BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions
Figures
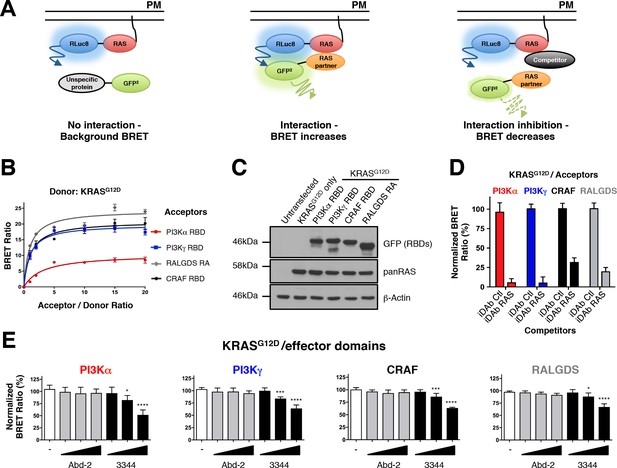
RAS-effector BRET biosensors and interference of KRAS-effector interactions by a RAS-binding compound.
An outline of the BRET2-based RAS biosensor system is shown in A. RAS bound to the plasma membrane (PM) is fused at its amino terminal end to the RLuc8 moiety (donor). When a protein fused to the GFP2 moiety (acceptor) does not bind to RAS, it only produces a background BRET signal. However, when an acceptor binds to RAS, it induces a BRET signal, if the luciferase and GFP domains are within 100 Å. The BRET signal can be decreased by addition of a competitor (either by a macrodrug or a small molecule inhibitor). The interaction titration of full-length KRASG12D-CAAX (for simplicity, the CAAX motif is omitted in all the RAS constructs described hereafter) with the four effector acceptor proteins and the effect on intracellular protein levels are shown in B and C. Competition assays show the specificity of the RAS biosensors in D (iDAb) and E (RAS-binding compounds). In D, the non-relevant anti-LMO2 iDAb (called hereafter iDAb control, Ctl) serves as a negative control and anti-RAS iDAb (herein named iDAb RAS) serves as a positive control. In E, 3344 (black bars) decreases KRASG12D/effector domain interactions in a dose-dependent manner showing its broad range of inhibition. Cells were treated with 5, 10 and 20 μM of 3344 (black bars), Abd-2 (grey bars) or DMSO alone (white bars) as the negative control. Statistical analysis was performed with a one-way ANOVA followed by Dunnett’s post-hoc tests (*p<0.05, ***p<0.001, ****p<0.0001). Each experiment was repeated three (B, D) or four times (E). Where error bars are presented, these correspond to mean values ± SD of biological repeats (B, D–E). See also Figure 1—figure supplement 1, Figure 1—figure supplement 2, Figure 1—figure supplement 3 and supplementary file 1.
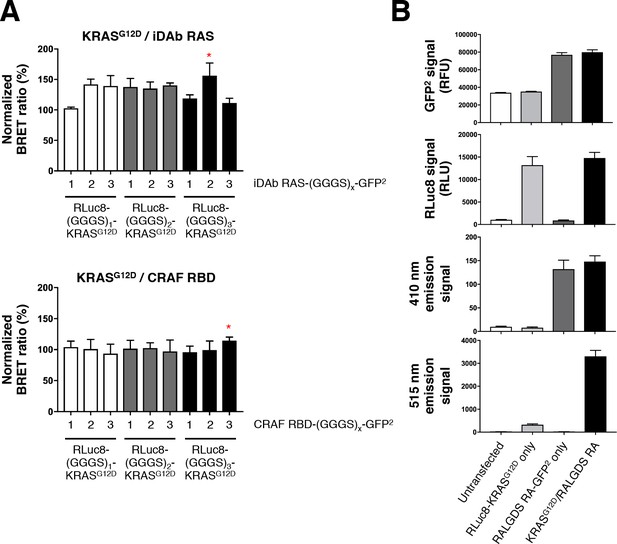
Optimization of the RAS biosensors.
(A) Optimization of the donor and acceptor linker length. Top panel shows KRASG12D/iDAb RAS optimization and the bottom panel shows KRASG12D/CRAF RBD optimization. The red stars indicate the linker length chosen for the study: all RLuc8-RAS constructs bear a (GGGS)3 linker, the iDAb-GFP2 fusions a (GGGS)2 linker and all effectors fused to the GFP2 moiety a (GGGS)3 linker. (B) Background analysis with total GFP2 and RLuc8 levels, emission signal at 410 nm and at 515 nm upon coelenterazine 400a addition from untransfected cells, RLuc8-KRASG12D transfected cells only, RALGDS RA-GFP2 transfected cells only and cells transfected with the BRET pair KRASG12D/RALGDS RA. Each experiment was repeated twice (A–B). Where error bars are presented, they correspond to mean values ± SEM of biological repeats.
