Species-specific maturation profiles of human, chimpanzee and bonobo neural cells
Figures
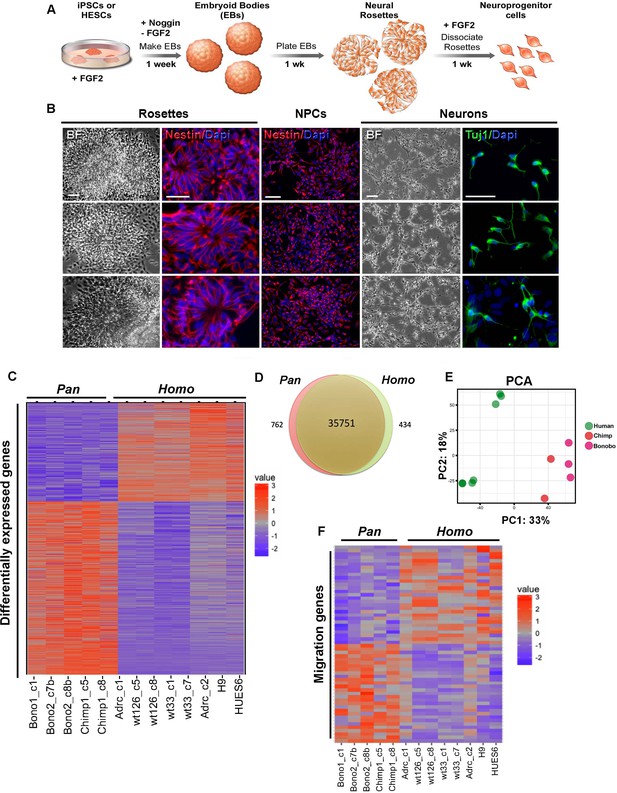
Characterization of neural progenitor cells (NPCs) derived from human (Homo), chimpanzee and bonobo (Pan) iPSCs.
(A) Scheme showing the main steps and timeline involved in the generation of NPCs from iPSCs. (B) Morphological aspects of neural rosettes derived from embryoid bodies (EBs) (BF: Bright Field). Neural rosettes are positive for the early neural marker Nestin (red). In the presence of FGF2, dissociated rosettes form a homogeneous monolayer population of proliferating Nestin-positive NPCs. Upon FGF2 withdrawal, NPCs differentiate readily in Tuj-1-positive neurons (green). Scale bars: 50 µm. (C) Heat map representation of mapped reads corresponding to protein coding genes. Chimpanzee and bonobo NPCs clustered closer to each other than to human NPC lines and to human embryonic stem cell-derived NPCs (HUES6 and H9 ES lines). (D) Venn diagram showing pairwise comparison of protein-coding genes. Red or green: significantly differentially expressed genes (FDR < 0.05 and fold change >2). Red and green: total of expressed genes with no significant differences in mRNA levels between compared species. (E) Principal component analysis (PCA) of human, chimpanzee and bonobo NPCs showing human samples (green circles) clustering separately from NHPs (red and pink circles) on the first principal component (33% of the variance). (F) Heat map showing comparison between Homo and Pan NPC expression profiles of genes related to cell migration.

Similar expression of cortical progenitor markers and similar spatial enrichment of differentially regulated genes in Homo and Pan.
(A–B) Representative images of NPCs from human, chimpanzee and bonobo stained for cortical progenitor transcription factors (A) FOXG1 and (B) PAX6. Scale bar: 50 μm. (C) Quantification of the percentage of cells expressing FOXG1 or PAX6 in human NPCs was comparable to chimpanzee and bonobo percentages. (D) Spatial enrichment of genes in Homo (top) or Pan (bottom) NPCs was calculated using the BrainImageR tool (Linker et al., 2019). Genes upregulated in human NPCs exhibited similar regional enrichment to genes upregulated in Pan (chimpanzee and bonobo) NPCs, indicating that the differences observed in bulk RNA sequencing are not influencing broad fate specification between NPCs from Homo versus Pan. (E) Scheme of differentially expressed (Diff Exp) genes in human compared to chimpanzee and bonobo (nonhuman primates, NHP). The scheme shows that, of the 1,196 Diff Exp genes between humans and NHPs, 52 were classified in the migration category after gene ontology analysis (Fisher’s exact test, p<1.07e-24).

Single cell analysis on human and Pan NPCs.
