The plant pathogen Pseudomonas aeruginosa triggers a DELLA-dependent seed germination arrest in Arabidopsis
Figures
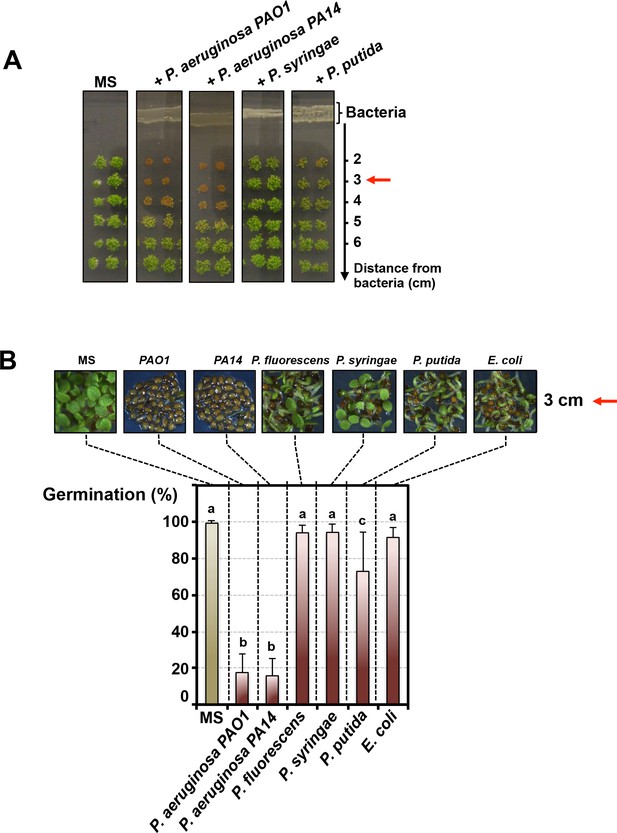
Pseudomonas aeruginosa releases a germination repressive activity (GRA).
(A) Pictures show Arabidopsis plants 4 days after sowing WT seeds in germination medium lacking bacteria (MS) or containing a given Pseudomonas species as indicated. Escherichia coli was included as a Gram-negative non-Pseudomonas species control. Note that seeds in close proximity of P. aeruginosa strains mostly did not germinate. Red arrows indicate distance used to calculate germination percentage in B. (B) Same experiment as in A. Representative pictures of plants 4 days after sowing seeds in germination medium in absence (MS) or presence of bacteria as indicated. Histograms show seed germination percentage 4 days upon sowing seeds in absence (MS) or presence of bacteria as indicated. Data represent mean ± standard deviation (nine replicates, n = 300–350). Lower case letters above histograms are used to establish whether two seed germination percentage values are statistically significantly different as assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05): different letters denote statistically different values.
-
Figure 1—source data 1
Germination percentage for each replicate.
- https://doi.org/10.7554/eLife.37082.005
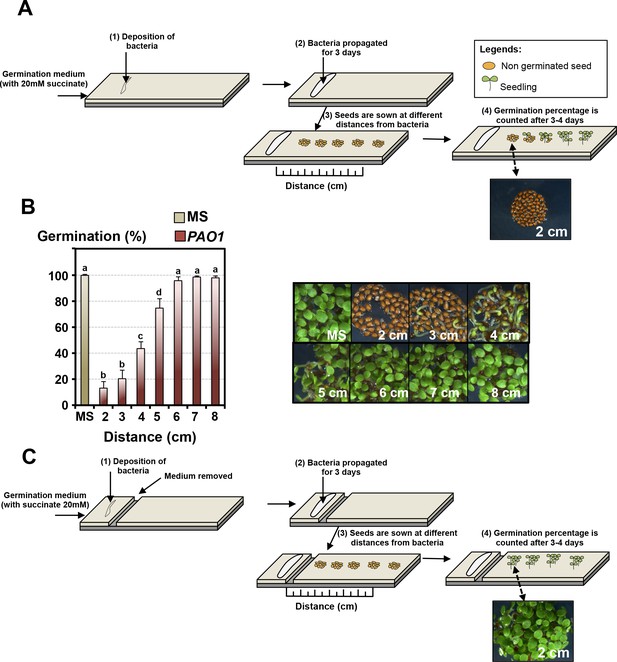
Pseudomonas aeruginosa releases a diffusible GRA in the germination medium.
(A) Experimental procedure to assess germination of Arabidopsis WT seeds exposed to Pseudomonas. (1) Bacteria are streaked on germination medium supplemented with 20 mM succinate as a carbon source, (2) after letting bacteria grow for 3 days, (3) seeds are sown at different distances from the bacteria in order to (4) assess their germination thereafter. (B) Histogram depicts germination percentage 3 days after sowing WT seeds using the procedure described in A. Representative pictures of plant material 3 days after sowing are shown for each distance as indicated. Data represent mean ± standard deviation (four replicates, n = 200–250) and statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05). (C) Same experimental procedure as in A except that the medium separating bacteria and seeds is initially removed.
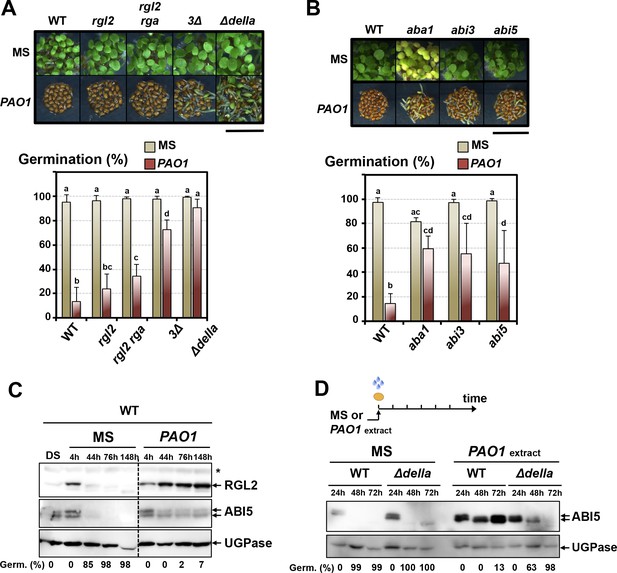
The GRA released by P.aeruginosa stimulates GA and ABA signaling pathways to repress seed germination.
(A) Representative pictures of Arabidopsis plants 3 days after sowing WT and GA signaling mutant seeds in germination medium lacking bacteria (MS) or containing P. aeruginosa (PAO1). Arabidopsis plants include WT (Col-0) seeds and rgl2, rgl2/rga, rgl2/rga/gai (3∆) and rgl2/rga/gai/rgl1/rgl3 (∆della) mutant seeds as indicated. Seeds and P. aeruginosa cells were separated by 2 cm. Histograms show seed germination percentage 3 days upon sowing seeds in absence (MS) or presence (PAO1) of bacteria as shown. Seeds and P. aeruginosa cells were separated by 2 cm. Data represent mean ± standard deviation (five replicates, n = 200–250). Statistical treatment and lower case letters as in Figure 1B. (B) Same as in A. using ABA synthesis and signaling mutant seeds. Mutants included aba1 (deficient in ABA synthesis), abi3 and abi5 as indicated. Histograms show seed germination percentage 3 days upon sowing seeds in absence (MS) or presence (PAO1) of bacteria as indicated. Seeds and P. aeruginosa cells were separated by 2 cm. Data represent mean ± standard deviation (four replicates, n = 150–200). Statistical treatment and lower case letters as in Figure 1B. (C) Protein gel blot analysis of a time course of RGL2 and ABI5 protein levels upon WT (Col-0) seed imbibition in the absence (MS) or presence of P. aeruginosa (PAO1). Seeds and P. aeruginosa cells were separated by 2 cm. DS, dry seeds. UGPase protein levels are used as a loading control. Germination percentage at each time point is indicated. (D) Protein gel blot analysis of a time course of ABI5 protein levels upon WT (Col-0) and Δdella seed imbibition in the absence (MS) or presence of WT P. aeruginosa (PAO1) extract. UGPase protein levels are used as a loading control. Germination percentage at each time point is indicated. The asterisk (*) represents and unspecific banc detected by the RGL2 antibody.
-
Figure 2—source data 1
Germination percentage for each replicate.