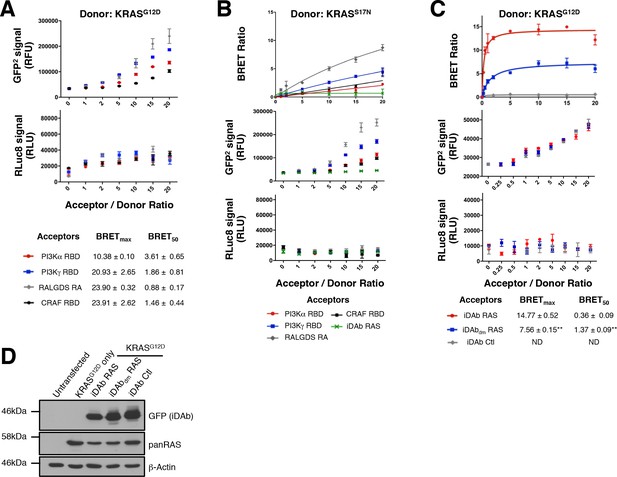
Validation of the RAS biosensors with the anti-iDAb RAS.
(A) Total GFP2 and RLuc8 levels from the BRET titration curves in Figure 1B. (B) Representative BRET titration curves of KRASS17N and RAS binders (RBDs and iDAb RAS) with total GFP2 and RLuc8 controls. (C) BRET titration curves of KRASG12D and iDAbs with total GFP2 and RLuc8 controls. Statistical analyses were performed using an unpaired Student’s test (**p<0.01). (D) Western blots for assessment of the expression levels of each KRASG12D-iDAb BRET pair. Each experiment was repeated three times (A–C). Where error bars are presented, they correspond to mean values ± SD of biological repeats.

3344 inhibits RAS-RBD interactions.
(A) Chemical structure of 3344. (B) 1H NMR and (C) 13C NMR spectra of 3344 were recorded on a Bruker Avance spectrometer (600 MHz) at room temperature in a solution of the deuterated solvent (CDCl3). The field was locked by external referencing to the relevant deuteron resonance. Chemical shifts are reported in parts per million (ppm). (D) NMR Carr-Purcell-Meiboom-Gill (CPMG) evaluation of 3344 Kd. Dose-dependent CPMG spectra of 3344 (at a fixed concentration of 55 μM) were recorded on a Bruker Avance spectrometer (700 MHz) at room temperature against an array of concentration of GST-KRASG12V(0 to 20 μM, left hand panel). The amount of protein was increased from 0 μM until the signals of the compound completely disappear in the proton NMR (here 20 μM). The integrations of the proton acquired were all compared to the compound alone (0 μM of protein) in order to obtain a percentage of decrease for each concentration of GST-KRASG12V. Concentration and percentage of decrease were plotted and Kd fitting was run on the generated binding curve using Origin® software (right hand panel, see Materials and methods for details). (E) WaterLOGSY spectra of 3344 interacting with GST-KRASG12V-GppNHp. The proton NMR of 3344 is the lower spectrum (blue), the spectrum of 3344 with KRAS is shown in the top (green) and the inhibitory effect of added anti-RAS scFv on 3344 binding to KRAS is shown in the middle spectrum (red). (F) Chemical structure of Abd-2. (G–I) 3344 decreases KRASG12D-iDAbdm RAS interaction in a dose-dependent manner and not with iDAb RAS or with a negative BRET-biosensor LMO2-iDAbdm LMO2. Statistical analyses were performed using a one-way ANOVA followed by Dunnett’s post-tests (*p<0.05, ****p<0.0001). (J) Total GFP2 and RLuc8 levels from the BRET competition assay shown in G-I and Figure 1E. Each experiment was repeated four times (G–I). Where error bars are presented, they correspond to mean values ± SD of biological repeats.
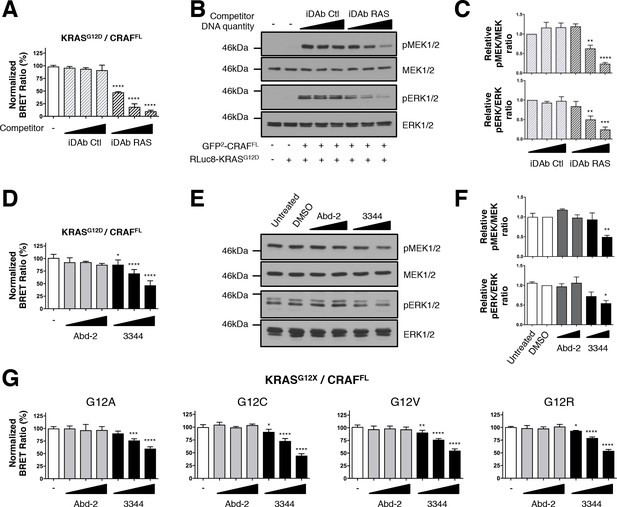
BRET biosensors of KRASG12 mutants and full-length CRAF are inhibited by compound 3344.
