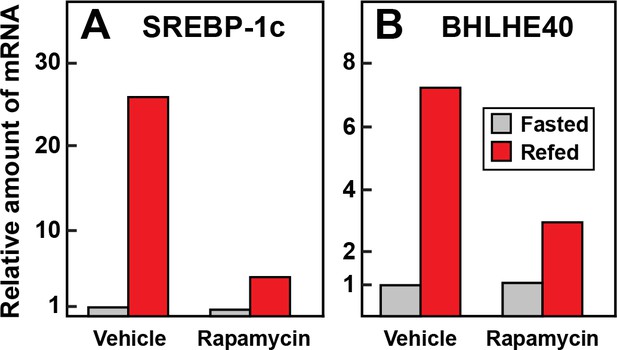
Requirement for mTOR in induction in mRNAs encoding SREBP-1c (A) and BHLHE40 (B) in livers of rats subjected to fasting and refeeding.
Sixteen male rats (age 2–3 months) were fasted for 48 hr. Six hours prior to sacrifice, eight rats received an intraperitoneal injection of 20 mg/kg rapamycin, and the other eight received vehicle. Four of the animals in the treated and untreated groups continued to fast, and the other four rats were refed a high-carbohydrate diet as described in Materials and methods. After 6 hr, the rats were sacrificed, and livers were homogenized. Equal amounts of total RNA from the livers of the four rats in each group were pooled and subjected to quantitative RT-PCR. Each bar represents the amount of mRNA relative to that of the vehicle-treated fasted group, which was defined as 1.0. Ct values for BHLHE40 and SREBP-1c in the fasted and vehicle treated group were 24.0 and 23.7, respectively.
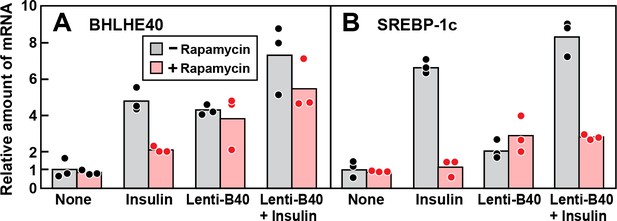
Overexpression of BHLHE40 fails to overcome rapamycin-mediated inhibition of SREBP-1c induction by insulin in primary rat hepatocytes.
Rat hepatocytes were prepared and plated on day 0 as described in Materials and methods. At 2 hr after plating, cells were infected with or without lentivirus expressing rat BHLHE40 (Lenti-B40; 5 × 105 TU/well) as indicated. On day 1, cells were pretreated with or without 100 nM rapamycin for 30 min, after which the cells received either no insulin or 100 nM insulin for 6 hr as indicated. The cells were then harvested, and the levels of mRNA encoding BHLHE40 (A) and SREBP-1c (B) were determined by RT-PCR. Each dot represents a value from an individual dish. Mean Ct values for BHLHE40 and SREBP-1c in the untreated control groups were 22.8 and 25.6, respectively.
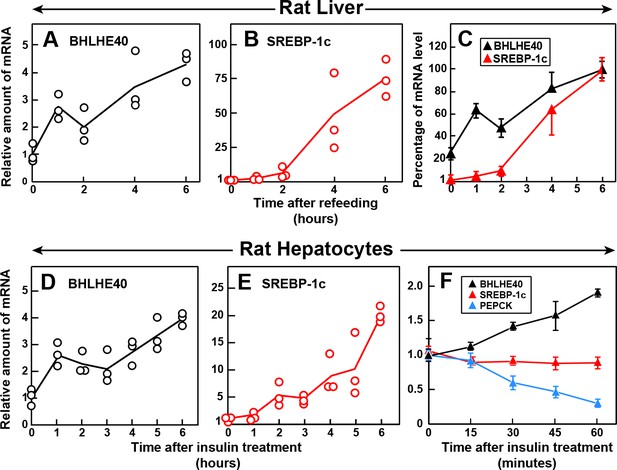
Time course of induction of BHLHE40 and SREBP-1c mRNA in livers of refed rats (A–C) and in primary rat hepatocytes treated with insulin (D–F).