- https://doi.org/10.7554/eLife.37082.009
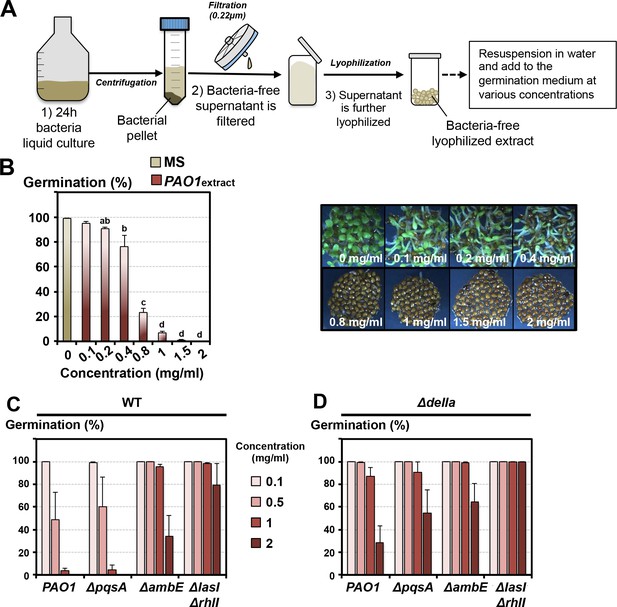
Lyophilized extracts of P. aeruginosa liquid culture medium elicit germination arrest responses similar to those observed with P. aeruginosa in germination plates.
(A) Experimental procedure to produce bacteria-free lyophilized extracts (detailed procedure is described in Materials and methods). (1) P. aeruginosa cells are cultured for 24 hr in liquid solution containing 0.2% MS salts and 20 mM succinate. (2) After centrifugation the liquid solution is collected, filtered and lyophilized. (3) The lyophilizate is resuspended in water, and added to the germination medium to assess germination repressive activity. (B) Histogram depicts germination percentage 4 days after sowing WT seeds in germination plates made as described in A. Different amounts of lyophilizate per ml of germination agar medium were used as indicated. Representative pictures of plant material 4 days after sowing are shown for each concentration as indicated. Data represent mean ± standard deviation (eight replicates, n = 250–300) and statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05). (C) Histograms depict germination percentage 3 days after sowing WT seeds in germination plates made as described in A. Different amounts of lyophilizate per ml of germination agar medium were used as indicated. Note the lower GRA present in extracts obtained from ΔambE and ΔlasIΔrhlI P. aeruginosa strains. Data represent mean ± standard deviation (three replicates, n = 200–250). (D) Same as in C using Δdella mutant seeds. Note the higher percentage of germination of Δdella seeds relative to WT seeds for a given extract concentration in the germination medium. Data represent mean ±standard deviation (three replicates, n = 200–250).
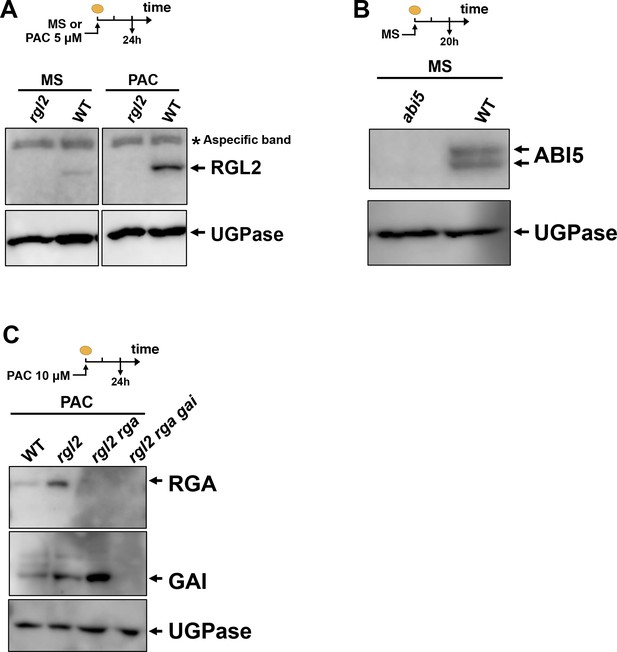
RGL2, GAI, RGA and ABI5 antibodies specificity.
Protein gel blot analysis using antibodies to RGL2 (A), ABI5 (B), GAI and RGA (C) as indicated. Protein extracts from Col-0 (WT), rgl2-13 (rgl2), rgl2-SK54 rga-28 (rgl2 rga), rgl2-SK54 rga-28 gai-t6 (rgl2 rga gai) and abi5-3 (abi5) mutant seeds harvested after seed imbibition in presence or absence of PAC (PAC) as indicated. UGPase protein levels are used as a loading control.
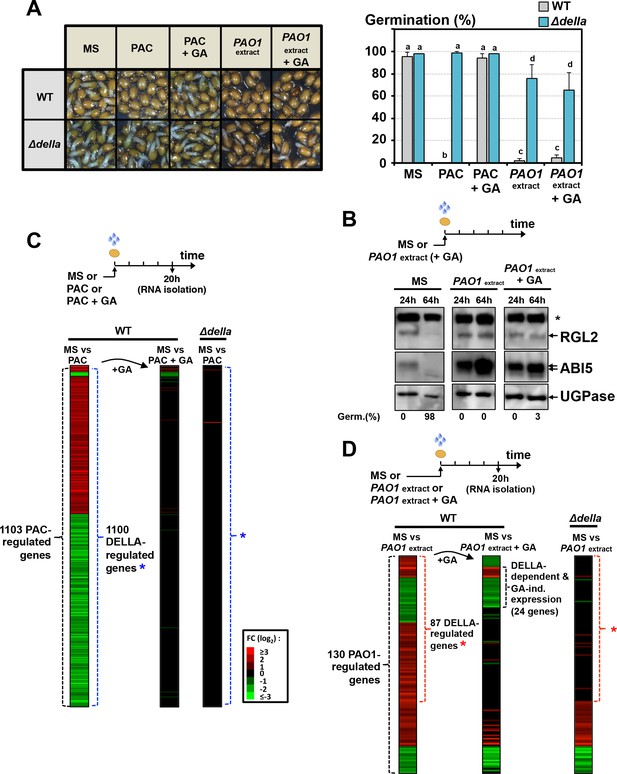
The GRA released by P. aeruginosa elicits DELLA-dependent responses in a GA-independent manner.
(A) Representative pictures of Arabidopsis plants 48 hr after sowing WT and Δdella seeds in absence (MS) or presence of 5 µM paclobutrazol (PAC) or 5 µM PAC and 10 µM GA (PAC + GA) or WT P. aeruginosa extract without (PAO1 extract) or with 10 µM GA (PAO1extract + GA) as indicated. 0.7 mg/ml of WT P. aeruginosa extract was used. Histograms show seed germination percentage 48 hr upon sowing seeds under conditions as indicated. Data represent mean ± standard deviation (three replicates, n = 150–200). Statistical treatment and lower case letters as in Figure 1B. (B) Protein gel blot analysis of RGL2 and ABI5 levels in WT (Col) seeds 24 hr and 64 hr after imbibition in the absence (MS) or presence of WT P. aeruginosa extract without (PAO1 extract) or with 10 µM GA (PAO1extract + GA) as indicated. UGPase protein levels are used as a loading control. Germination percentage at each time point is indicated. The asterisk (*) represents and unspecific banc detected by the RGL2 antibody. (C) Diagram describes the procedure to isolate seed RNA in order to compare early transcriptomes of WT and ∆della seed exposed to PAC or PAC with GA. Total RNA isolated from WT (Col-0) and Δdella seeds imbibed for 20 hr in the absence (MS) or presence of 5 µM paclobutrazol without (PAC) or with 10 µM GA (PAC + GA) was used for RNAseq analysis (two replicates). The red and green horizontal lines represent individual genes whose mRNA expression is significantly upregulated (red) or downregulated (green) by at least twofold under different seed treatments as follows: left column represents the 1103 genes whose expression is either upregulated or downregulated in PAC-treated seeds relative to non-treated seeds (MS) (blue asterisk indicates the genes whose expression does not change in PAC-treated Δdella seeds); middle column represents the expression of the 1103 genes in seeds treated with PAC and GA (PAC + GA) relative to non-treated WT seeds (MS); right column represents the expression of the 1103 genes in Δdella seeds treated with PAC relative to non-treated seeds (MS). The scale bar relates color with absolute fold changes. Black color represents no change in gene expression. (D) Same experiment as in C using WT and ∆della seed exposed to WT P. aeruginosa extract without (PAO1 extract) or with 10 µM GA (PAO1extract + GA) as indicated. 0.7 mg/ml of WT P. aeruginosa extract was used. Red asterisk indicates the genes whose expression does not change in Δdella seeds treated with PAO1 extract.
-
Figure 3—source data 1
Germination percentage for each replicate.