A biosensor for the full-length CRAFS257L (CRAFFL) protein was made and tested for interaction with mutants of KRAS glycine 12. For A and B, the plasmids expressing BRET pair KRASG12D/CRAFFL was transfected into HEK293T cells and competed with iDAb expression as indicated; the BRET ratios are shown in A and western blot data in B. The iDAb RAS inhibition of phosphorylation of ERK and MEK signals are quantified in C. The β-actin loading control, iDAbs and BRET pair expression controls are shown in Figure 2—figure supplement 1. In D, the BRET ratio of KRASG12D/CRAFFL interaction was measured in the presence of an increasing dose of compound 3344. This induces a dose-dependent decrease of MEK and ERK kinase phosphorylation (E) after cells expressing the KRASG12D/CRAFFL biosensor pair were treated 20 hr with DMSO, 10 and 20 μM of Abd-2 and 3344 compounds or not treated (untreated lane). The β-actin loading control and BRET pair expression controls are shown in Figure 2—figure supplement 1. Quantification of the relative levels of pMEK1/2 and pERK1/2, normalized to total MEK1/2 and ERK1/2 respectively, are shown in F. The RAS biosensor toolkit includes KRAS G12A, G12C, G12V and G12R, in addition to KRAS G12D. In G, each was expressed with CRAFFL and BRET ratios determined at 0, 5, 10 and 20 μM Abd-2 or 3344. Statistical analyses in C were performed using a one-way ANOVA followed by Sidak’s post-hoc tests and in A, D, F and G using a one-way ANOVA followed by Dunnett’s post-tests (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Each experiment was repeated twice (E–F), three times (B–D), four times (A) or five times (G). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D, G) or correspond to mean ±SEM of biological repeats (C, F). See also Figure 2—figure supplement 1.
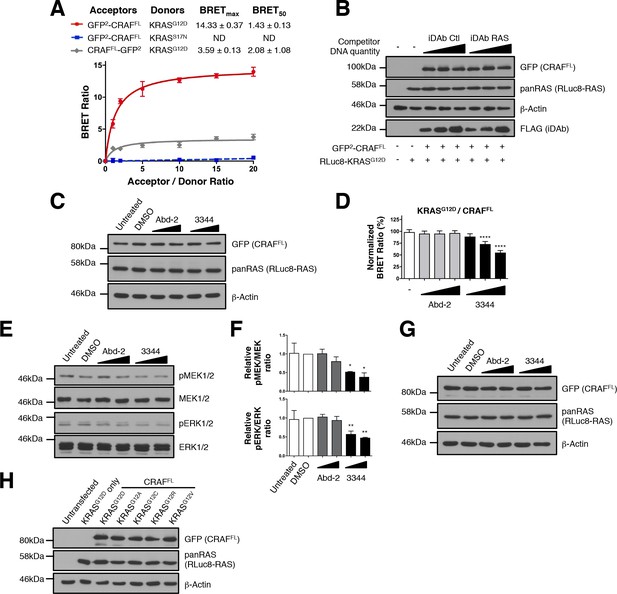
Interactions of KRASG12X mutants and full-length CRAF are inhibited by 3344.
(A) BRET titration curves of KRAS mutants with full-length CRAFS257L (CRAFFL). KRASG12D interacts with GFP2-CRAFFL while it gives a low BRET ratio with CRAFFL-GFP2. The dominant negative KRASS17N does not interact with GFP2-CRAFFL showing the accuracy and optimization of this biosensor. (B) Controls from Figure 2B. The expression level of the BRET pair was assessed by western blot with the GFP (for CRAFFL) and pan-RAS (for RLuc8-KRASG12D) antibodies. iDAb expression was revealed using anti-FLAG antibody; anti-β-actin binding was used as the loading control. (C) Controls from Figure 2E. The expression level of the BRET pair was assessed with the GFP (for CRAFFL) and pan-RAS (for RLuc8-KRASG12D) antibodies, anti-β-actin binding was used as the loading control. (D–F) Short-term incubation of the compounds (3 hr) on cells transfected with the KRASG12D/CRAFFL biosensor. The BRET ratio was measured in the presence of an increasing dose of compound 3344 (D). This induces a dose-dependent decrease of MEK and ERK kinase phosphorylation (E) after cells expressing the KRASG12D/CRAFFL biosensor pair were treated 3 hr with DMSO, 10 and 20 μM of Abd-2 and 3344 compounds or not treated (untreated lane). Quantification of the relative levels of pMEK1/2 and pERK1/2, normalized to total MEK1/2 and ERK1/2 respectively, are shown in panel F. (G) Controls from panel E. (H) Controls from Figure 2G. The expression level of each BRET pair was assessed with the GFP (for CRAFFL) and pan-RAS (for RLuc8-KRASG12X) antibodies. One-way ANOVA followed by Dunnett’s post-hoc tests were used to determine the statistical significance of BRET, pERK and pMEK modulations induced by the compounds (*p<0.05, **p<0.01, ****p<0.0001). Each experiment was repeated twice (A, E–F) or three times (D). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D) or correspond to mean ± SEM of biological repeats (F).