(A–C) Male rats (age, 2–3 months) were fasted for 48 hr and then refed with a high-carbohydrate diet for the indicated time, after which total RNA from the liver was subjected to quantitative RT-PCR. Each point in (A) and (B) represents the mRNA level from a single animal relative to the mean value from three fasted rats (i.e. zero-time values). Zero-time Ct values for BHLHE40 and SREBP-1c were 23.5 and 25.7, respectively. Values in (C) represent the mean ± SEM of the values from (A) and (B) plotted as the percentages of the 6 hr value, which is defined as 100%. (D–F) Hepatocytes from nonfasted male rats (age, 2–3 months) on a chow diet were prepared and plated on day 0. On day 1, the cells received either no insulin or 100 nM insulin for the indicated time, after which the cells were harvested for measurement of total RNAs by quantitative RT-PCR. Each value in (D) and (E) (6 hr time course) represents the amount of mRNA from a single dish relative to that of the mean value from the three dishes at zero-time, which is defined as 1.0. Mean Ct values (zero-time) for BHLHE40 and SREBP-1c in the absence of insulin were 23.6 and 26.4, respectively. The values in (F) (1 hr time course) represent the mean ±SEM of the values from three dishes. Mean Ct values (zero-time) for BHLHE40, SREBP-1c, and PEPCK in the absence of insulin were 23.6, 26.3, and 20.0, respectively.
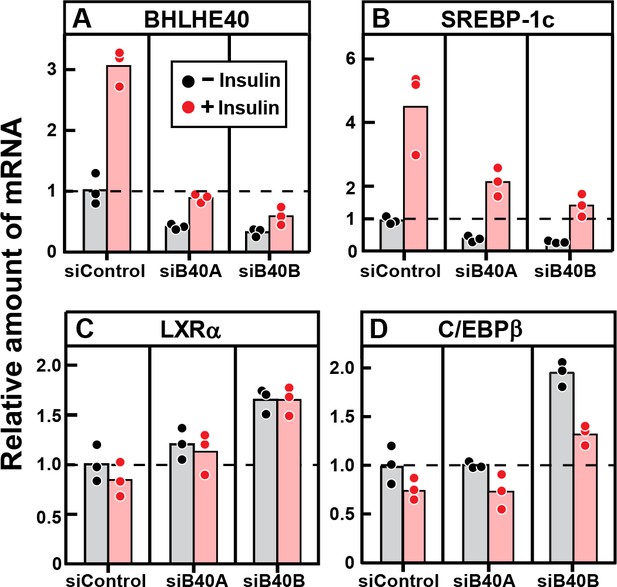
Knockdown of BHLHE40 mRNA in rat hepatocytes reduces insulin-induced mRNA for SREBP-1c, but not mRNA for LXRα or C/EBPβ.
Hepatocytes from non-fasted male rats (age, 2–3 months) on a chow diet were prepared and plated on day 0. After attachment for 2 hr, cells were transfected with either 50 nM control siRNA (siControl) or one of two siRNAs targeting BHLHE40 (siB40-A or siB40-B). On day 1, cells were treated with or without 100 nM insulin for 6 hr and then harvested for measurement of the indicated mRNA by quantitative RT-PCR. Each value represents an individual dish. Mean Ct values in the absence of insulin for BHLHE40, SREBP-1c, LXRα, and C/EBPβ were 21.8, 24.7, 25.5, and 22.2, respectively.
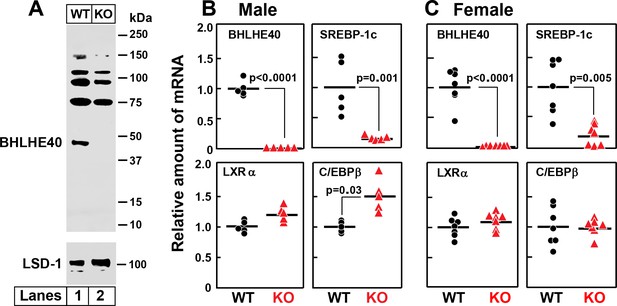
Knockout of Bhlhe40 gene in mice decreases SREBP-1c mRNA in livers of refed animals.
(A) Immunoblot analysis of liver nuclear extracts from wild type (WT) and Bhlhe40−/− (KO) mice. Nuclear extracts (20 µg protein) were subjected to 4–12% SDS-PAGE, followed by immunoblot analysis with 1.5 µg/ml of rabbit polyclonal anti-BHLHE40 (directed against amino acids 1–60 of rat BHLHE40 protein). LSD1 (lysine-specific demethylase 1) served as a loading control and was immunoblotted with a 1:1000 dilution of rabbit monoclonal anti-LSD1. Proteins were detected with the LI-COR Odyssey Infrared Imaging System using a 1:5000 dilution of anti-rabbit IgG conjugated to horseradish peroxidase. (B and C) WT and Bhlhe40 KO mice were fasted overnight and then refed a high-carbohydrate diet for 4 hr, after which total liver RNA was prepared and subjected to quantitative RT-PCR. Each circle (WT) or triangle (KO) represents an individual mouse. The mean value for WT mice is defined as 1.0. (B) mRNAs in livers of male WT and KO mice (age 6 wk; 5 mice/group). Mean Ct values for BHLHE40, SREBP-1c, LXRα and C/EBPβ in WT mice were 21.2, 22.2, 21.7, and 22.1, respectively. (C) mRNAs in livers of female WT and KO mice (age 7 wk; 7 mice/group). Mean Ct values for BHLHE40, SREBP-1c, LXRα and C/EBPβ in WT mice were 21.5, 21.0, 22.3, and 22.5, respectively. p-Values calculated using Student t-test.