- https://doi.org/10.7554/eLife.37082.012
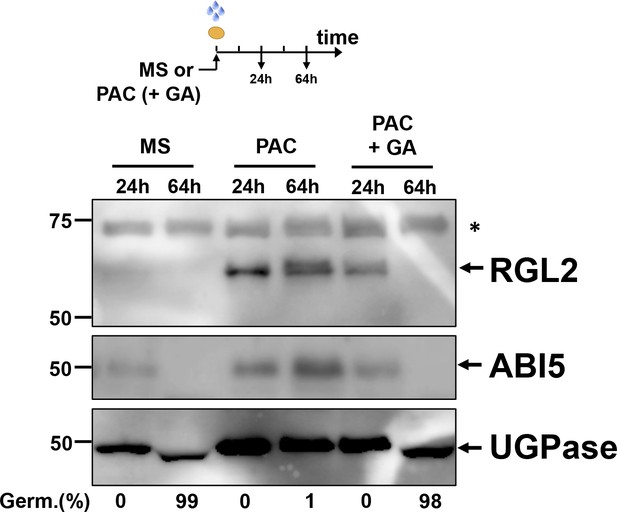
Effect of exogenous GA on RGL2 and ABI5 protein level under low GA condition.
Protein extracts from WT (Col) seeds harvested at 24 hr and 64 hr after imbibition in absence (MS), presence of 5 µM PAC (PAC) or presence of 5 µM PAC and 10 µM GA (PAC + GA). UGPase protein levels are used as a loading control.
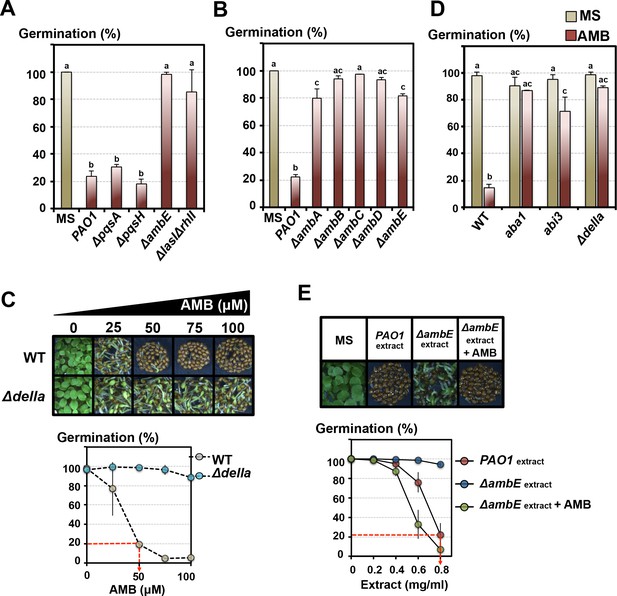
The GRA is under the control of the P. aeruginosa LASI/RHLI and IQS QS systems.
L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) is the main GRA released by P. aeruginosa. (A) Histograms show seed germination percentage 4 days upon sowing seeds in absence (MS) or presence of WT P. aeruginosa (PAO1) or mutant P. aeruginosa strains as indicated. PAO1 is the reference WT P. aeruginosa strain from which all the QS mutant strains were derived. Seeds and P. aeruginosa cells were separated by 2 cm. Data represent mean ± standard deviation (four replicates, n = 150–200). Statistical treatment and lower case letters as in Figure 1B. (B) Same experiment as in A. using P. aeruginosa ΔambA-E mutants, as indicated, each affected in individual AMB operon genes. Data represent mean ± standard deviation (three replicates, n = 100–150). Statistical treatment and lower case letters as in Figure 1B. (C) AMB represses Arabidopsis seed germination in a DELLA-dependent manner. Pictures show representative Arabidopsis plants 4 days after sowing WT and Δdella seeds in presence of different AMB concentrations as indicated. The graph shows quantification of the germination percentage of WT and Δdella seeds exposed to AMB. The red dashed line indicates the concentration of synthetic AMB (50 µM) having the same GRA as that present in germination plates containing 0.8 mg/ml of PAO1 extracts (see Figure 4E). Data represent mean ± standard deviation (three replicates, n = 150–200). (D) AMB represses Arabidopsis seed germination in an ABA-dependent manner. Histograms show germination percentage of WT, aba1, abi3 and Δdella 3 days after sowing seeds in absence (MS) or presence of 50 µM of synthetic AMB. Data represent mean ± standard deviation (two replicates, n = 100–150). Statistical treatment and lower case letters as in Figure 1B. (E) Synthetic AMB introduces a GRA in ΔambE extracts equivalent to that of PAO1 extracts. ΔambE extracts, which lack AMB, were supplemented with synthetic AMB so as to obtain the same amount of AMB naturally present in PAO1 extracts. Pictures show representative Arabidopsis plants 4 days after sowing WT seeds in absence (MS) or presence of WT P. aeruginosa extracts (PAO1extract) or ΔambE P. aeruginosa extracts (ΔambEextract) or AMB-supplemented ΔambE extracts (ΔambEextract + AMB). 0.8 mg/ml of WT P. aeruginosa extract was used. The graph shows quantification of the germination percentage of WT seeds exposed to the different extract concentrations as indicated. Note that the supplemented ΔambE extract (ΔambEextract + AMB) has a GRA equivalent, if not higher, than that of PAO1 extract. The red dashed line indicates the concentration of PAO1 extract (0.8 mg/ml) having the same GRA as that present in germination plates containing 50 µM synthetic AMB (see Figure 4C). Data represent mean ± standard deviation (six replicates, n = 250–300).
-
Figure 4—source data 1
Germination percentage for each replicate.
- https://doi.org/10.7554/eLife.37082.025
-
Figure 4—source data 2
Germination percentage for each replicate.
- https://doi.org/10.7554/eLife.37082.026
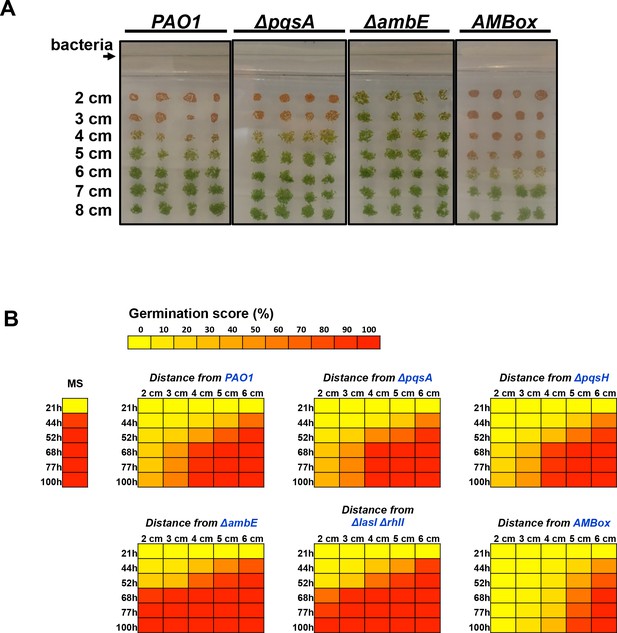
The GRA is under the control of the P. aeruginosa LASI/RHLI and IQS QS systems.
(A) Pictures show Arabidopsis plants 4 days after sowing WT seeds in germination medium containing a given Pseudomonas species as indicated. PAO1 is the reference WT P. aeruginosa strain from which the QS mutant (ΔpqsA and ΔambE) and AMB operon overexpressing (AMBox) strains were derived. (B) Same experiment as in A. Heatmap representation mean of the germination percentage of seeds according to time after sowing and distance from bacteria (four replicates, n = 150–200). Seeds were sowed in germination plates in absence (MS) or presence of P. aeruginosa strains as indicated in blue. PAO1 is the reference WT P. aeruginosa strain from which the QS mutant (ΔpqsA, ΔpqsH, ΔambE, ΔlasIΔrhlI) and AMB operon overexpressing (AMBox) strains were derived.