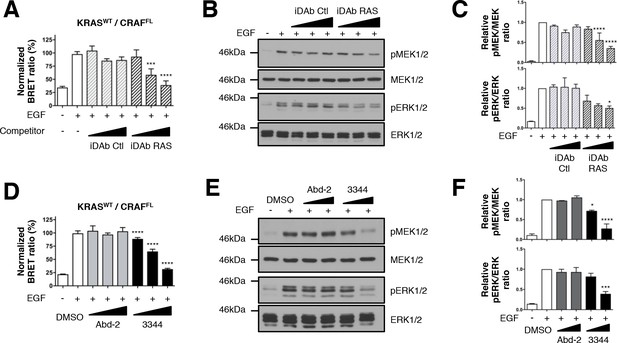
Wild-type KRAS and CRAF biosensor interaction-induced signaling is impaired by 3344.
The BRET KRASWT/CRAFFL pair was tested for interaction after EGF stimulation of HEK293T cells in presence of competitors. In A, cells were transfected with plasmids to express the KRASWT biosensor with or without iDAbs and stimulated by EGF (50 ng/mL). iDAb RAS shows an inhibition of KRASWT/CRAFFL interaction after EGF treatment in a dose-dependent manner. B is a western blot of the transfected cells from panel A showing the effect of the iDAbs on EGF-stimulated RAS-RAF-MEK-ERK signaling pathway (pMEK and pERK signals are quantified in C). β-actin loading control, iDAbs and BRET pair expression controls are shown in Figure 3—figure supplement 1. The effect on BRET2 signal of compounds Abd-2 (grey bars) and 3344 (black bars) on KRASWT/CRAFFL interaction after EGF treatment in a BRET competition experiment is shown in panel D. In panel E, HEK293T cells were transfected as in D with the plasmids expressing the BRET pair KRASWT/CRAFFL for 24 hr and serum starved 20 hr in the presence of DMSO, 10 and 20 μM of Abd-2 and 3344 compounds. Cells were treated 5 min with EGF (50 ng/mL), lysed and analyzed by western blot. The expression level of the BRET protein pair is shown in Figure 3—figure supplement 1 as well as the loading control β-actin for the western blot. The western blot data are quantified in panel F. One-way ANOVA followed by Dunnett’s post-hoc tests were used to determine the statistical significance of BRET, pERK and pMEK modulations induced by the compound or the iDAb (*p<0.05, ***p<0.001, ****p<0.0001). Each experiment was repeated twice (B–C) or three times (A, D–F). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D) or correspond to mean ±SEM of biological repeats (C, F). See also Figure 3—figure supplement 1.

3344 inhibits KRASWT/CRAFFL interaction induced by EGF treatment.
(A) BRET titration curves of KRASWT with CRAFFL. After EGF stimulation (50 ng/mL), KRASWT contacts CRAFFL as indicated by an increase of the BRETmax value. (B) Controls from Figure 3B. The expression level of the BRET pair was assessed with the GFP (for CRAFFL) and pan-RAS (for RLuc8-KRASWT) antibodies. iDAb expression is revealed by the CMYC tag antibody; anti-β-actin binding was used as the loading control. (C) Controls from Figure 3E. The expression level of the BRET pair was assessed with the GFP (for CRAFFL) and pan-RAS (for RLuc8-KRASWT) antibodies. Anti-β-actin binding was used as control. Panel D shows the short-term effect on BRET2 signal of compounds Abd-2 (grey bars) and 3344 (black bars) on KRASWT/CRAFFL interaction after EGF treatment in a BRET competition experiment (3 hr incubation of the compounds). In panel E, HEK293T cells were transfected with the plasmids expressing the BRET pair KRASWT/CRAFFL for 24 hr, serum starved 24 hr and then incubated for 3 hr with DMSO, 10 and 20 μM of Abd-2 and 3344 compounds. Cells were treated 5 min with EGF (50 ng/mL), lysed and analysed by western blot. Quantification of panel E is shown in panel F. (G) Controls from panel E. One-way ANOVA followed by Dunnett’s post-hoc tests were used to determine the statistical significance of BRET, pERK and pMEK modulations induced by the compounds (**p<0.01, ***p<0.001, ****p<0.0001). Each experiment was repeated twice (A, E–F) or three times (D). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D) or correspond to mean ± SEM of biological repeats (F).
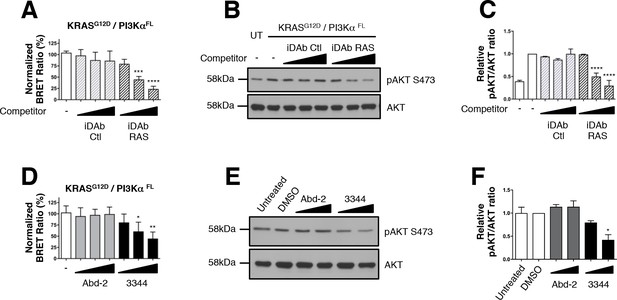
Interaction between mutant KRAS and full-length PI3Kα BRET pair interaction is impeded by 3344.