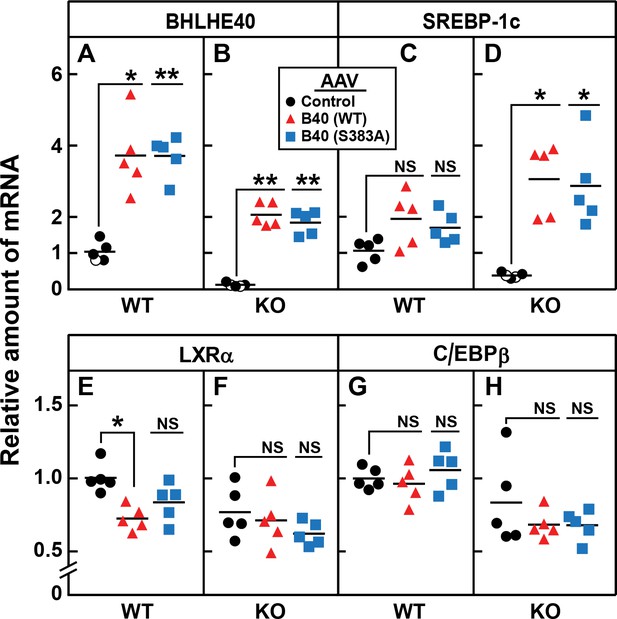
AAV-mediated injection of rat BHLHE40 restores the decrease of SREBP-1c mRNA in livers of refed Bhlhe40 knockout mice.
WT and KO mice (age 6 wk; 4 mice/group) received tail vein injections of AAV encoding a control mRNA (AAV-EGFP), wild-type BHLHE40, or mutant BHLHE40(S383A). Three weeks after injection, mice were fasted overnight and then refed with a high-carbohydrate diet for 4 hr, after which total liver RNA was prepared and subjected to quantitative RT-PCR. Each circle, triangle, or square represents an individual mouse. The mRNA expression is plotted as the amount relative to livers of WT mice injected with the control AAV-EGFP, which is assigned a value of 1.0. Mean Ct values for BHLHE40, SREBP-1c, LXRα and C/EBPβ in the livers of WT mice receiving AAV-EGFP were 22.3, 25.4, 22.4, and 22.7, respectively. *p<0.01; **p<0.001; NS, not significant. p-Values calculated using Student t-test.
Tables
Primer sequence for mRNA measurements
Primer sequences were custom synthesized by Integrated DNA Technologies, Coraville, IA.
| Gene (species) | Sequences for forward and reverse primers (5'–3') |
|---|---|
| 36B4 (rat) | Forward: TTCCCACTGGCTGAAAAGGT Reverse: CGCAGCCGCAAATGC |
| Srebp-1c (rat) | Forward: GACGACGGAGCCATGGATT Reverse: GGGAAGTCACTGTCTTGGTTGTT |
| C/Ebpβ (rat) | Forward: AAGCTGAGCGACGAGTACAAGA Reverse: GTCAGCTCCAGCACCTTGTG |
| Lxrα (rat) | Forward: TTCCCACGGATGCTAATGAA Reverse: GAATGGACGCTGCTCAAAGT |
| Bhlhe40 (rat) | Forward: GCTTCCAGGAAACCATTGGA Reverse: GGCTAGGAAGCTGGGCTTCT |
| Apob (mouse) | Forward: CGTGGGCTCCAGCATTCTA Reverse: TCACCAGTCATTTCTGCCTTTG |
| Srebp-1c (mouse) | Forward: GGAGCCATGGATTGCACATT Reverse: GGCCCGGGAAGTCACTGT |
| Bhlhe40 (mouse) | Forward: TGGTGATTTGTCGGGAAGAAA Reverse: ACGGGCACAAGTCTGGAAAC |
| Lxrα (mouse) | Forward: TCTGGAGACGTCACGGAGGTA Reverse: CCCGGTTGTAACTGAAGTCCTT |
| C/Ebpβ (mouse) | Forward: AAGCTGAGCGACGAGTACAAGA Reverse: GTCAGCTCCAGCACCTTGTG |
| Cyclophilin (mouse) | Forward: TGGAGAGCACCAAGACAGACA Reverse: TGCCGGAGTCGACAATGAT |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36826.013