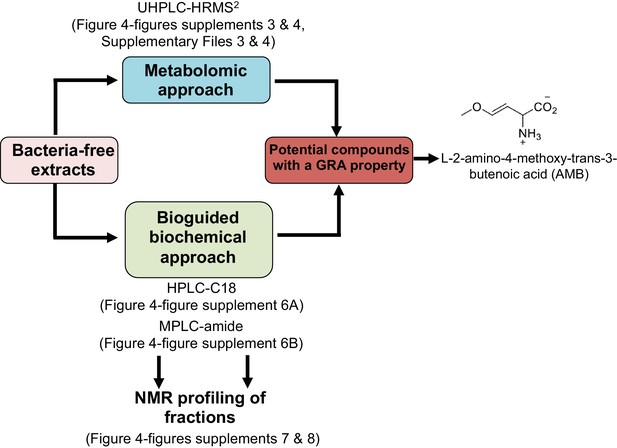
Approaches used in this study to identify compounds released by P. aeruginosa with a germination repressive activity (GRA).
https://doi.org/10.7554/eLife.37082.015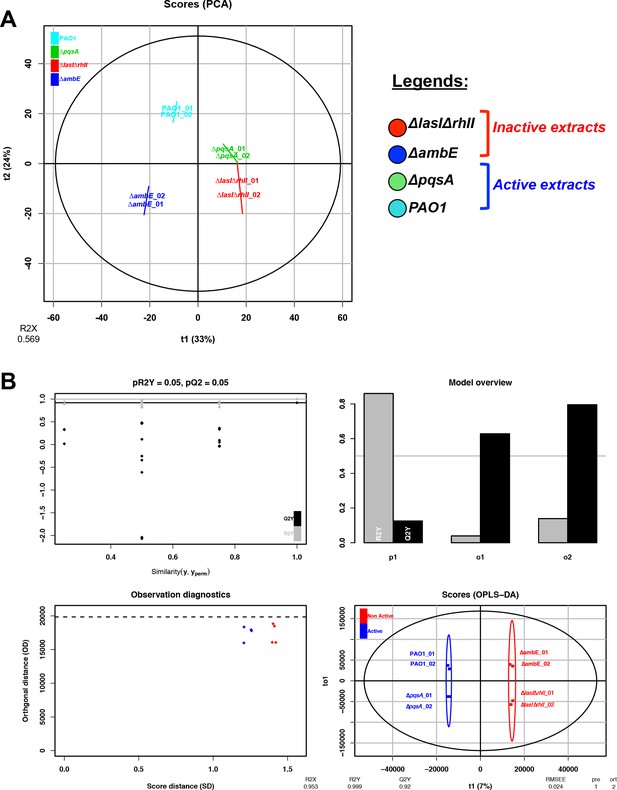
Unsupervised and OPLS-DA results from metabolomic analysis 1.
(A) PCA analysis using HRMS2 data from four different extracts: PAO1, ΔpqsA, ΔambE and ∆lasI∆rhlI (two replicates). (B) Scores and model overview of the OPLS-DA realized between strains releasing a GRA (PAO1 and ∆pqsA), classified as a group named ‘Active’ (blue color), or not releasing a GRA (∆ambE and ∆lasI∆rhlI), classified as a group named ‘Not Active’ (red color). All features are reported in Supplementary file 3, sheet ‘MS-MS analysis 1’.
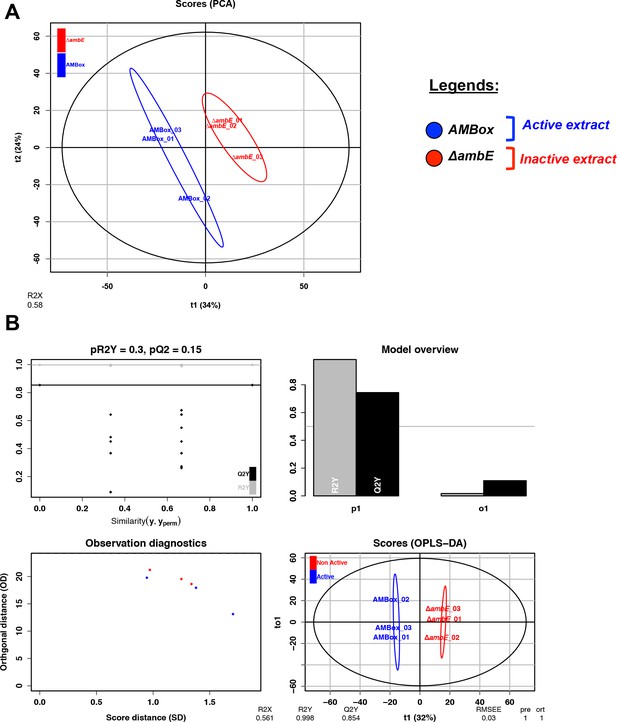
Unsupervised and OPLS-DA results from metabolomic analysis 2.
(A) PCA analysis using HRMS2 data from three different extracts: PAO1, AMBox and ΔambE (three replicates). (B) Scores and model overview of the OPLS-DA realized between strains releasing a GRA (AMBox), classed as ‘Active’ (blue color) or not releasing a GRA (∆ambE), classed as ‘Not Active’ (red color). All features are reported in Supplementary file 3, sheet ‘MS-MS analysis 2’.
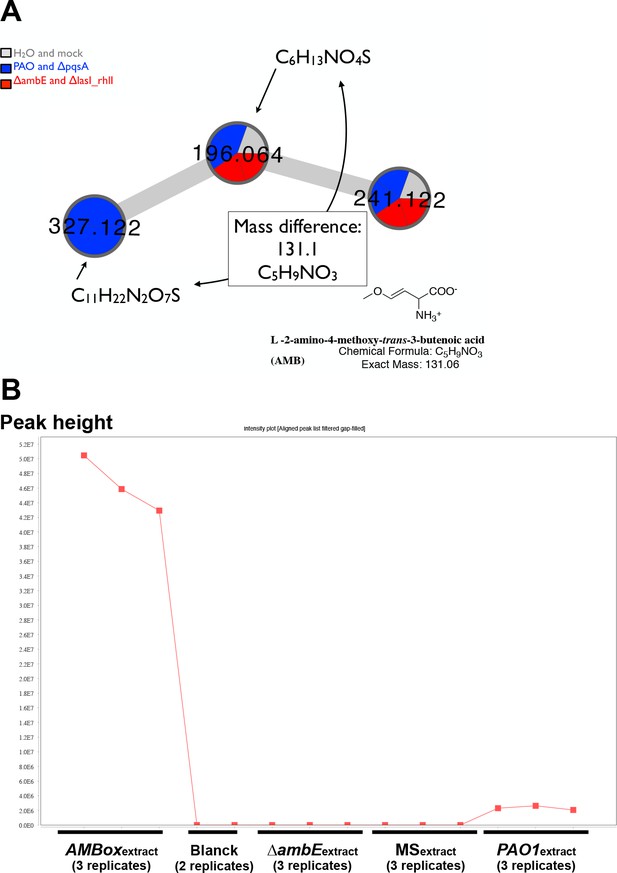
m/z 327.12 is the Mass Spectrometry (MS) signature of AMB.
(A) Specific cluster of the molecular network realized on UHPLC-HRMS2 data acquired on extracts from strains releasing (PAO1 and ∆pqsA) or not releasing a GRA (∆ambE and ∆lasI∆rhlI). Color mapping grouped the MS/MS spectrum as following: blue if acquired from WT (PAO1) or ∆pqsA (∆pqsA) P. aeruginosa extracts, red if acquired from ∆ambE (∆ambE) or ∆lasI∆rhlI (∆lasI∆rhlI) mutant P. aeruginosa extracts and grey if acquired from analytical blanck (H2O) or culture media (MS) extracts. (B) Peak intensities visualization plot of the extracted ion trace from feature m/z 327.12 at 0.46 min. This plot shows the intensity of this particular feature across samples of AMBox [n = 3], analytical blanck [n = 2], ∆ambE [n = 3], culture media extract [n = 3] and PAO1 [n = 3], in this respective order.
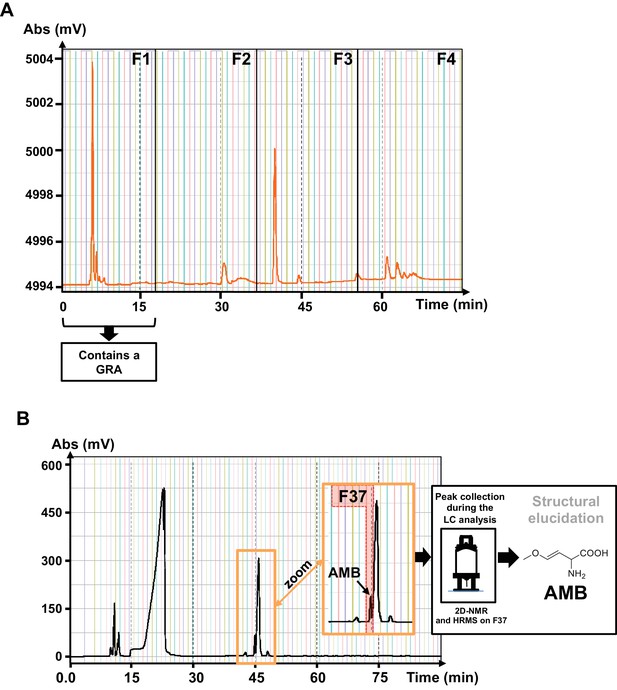
Isolation of the GRA released by P.aeruginosa.