The BRET signal produced from the interaction of the KRASG12D and full-length PI3Kα (PI3KαFL) was obtained by transfecting HEK239T cells with plasmids encoding this BRET pair. In A, cells were co-transfected with the biosensor and increasing levels of competitor plasmids encoding iDAbs RAS (black striped bars) or iDAb control (grey striped bars) or biosensor alone (white bar). iDAb RAS impedes KRASG12D/PI3KαFL interaction and this inhibition causes a decrease of pAKT at serine 473 as shown by western blot in B and its quantification in C. UT is for untransfected cells. In D, HEK293T cells transfected with the BRET biosensor KRASG12D/PI3KαFL were treated for 20 hr with DMSO (white bar), 5, 10 and 20 μM of Abd-2 (grey bars) and 3344 (black bars) compounds and the BRET signal of the biosensor was assessed. In panel E, the cells were transfected and treated as in D but with 10 and 20 μM of Abd-2 and 3344 compounds. 20 hr after the treatment, cells were lysed and analysed by western blot using anti-pAKT (Ser 473) or anti-pan-AKT antibody. The signal in the western blot is quantitated in F. Related controls are shown on Figure 4—figure supplement 1. One-way ANOVA followed by Dunnett’s post-hoc tests were used to determine the statistical significance of BRET and pAKT modulations induced by the compound or the iDAb (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Each experiment was repeated twice (E–F) or three times (A–D). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D) or correspond to mean ±SEM of biological repeats (C, F). See also Figure 4—figure supplement 1.

Interaction of KRASG12D with PI3KαFL is inhibited by 3344.
(A) BRET titration curves of KRASG12D and KRASS17N mutants with full-length PI3Kα (PI3KαFL). KRASG12D interacts with PI3KaFL when the full-length regulatory subunit p85a is co-expressed along with the BRET pair but not KRASS17N. The optimal BRET signal is obtained with the following pair: RLuc8-KRASG12D/PI3KαFL-GFP2. (B) Controls from Figure 4B. The expression level of the BRET pair was assessed with the GFP (for PI3KαFL) and pan-RAS (for RLuc8-KRASG12D) antibodies. iDAb and p85αFL expression was revealed by the CMYC tag antibody, β-actin was used as the loading control. (C) Controls from Figure 4E. The expression level of the BRET pair was assessed with the GFP (PI3KαFL) and pan-RAS (RLuc8-KRASG12D) and CMYC (p85αFL) antibodies. Anti-β-actin was used as the loading control. In panel D, HEK293T cells transfected with the BRET biosensor KRASG12D/PI3KαFL were treated for 3 hr with DMSO (white bar), 5, 10 and 20 μM of Abd-2 (grey bars) and 3344 (black bars) compounds and the BRET signal of the biosensor was assessed. In panel E, the cells were transfected and treated as in panel D but with 10 and 20 μM of Abd-2 and 3344 compounds. 3 hr after the treatment, cells were lysed and analyzed by western blot using anti-pAKT (Ser 473) or anti-pan-AKT antibody. The signal in the western blot is quantitated in panel F. (G) Controls from panel E. One-way ANOVA followed by Dunnett’s post-hoc tests were used to determine the statistical significance of BRET and pAKT modulations induced by the compounds (**p<0.01, ****p<0.0001). Each experiment was repeated twice (A, E–F) or three times (D). Where error bars are presented, they correspond to mean values ± SD of biological repeats (A, D) or correspond to mean ± SEM of biological repeats (F).
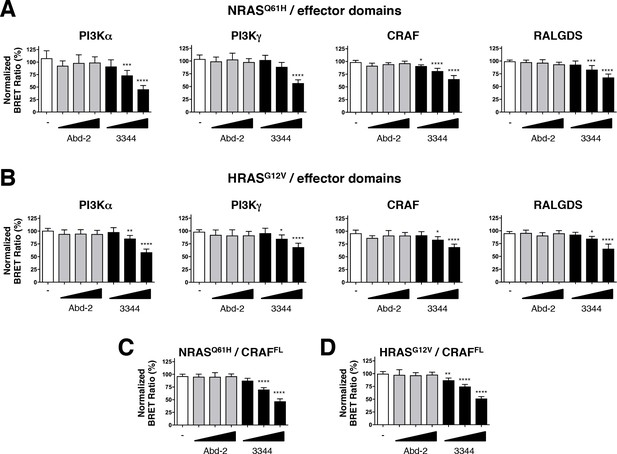
Compound 3344 inhibits NRAS and HRAS-effector BRET-based biosensors.
HEK293T cells were transfected 24 hr with plasmids expressing the NRASQ61H (A, C) and HRASG12V (B, D) biosensors together with the indicated RBDs of PI3K, CRAF and RALGDS (A, B) or full-length CRAF (C, D). These were treated with 5, 10 and 20 μM of Abd-2 (grey bars) or 3344 (black bars) compounds for 20 hr. DMSO (white bar) was used as the negative control. Statistical analyses were performed using a one-way ANOVA followed by Dunnett’s post-tests (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Each experiment was repeated at least four times. Where error bars are presented, they correspond to mean values ± SD of biological repeats (A–D). See also Figure 5—figure supplement 1.