(A) Proportion of cells expressing known markers for different cortical layers as well as hindbrain markers. Proportions are expressed out of the total number of NPCs expressing at least one marker. (B) T-SNE analysis of NPCs from human (top; green), bonobo (middle; orange), and chimpanzee (bottom; red). We investigated whether HOXA7 expression was indicative of NPCs being differentially primed toward the hindbrain or spinal cord fates across species. Using PAX6 and EN2 as markers of cortical and hindbrain NPCs, respectively, we noted that HOXA7 cells preferentially also expressed PAX6 (bonobo = 6.0%, chimpanzee = 8.2%) in comparison to EN2 (bonobo = 0%, chimpanzee = 0.7%). Importantly, the existence of NPCs that were double positive for HOXA7 and PAX6 in bonobos and chimpanzees and the striking absence of HOXA7 expression in any human NPCs further supported that HOXA7 expression was an intrinsic characteristic of Pan NPCs, independent of regional identity. Populations that expressed SOX2, SOX2 and HOXA7, or all four markers, are noted in dashed circles. Percentage of PAX6+ or EN2+ cells out of all HOXA7-expressing NPCs are noted below the respective ovals. (C) Percentage of NPCs expressing known cortical or hindbrain markers for each species (human = green, bonobo = orange, chimpanzee = red).
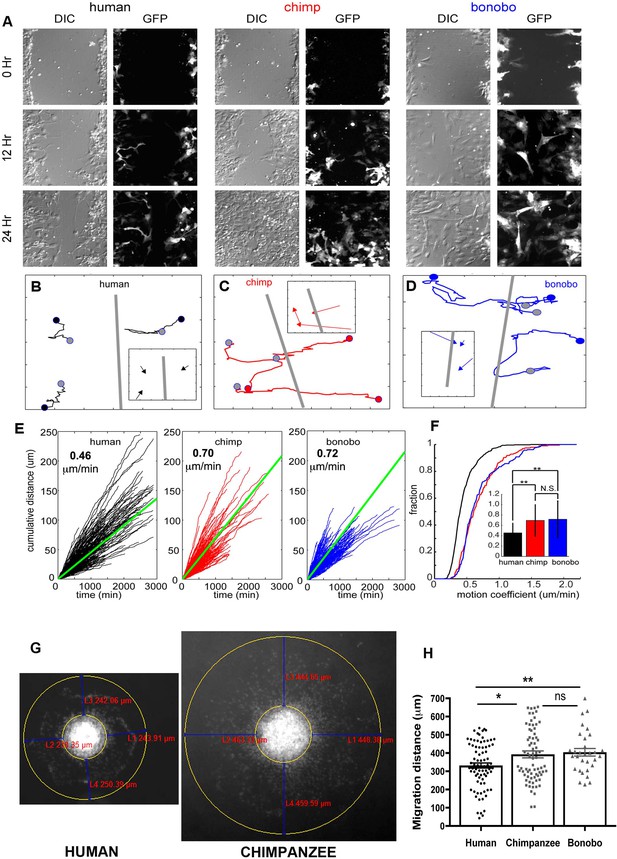
NPC differential migration patterns in humans, chimpanzees and bonobos.
(A) DIC (differential interference contrast) (left) and fluorescent (GFP) (right) images of time-lapse NPC migration from humans, chimpanzees, and bonobos at 0 hr (top row), 12 hr (middle row), and 24 hr (bottom row) following a scratch made with a glass pipette in the center of the field of view (at 0 hr). (B–D) Individual tracks with NPC origin marked with a black circle for human NPCs (B), red circle for chimpanzee NPCs (C), and blue circle for bonobo NPCs (D). Migration paths for each NPC are marked with a line and the end of the migration is marked with a gray circle. The scratch location is marked with a gray line. Inserts contain vectors for each cell showing net migration (from beginning of the track to the end – colored arrows for each NPC) relative to the scratch (gray line). (E) Cumulative distance from each NPC over time from all human (black lines), chimpanzee (red lines) and bonobo (blue lines). Also shown is the mean cumulative distance for each species (green line). (F) Cumulative histogram of motion coefficients of NPCs from three species and mean motion coefficient for NPCs from three species (inset) (**p<0.01, Mann-Whitney U test), showing that chimpanzee and bonobo NPCs migrate longer distances at higher speeds compared to human NPCs. (G–H) Migration analysis using neurosphere outgrowth assay. (G) Representative images of iPSC forebrain neurospheres from human and chimpanzee showing differential outgrowth migration of NPCs. The average distance between the radius of the inner neurosphere and outer circumference of cells was calculated using (DAPI)-stained nuclei (blue). Four measurements were taken and averaged for each neurosphere (examples of distances shown in red). (H) Graph showing quantification of distances migrated by human, chimpanzee and bonobo NPCs. The data confirmed that NPCs from chimpanzees and bonobos migrate longer distances than human NPCs. Comparative statistics as follows: Human versus Chimpanzee, p=0.0058; Human versus Bonobo, p=0.0108; Chimpanzee versus Bonobo, p=0.6831. *p<0.05, **p<0.01, Welch’s t test).