(A) Semi-prep HPLC-UV (scan 210–600 nm) chromatogram of the fractionation of the active PAO1 extract. Fraction F1 presented strong inhibitory seed germination activity. (B) Semi-prep HPLC-ELSD chromatogram of the isolation of the active principle from the AMBox extract localized in fraction 37 (F37). The structure of the AMB was determined by nuclear magnetic resonance (NMR) and high-resolution mass spectrometry HRMS.
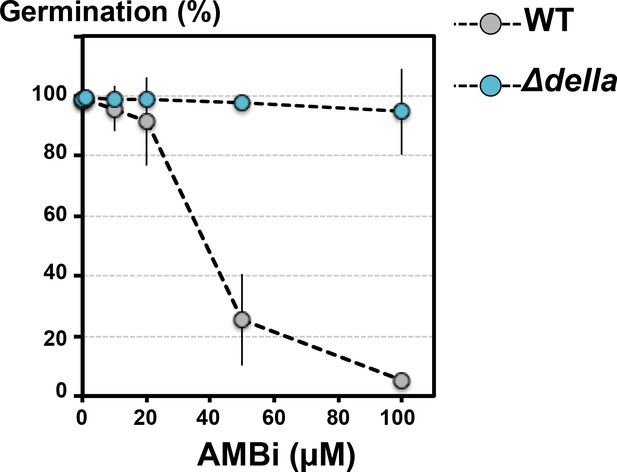
Fraction F37 contains AMB and a GRA.
Fraction F37 was solubilized in water and its AMB content (referred as AMBi) was measured by NMR, which allowed to prepare a stock solution containing 10 mM AMBi. Dose response of WT (Col-0) and Δdella seeds treated with different concentrations of AMBi as indicated. Data represent mean ± standard deviation (2 to 5 replicates, n = 150–300).
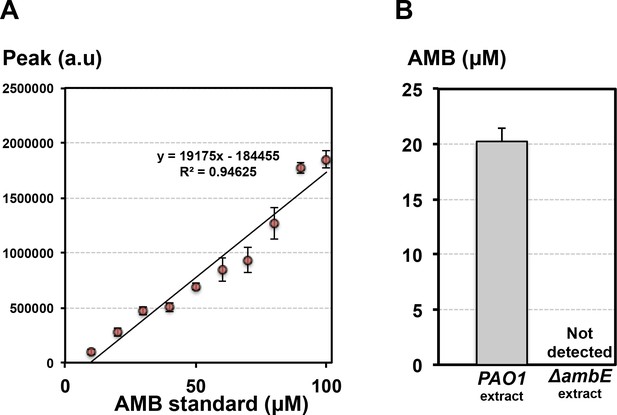
Quantification of AMB in Pseudomonas extracts.
(A) Calibration curve using synthetic AMB used for quantification. Data represent mean ± standard deviation (three replicates). (B) Quantification of AMB in Pseudomonas extracts at 1 mg/mL. Data represent mean ±standard deviation (three replicates). Bioactivity of the same extracts were represented in Figure 4E.
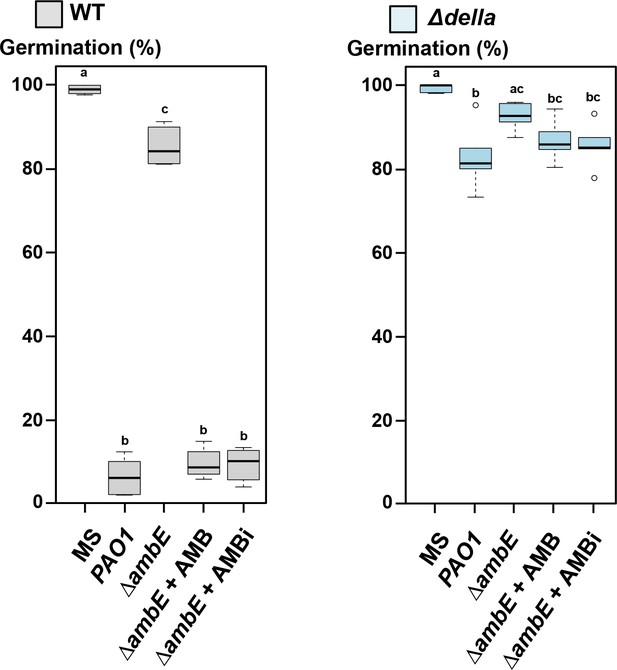
∆ambE extracts supplemented with AMBi or synthetic AMB contain the same GRA.
1 mg/ml of PAO1 extract (containing 20 μM of AMB) strongly inhibits germination of WT (Col-0) seeds (grey boxplot) but not that of Δdella mutant seeds (blue boxplot). 1 mg/ml of ΔambE extract does not inhibit germination. An ΔambE extract supplemented with 20 μM of synthetic AMB (ΔambE + AMB) or AMB isolated from P. aeruginosa (ΔambE + AMBi) has the same GRA as an extract isolated from P. aeruginosa (PAO1). Statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05, six replicates for each conditions, n = 250–300).
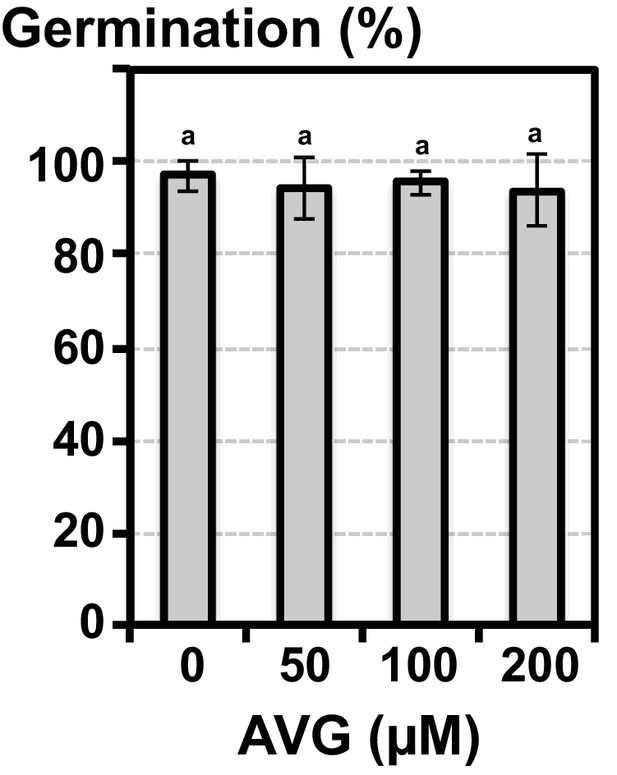
The oxyvinylglycine AVG does not inhibit Arabidopsis seed germination.
Histograms show germination percentage of Arabidopsis seeds sown in absence or presence of different concentrations of AVG, as indicated. Data represent mean ± standard deviation (three replicates, n = 150–200) and statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05).
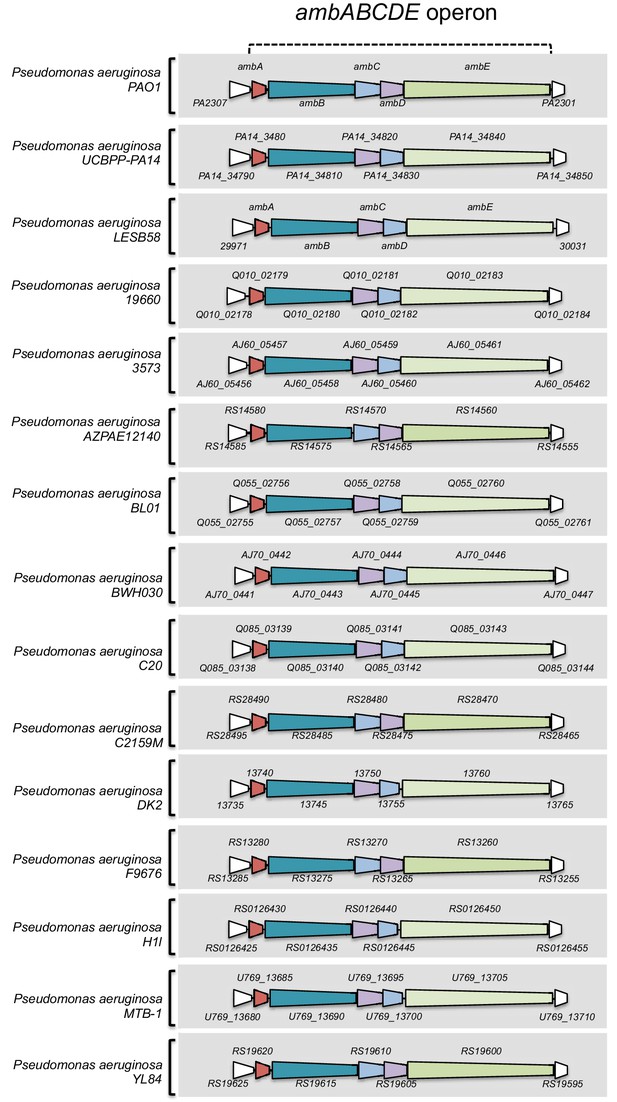
Presence of the ambABCDE operon in at least 15 other P. aeruginosa strains.