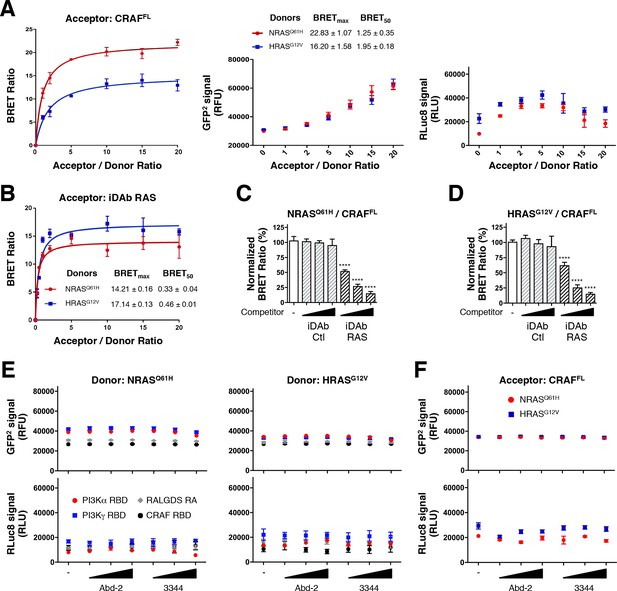
iDAb RAS inhibits mutant NRAS and HRAS interaction with CRAFFL.
(A) BRET titration curves of NRASQ61H and HRASG12V with CRAFFL with total GFP2 and RLuc8 controls. (B) BRET titration curves of NRASQ61H and HRASG12V with iDAb RAS. (C, D) Competition assays show the inhibition of NRASQ61H/CRAFFL interaction (C) and HRASG12V/CRAFFL interaction (D) by iDAb RAS (black striped bars) in a dose-dependent manner compared to the non-relevant iDAb control (grey striped bars) and the no competitor control (-, white bar). (E, F) Total GFP2 and RLuc8 levels from the BRET competition assay shown in Figure 5A–D. Statistical analyses in C and D were performed using a one-way ANOVA followed by Dunnett’s post-hoc tests (****p<0.0001). Each experiment was repeated twice (A, B) or four times (C, D). Where error bars are presented, they correspond to mean values ± SD biological repeats.

Compound 3344 interacts in a pocket close to the switch regions of KRAS.
The interaction of mutant KRAS with compound 3344 was analyzed by X-ray crystallography. (A) KRASQ61H crystals were soaked with 3344 compound and crystal structures obtained from X-ray diffraction. The compound is shown binding in the hydrophobic pocket near switch I (shown in red) and switch II (shown in blue). The electron density map of the compound (2Fo-Fc) is shown as green mesh, and contoured at 1.0 rms. (B) We have modeled the potential interactions that could prevent 3344 and a RAS effector binding simultaneously to the same RAS molecule by overlaying our structure of the KRAS-3344 complex onto the published structures of top panel: HRAS-CRAF RBD (PDB 4G3X), middle panel: HRAS-RALGDS RA (PDB 1LFD), bottom panel: HRAS-PI3Kγ RBD (PDB 1HE8). (C, D) Two human mutant KRAS expressing lines (C: DLD-1 and D: H358) were serum-starved for 24 hr and treated 3 hr with different concentrations of 3344 (2, 5, 10 and 20 μM) before stimulation with EGF (50 ng/mL) for 10 min. Cells were harvested, proteins extracted and separated by SDS-PAGE for western blot analysis. Western membranes were treated with anti-pAKT S473; anti-pan AKT; anti-pERK1/2 and anti-ERK1/2 as indicated. Statistical analyses of pERK/ERK and pAKT/AKT quantifications were performed using a one-way ANOVA followed by Dunnett’s post-tests (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Where error bars are presented, they correspond to mean values ± SEM of biological repeats (C–D). Each experiment was performed twice (C–D).

Tables
Data processing and refinement statistics.
https://doi.org/10.7554/eLife.37122.016| Structure | KRASQ61H-3344 |
|---|---|
| Data collection | |
| PDB ID | 6F76 |
| Diffraction source | ID30A-1, ESRF |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.966 |
| Rotation range per image (°) | 0.05 |
| Exposure time per image (s) | 0.092 |
| Space group | P 212121 |
| Molecules/asymmetric unit | 6 |
| Unit cell dimensions | |
| a, b, c (Å) | 63.17, 118.19, 155.95 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 77.98–2.20 (2.16–2.20)* |
| Total no. of reflections | 295785 (13854) |
| Unique reflections | 65992 (2888) |
| Completeness (%) | 99.2 (87.3) |
| Multiplicity | 4.5 (4.8) |
| Rmeas(I)† | 0.193 (0.997) |
| Rmerge‡ | 0.151 (0.780) |
| Rpim(I)§ | 0.119 (0.612) |
| I/sigma | 5 (1.8) |
| CC1/2 (%)# | 0.985 (0.513) |
| Refinement | |
| No. of reflections, working set | 62692 (2744) |
| No. of reflections, test set | 3300 (144) |
| Rwork/Rfree | 22.7/25.0 |
| No. of atoms | |
| Protein | 8400 |
| Water | 57 |
| Average B factors (Å2) | |
| Protein | 46.8 |
| Ligand GTP | 31.9 |
| Water | 30.1 |
| RMSD | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.67 |
| Ramachandran plot | |
| Favoured regions (%) | 97.1 |
| Additionally allowed (%) | 2.9 |
| Outliers | 0 |
| MolProbity statistics | |
| Overall score | 1.11 |
| Clash score | 1.22 |
| Rotamer outliers (%) | 1.4 |
-
a*Values in parentheses are for data in the highest resolution shell.