Non-exhaustive list of 15 different P. aeruginosa strains containing ambABCDE genes in there genomes. Ortholog genes and structural operon organization where defined based on Pseudomonas database (http://www.pseudomonas.com/).
-
Figure 4—figure supplement 11—source data 1
Description of 15 different Pseudomonas aeruginosa strains containing the ambABCDE operon in their genomes.
- https://doi.org/10.7554/eLife.37082.027
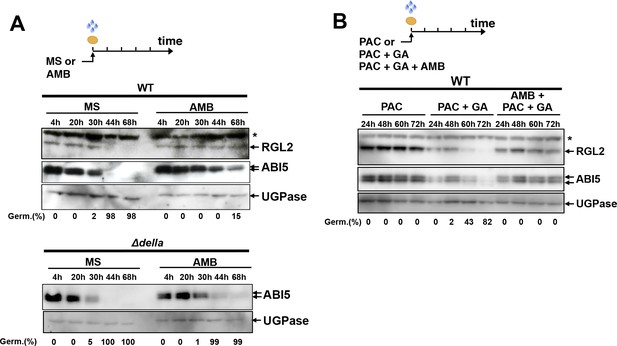
AMB induces changes in RGL2 and ABI5 accumulation in a manner similar to that observed with P. aeruginosa or WT PAO1 extracts.
(A) Protein gel blot analysis of a time course of RGL2 and ABI5 protein levels upon seed imbibition in the absence (MS) or presence of 50 µM AMB (AMB). UGPase protein levels are used as a loading control. Top panel: RGL2 and ABI5 protein levels in WT seeds. Bottom panel: ABI5 protein levels in Δdella seeds. Germination percentage at each time point is indicated. The asterisk (*) represents and unspecific banc detected by the RGL2 antibody. (B) Same as in A using seeds imbibed in the presence of 10 µM PAC, 1 µM GA or 100 µM AMB as indicated. Germination percentage at each time point is indicated.
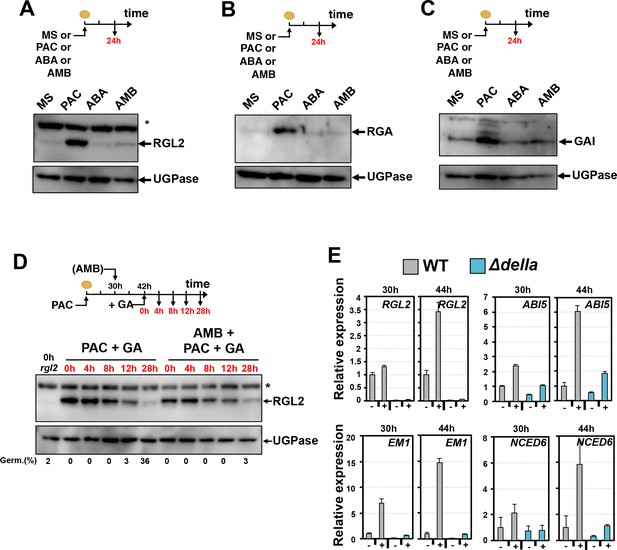
AMB and PAC induce different changes on DELLA protein accumulation.
AMB promotes DELLA factor activity to stimulate ABA signaling. (A-C) Protein gel blot analysis using antibodies to RGL2 (A), RGA (B) and GAI (C) as indicated. Protein extracts from WT (Col seeds harvested 24 hr after seed imbibition in absence (MS) or presence of 10 µM PAC (PAC), 5 µM ABA (ABA) or 50 µM AMB (AMB). UGPase protein levels are used as a loading control. (D) AMB does not interfere with GA-dependent RGL2 protein downregulation. WT seeds are imbibed in presence of 5 µM PAC for 30 hr to trigger high RGL2 accumulation. Thereafter, seeds are transferred to germination plates containing 5 µM PAC or containing 5 µM PAC and 50 µM AMB. After 12 hr, 1 µM GA is further added and RGL2 protein levels are followed by protein gel blot analysis over the time points indicated in red. UGPase protein levels are used as a loading control. Germination percentage at each time point is indicated. The asterisk (*) represents and unspecific banc detected by the RGL2 antibody. (E) AMB stimulates DELLA activity to promote ABA-dependent responses. Histograms show RGL2, ABI5, EM1 and NCED6 mRNA accumulation in WT (Col) and Δdella seeds treated as described in A. For each time point, mRNA levels are normalized to mRNA levels in WT seeds sown in absence of AMB. Data represent mean ±standard deviation (three replicates).
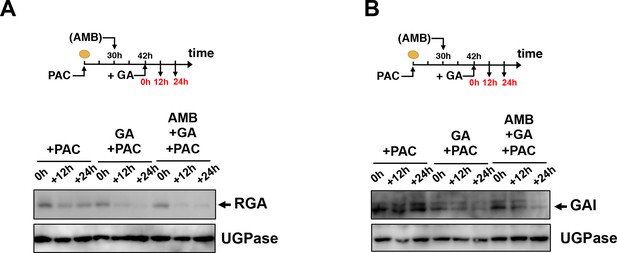
AMB does not interfere with GA-dependent GAI and RGA protein downregulation.
WT seeds are imbibed in presence of 5 µM PAC for 30 hr to trigger high DELLA accumulation. Thereafter, seeds are transferred to germination plates containing 5 µM PAC or containing 5 µM PAC and 50 µM AMB. After 12 hr, 1 µM GA is further added and RGA (A) and GAI (B) protein levels are followed by protein gel blot analysis over the time points indicated in red as indicated. UGPase protein levels are used as a loading control.
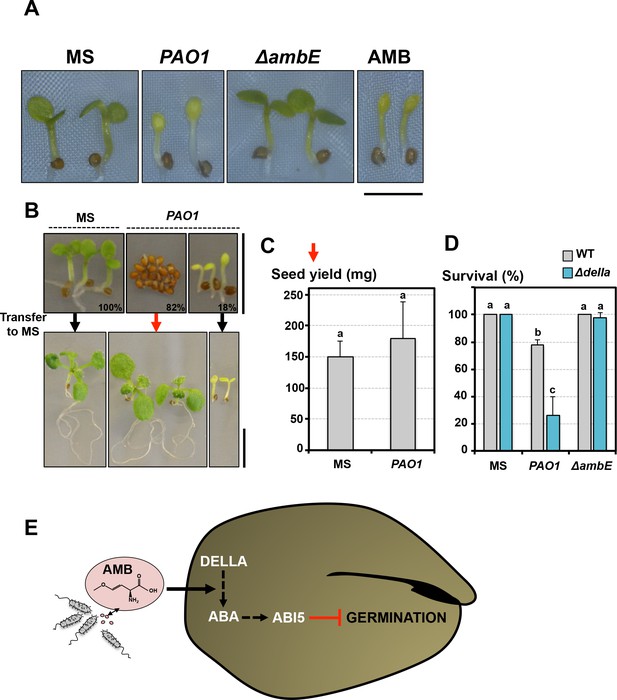
Germination-arrested seeds are protected from developmental abnormalities triggered by AMB.
(A) Representative pictures of 4-day-old seedlings produced by seeds that germinated in absence (MS) or presence of WT P. aeruginosa (PAO1) or ΔambE mutant P. aeruginosa (ΔambE) or 50 µM AMB (AMB). Seeds and bacteria were separated by 2 cm. Scale bar: 2 mm. (B) Seeds were sown in absence (MS) or presence of P. aeruginosa (PAO1). Seeds and bacteria were separated by 2 cm. Top panel: representative pictures of plants 3 days after sowing. All the seeds sowed in absence of P.aeruginosa germinated 3 days after sowing (100%). Plants produced by seeds sown in presence of P. aeruginosa are shown according to their germination status 3 days after sowing: 82% (82%) did not germinate and 18% (18%) germinated. 3 days after sowing, plant material was transferred to a medium lacking P. aeruginosa (MS) and cultured for 1 week. Bottom panel shows representative pictures of plants 1 week after the transfer. (C) Histograms show the seed yield (expressed in mg) from plants produced by seeds never exposed to P. aeruginosa (MS) or from plants produced by seeds that did not germinate for 3 days in presence of P. aeruginosa and then transferred to medium lacking P. aeruginosa. Average seed yield was calculated from seeds produced by five and nine individual plants from seeds sown in absence or presence of P. aeruginosa, respectively. Data represent mean ± standard deviation. Statistical treatment and lower case letters as in Figure 1B. (D) WT and Δdella seeds were exposed for three days to WT P. aeruginosa (PAO1) prior to transfer to a bacteria-free medium. Histograms show percent of survival one week after transfer. Data represent mean ± standard deviation (three replicates, n = 60). Statistical treatment and lower case letters as in Figure 1B. (E) Model describing AMB mode of action in seeds.
-
Figure 7—source data 1
Seed yield per plant (mg).
- https://doi.org/10.7554/eLife.37082.038
-
Figure 7—source data 2
Survival percentage.
- https://doi.org/10.7554/eLife.37082.039
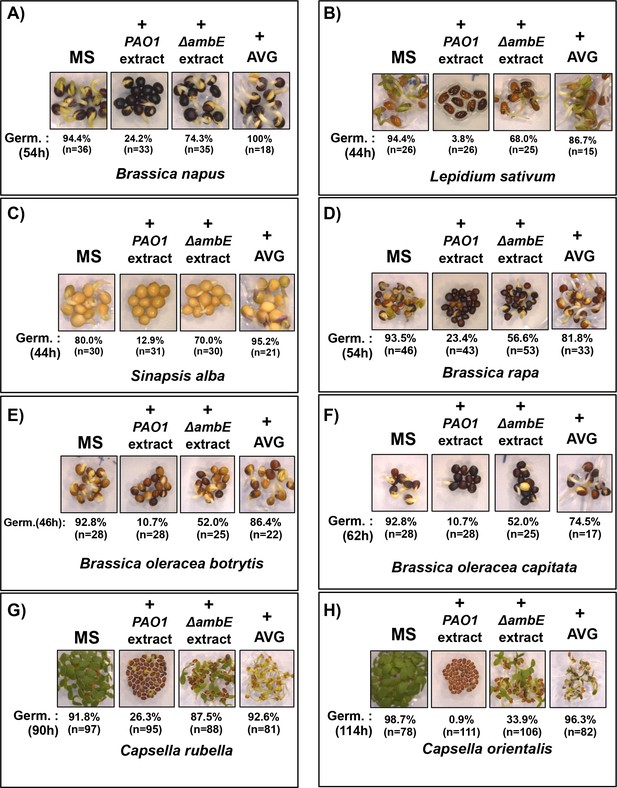
Effect of WT PAO1 extracts, ∆ambE extracts and 250 µM AVG on the germination of various brassicaceae seeds, as indicated (A to H).
Seeds were sown on normal germination medium without (MS) or with 2 mg/mL of PAO1 extract (+PAO1 extract) or with 2 mg/mL of ΔambE extract (+ ΔambE extract) or with 250 µM of AVG (AVG) as indicated. Germination (Germ.) percentage (%) was scored after seed imbibition at the time indicated.
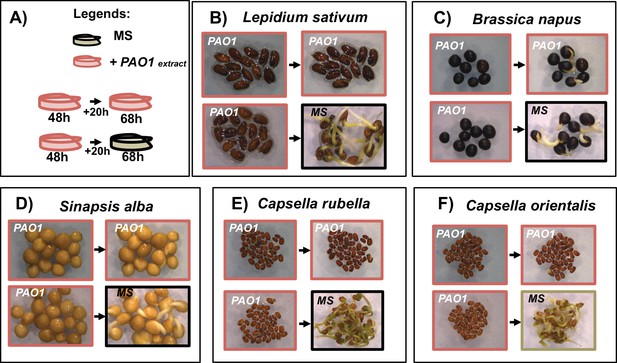
Arrested brassicaceae seeds germinate after transfert to MS.
Brassicaceae seeds were placed 2 days on plate containing 2.5 mg/mL of PAO1 extract. Arrested seeds were then transfert to normal condition (MS) or plate containing PAO1 extract (2.5 mg/mL) for 20 hr as indicate on panel A). Different brassicaceae species were tested as indicated (B to F).
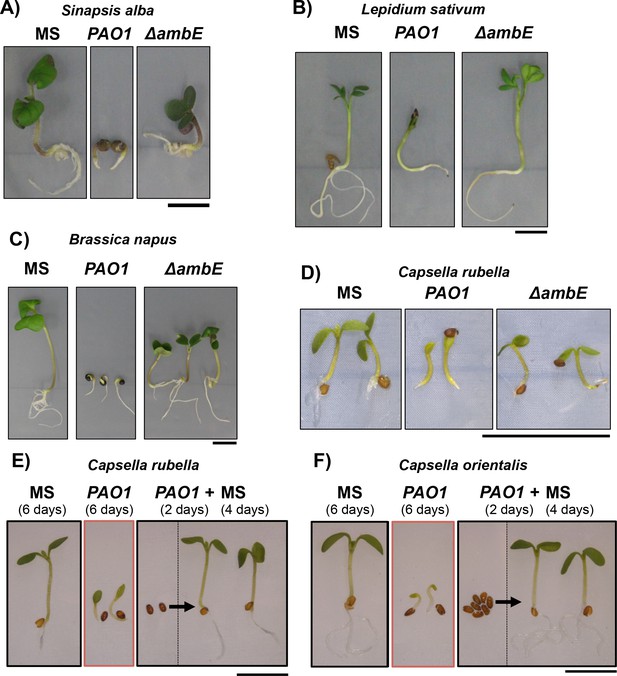
Representative pictures of brassicaceae seedlings in response to PAO1 or ΔambE extract.
(A–D) Representative pictures of 4 days old brassicaceae seedlings produced by seeds that germinated in experiment shown in Figure 7—figure supplement 1 in absence (MS) or presence of WT P. aeruginosa (PAO1) extracts (2 mg/mL) or ΔambE (ΔambE) mutant extracts (2 mg/mL). (E and F) Representative Capsella rubella (E) and Capsella orientalis (F) developing seedlings 6 days on MS (MS) or on PAO1 extract (2.5 mg/mL). 2 days arrested seeds on presence of PAO1 extract (2.5 mg/mL) were transferred to MS for 4 days (PAO1 + MS). Scale bar: 5 mm.
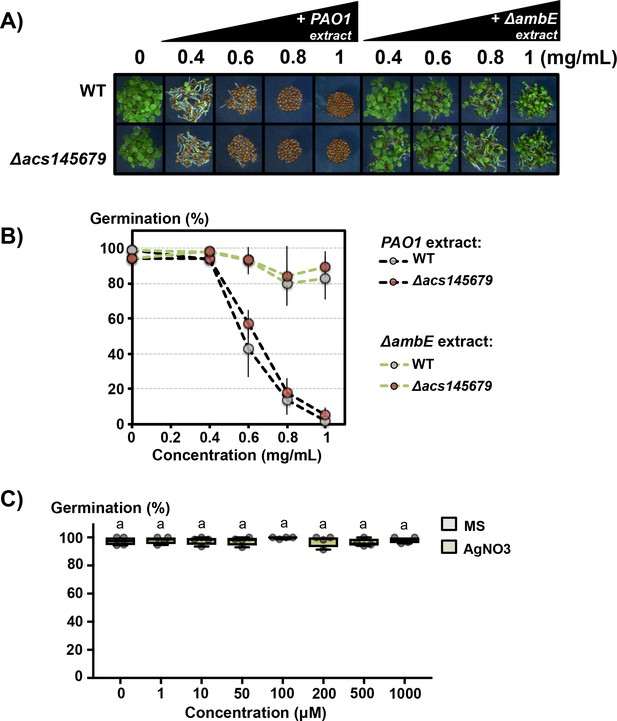
Effect of AgNO3, PAO1 and ΔambE extracts on seed germination.