†Rmeas = Σhkl{N(hkl)/[N(hkl)−1]}1/2 Σi|Ii(hkl)- < I(hkl)>|/ Σhkl ΣiIi(hkl), where Ii(hkl) is the intensity of reflection hkl. Σi is the sum over all i measurements of reflection hkl and N(hkl) is the multiplicity of reflection hkl.
-
‡Rmerge = Σhkl Σi | Ii (hkl)–<I(hkl)>| / Σhkl Σi Ii (hkl), where Ii (hkl) is the intensity of reflection hkl and Σi is the sum over all I measurements of reflection hkl.
§Rpim= Σhkl{1/[N(hkl)−1]} 1/2 Σi|Ii(hkl)- < I (hkl)> |/ Σhkl ΣiIi(hkl), where Ii(hkl) is the intensity of reflection hkl, Σi is the sum over all i measurements of reflection hkl and N(hkl) is the multiplicity of reflection hkl.
-
#CC1/2 is Pearson’s correlation coefficient between random half data sets.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | HEK293T | ATCC | Cat#CRL-3216 RRID:CVCL_0063 | |
| Cell line (human) | DLD-1 | ATCC | Cat#CCL-221 RRID:CVCL_0248 | |
| Cell line (human) | H358 | ATCC | Cat#CRL-5807 RRID:CVCL_1559 | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASG12D-CAAX plasmid | This paper | N/A | DNA/protein sequences provided in the Supplementary file 1 |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASG12A-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASG12C-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASG12V-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASG12R-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- NRASQ61H-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- HRASG12V-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASS17N-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-RLuc8-(GGGS)3- KRASWT-CAAX plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-GFP2-(GGGS)3- CRAFS257LFL plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-PI3KαFL-(GGGS)3- GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-CRAF RBD (aa 1–149)- (GGGS)3-GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-PI3Kα RBD (aa 161–315)- (GGGS)3-GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-PI3Kγ RBD (aa 190–315)- (GGGS)3-GFP2 plasmid | This paper | N/A | DNA/protein sequences provided in the Supplementary file 1 |
| Transfected construct (human) | pEF-iDAb RAS-(GGGS)2- GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-iDAbdm RAS-(GGGS)2- GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-iDAb control-(GGGS)2- GFP2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-LMO2-(GGGS)2- RLuc8 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-GFP2-(GGGS)3- iDAbdm LMO2 plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-memb-FLAG-iDAb RAS plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-memb-FLAG-iDAb control plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-iDAb RAS-myc plasmid | This paper | N/A | |
| Transfected construct (human) | pEF-iDAb control-myc plasmid | This paper | N/A | |
| Transfected construct (human) | pcDNA3.1-myc-p85αFL plasmid | A gift from R. Williams and O. Perisic | N/A | |
| Transfected construct (mouse) | pEF-RALGDS RA (aa 788–884)- (GGGS)3-GFP2 plasmid | This paper | N/A | The RALGDS RA domain corresponds to the mouse sequence |
| Antibody | Phospho-ERK 1/2 Rabbit antibody | Cell Signaling Technology | Cat#9101S RRID:AB_331646 | |
| Antibody | Total ERK 1/2 Rabbit antibody | Cell Signaling Technology | Cat#9102S RRID:AB_330744 | |
| Antibody | Phospho-MEK 1/2 Rabbit antibody | Cell Signaling Technology | Cat#9154S RRID:AB_2138017 | |
| Antibody | Total MEK 1/2 Mouse antibody | Cell Signaling Technology | Cat#4694S RRID:AB_10695868 | |
| Antibody | Phospho-AKT S473 Rabbit antibody | Cell Signaling Technology | Cat#4058S RRID:AB_331168 | |
| Antibody | Total AKT Rabbit antibody | Cell Signaling Technology | Cat#9272S RRID:AB_329827 | |
| Antibody | Pan-RAS Mouse antibody | Millipore | Cat#OP40 RRID:AB_213400 | |
| Antibody | GFP Mouse antibody | Santa Cruz Biotechnology | Cat#sc-9996 RRID:AB_627695 | |
| Antibody | β-Actin Mouse antibody | Sigma-Aldrich | Cat#A1978 RRID:AB_476692 | |
| Antibody | CMYC HRP-linked Goat antibody | Novus Biologicals | Cat#NB600-341 RRID:AB_10000717 | |
| Antibody | Anti-Mouse IgG HRP-linked antibody | Cell Signaling Technology | Cat#7076S RRID:AB_330924 | |
| Antibody | Anti-Rabbit IgG HRP-linked antibody | Cell Signaling Technology | Cat#7074S RRID:AB_2099233 | |
| Recombinant DNA reagent | pEF-myc-cyto vector | Invitrogen | Cat#V89120 | |
| Recombinant DNA reagent | pRLuc8-N3 vector | A gift from J. Felce | Felce et al., 2017 | |
| Recombinant DNA reagent | pGFP2-N3 vector | A gift from J. Felce | Felce et al., 2017 | |
| Recombinant DNA reagent | pBABEpuro-CRAFS257L FL plasmid | Addgene | Addgene#51125 | |
| Peptide, recombinant protein | KRASQ61H | This paper | N/A | |
| Peptide, recombinant protein | KRASG12V | This paper | N/A | |
| Peptide, recombinant protein | Anti-RAS scFv | This paper | N/A | |
| Peptide, recombinant protein | Recombinant Human Epidermal Growth Factor (EGF) | Life Technologies | Cat#PHG0311 | |
| Chemical compound, drug | Coelenterazine 400a | Cayman Chemical | Cat#16157 | |
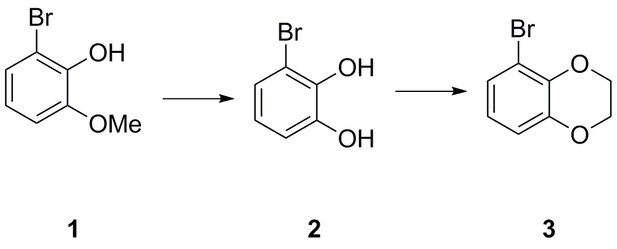
| Chemical compound, drug | 2-bromo-6-methoxyphenol | This paper | N/A | |
| Chemical compound, drug | 3-bromobenzene-1,2-diol | This paper | N/A | |
| Chemical compound, drug | 5-bromo-2,3- dihydrobenzo[b][1,4]dioxine | This paper | N/A | |
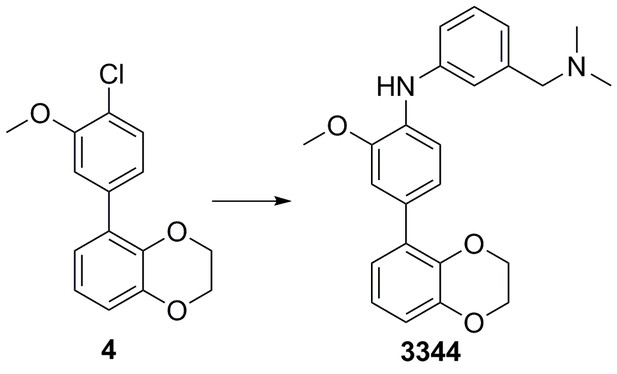
| Chemical compound, drug | 5-(4-chloro-3- methoxyphenyl)−2,3- dihydrobenzo[b][1,4]dioxine | This paper | N/A | |
| Chemical compound, drug | 4-(2,3-dihydrobenzo[b] [1,4]dioxin-5-yl)-N-(4- (dimethylamino) methyl)phenyl)-2- methoxyaniline | This paper | N/A | |
| Software, algorithm | Image J | National Institutes of Health | https://imagej.nih.gov/ij/download.html RRID:SCR_003070 | |
| Software, algorithm | Prism 7.0 c | GraphPad | https://www.graphpad.com/scientific-software/prism/ RRID:SCR_002798 | |
| Software, algorithm | PROCHECK | Laskowski et al. (1993a) | http://www.ccp4.ac.uk/html/procheck_man/index.html | |
| Software, algorithm | REFMAC | Murshudov et al. (1997) | http://www.ccp4.ac.uk/html/refmac5.html RRID:SCR_014225 | |
| Software, algorithm | MolProbity | Chen et al. (2010) | http://molprobity.biochem.duke.edu/ RRID:SCR_014226 | |
| Software, algorithm | Phenix | Adams et al. (2010) | https://www.phenix-online.org/ RRID:SCR_014224 | |
| Software, algorithm | PyMOL | Schrodinger | https://pymol.org/2/ RRID:SCR_000305 | |
| Other | Opti-MEM I Reduced Serum Medium, no phenol red | Thermo-Fisher | Cat#11058021 | |
| Other | ViewPlate, White 96-well plate, clear bottom for tissue culture | PerkinElmer | Cat#6005181 | |
| Other | BRET2 Dual Emission optical module | PerkinElmer | Cat#2100–8140 | |
| Other | Envision instrument, Multilabel Reader | PerkinElmer | Cat#2103 |
Additional files
-
Supplementary file 1
DNA and protein sequences of BRET biosensors constructs.
The list of the DNA and protein sequences from the different RAS BRET biosensor constructs used in this study.
- https://doi.org/10.7554/eLife.37122.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37122.021