(A) Effect of various concentrations, as indicated (in mg/mL), of PAO1 extract (+PAO1 extract) or ΔambE extract (+ ΔambE extract) on WT (WT) and acs1-1 acs2-1 acs4-1 acs5-2 acs6-1 acs7-1 acs9-1 (Δacs145679) seed germination. (B) Percentage of germination of seeds shown in A. Germination was scored at 3 days. Data represent mean ±standard deviation (three replicates, n = 150–200). (C) Effect of AgNO3 on WT seed germination. Germination was scored at 2 days (four replicates, n = 200–250). Statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05).
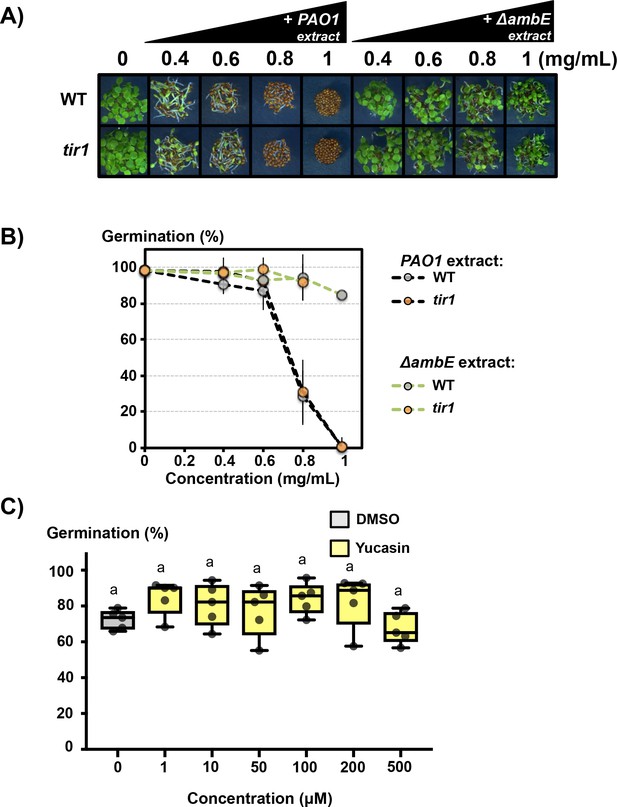
Effect of Yucasin, PAO1 and ΔambE extracts on seed germination.
(A) Effect of various concentrations, as indicated (in mg/mL), of PAO1 extract (+PAO1 extract) or ΔambE extract (+ ΔambE) on WT (Col) and tir1-1 (tir1) seed germination. (B) Percentage of germination of seeds shown in A. Germination was scored at 3 days. Data represent mean ± standard deviation (three replicates, n = 150–200). (C) Effect of yucasin on WT seed germination. Germination was scored at 2 days (five replicates, n = 250–300). Statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05).
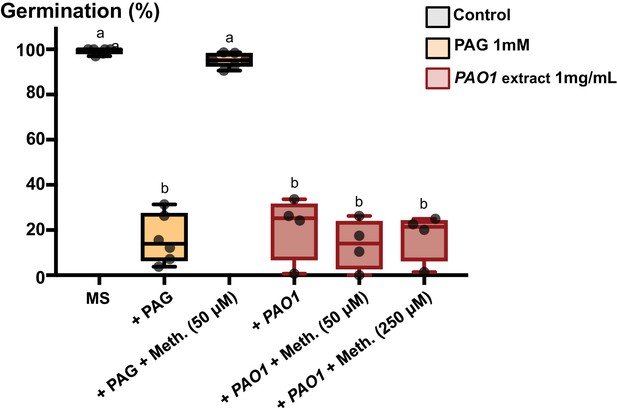
Effect of PAG, methionine and PAO1 extracts on seed germination.
DL-Propargylgylcine (PAG) and PAO1 extract (PAO1) were used at 1 mM and 1 mg/mL, respectively. Methionine (Meth.) was added as indicated (4 to 6 replicates, n = 250–300). Statistically significant differences were assessed by one-way ANOVA followed by a Tukey HSD test (p<0.05).
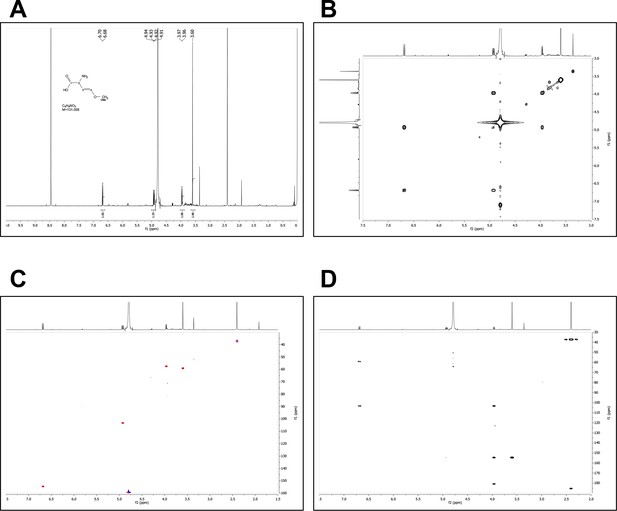
Characterization of the fraction F37 by NMR.
(A) 1H NMR spectrum of F37 at 600 MHz in D2O. (B) COSY NMR spectrum of F37 in D2O. (C) Edited HSQC NMR spectrum of F37 in D2O. (D) HMBC NMR spectrum of F37 in D2O.
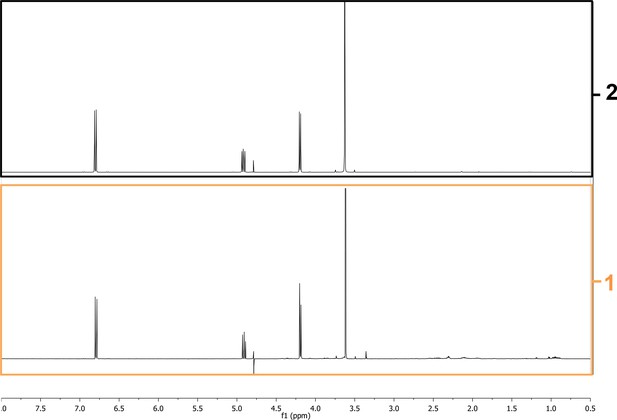
1H NMR spectra of F37 and synthetic AMB in D2O.
The 1H NMR spectra of AMB localized in F37 (1) is similar of that of synthetic AMB, demonstrating that F37 contains AMB.
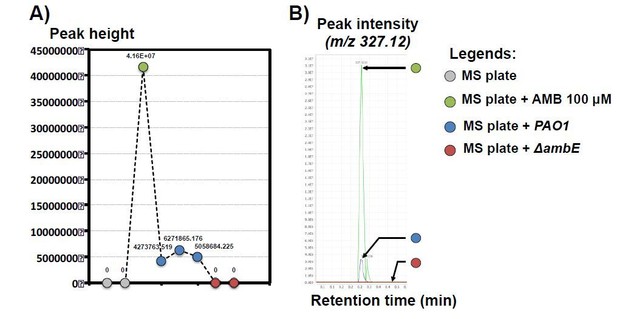
Identification of AMB in solid plate containing P. aeruginosa PAO1 by LC-MS/MS analysis.
A) Peak height of AMB isolated from different solid plates as indicated in legends. Each dot represents a biological replicate. The different labels above represent the exact value for each replicate. B) Representative peak intensity of AMB feature (m/z 327.12) in a representative replicate. Note that AMB signal in plate containing PAO1 growing bacteria is ≈7-8 fold lower compared to the control plate containing 100 μM AMB.
Additional files
-
Supplementary file 1
List of differentially expressed genes in WT seeds in response to PAC.
- https://doi.org/10.7554/eLife.37082.040
-
Supplementary file 2
List of differentially expressed genes in WT seeds in response to PAO1 extract.
- https://doi.org/10.7554/eLife.37082.041
-
Supplementary file 3
Peaklist of features detected by MS/MS in the different P. aeruginosa extracts.
- https://doi.org/10.7554/eLife.37082.042
-
Supplementary file 4
Peaklist of most discriminant features determined by OPLSDA analysis in active versus inactive extracts.
- https://doi.org/10.7554/eLife.37082.043
-
Supplementary file 5
Bacterial strains and oligonucleotides.
- https://doi.org/10.7554/eLife.37082.044
-
Supplementary file 6
- https://doi.org/10.7554/eLife.37082.045
-
Supplementary file 7
- https://doi.org/10.7554/eLife.37082.046
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37082.047