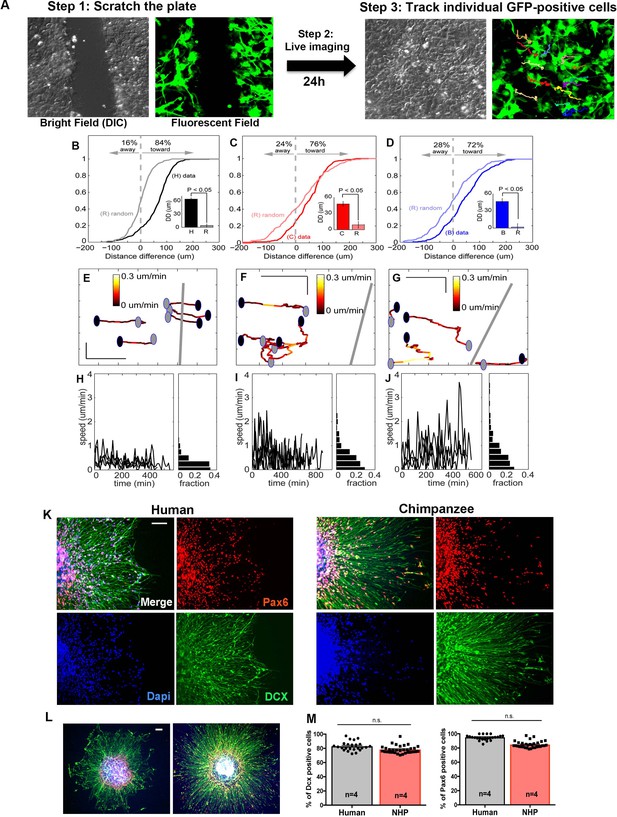
Characterization of migration patterns on NPCs.
(A) Scheme of the scratch assay and single cell tracking of GFP-positive cells 24 hr after the scratch. (B–D) Distribution of NPC distance differences for human (H, black line), chimp (C, red line), and bonobo (B, blue line) as compared to random movement (R) calculated for each species. The data show that the majority of the cells, independent of species, migrate towards the center of the scratch and the migration measure in this study is directional. (E–J) Migration coefficient differences for NPCs from different species. (E–G) Representative examples of a migration path of a human (E), chimpanzee (F), and bonobo (G) NPC with each time step color-coded by speed. Units = µm. (H–J) Histogram plots showing speed profiles over time for each representative cell in the panel above. (K–M) Migration analysis using neurosphere outgrowth assay. (K and L) Representative images of iPSC-derived forebrain neurospheres from human and chimpanzee showing differential outgrowth migration of NPCs. To evaluate if differential migratory properties were influenced by the presence of cells from other brain regions due to loss of cell purity during migration, we post-stained human and nonhuman primate cells after neurosphere migration experiment with cortical progenitor markers (PAX6, in red) and neuroblast migration marker (doublecortin, DCX, in green). (M) Percentages of cells expressing PAX6 or DCX were not significantly different between species. Scale bars: 10 µm.
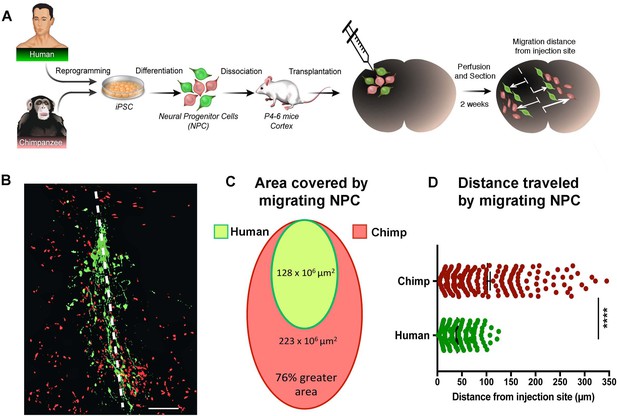
In vivo migration comparative analysis.
(A) Scheme of the transplantation experimental procedure to detect in vivo cell migration. Fibroblasts from human and chimpanzee were reprogrammed to iPSCs and differentiated to NPCs and then transduced with lentiviral vectors expressing either green fluorescent protein (GFP) or TdTomato. NPC lines from human and chimpanzee were mixed and grafted into the developing postnatal mouse cortex. Mice were sacrificed at two weeks post-transplantation. (B) Representative image of grafted human (green) and chimpanzee (red) NPCs migrating away from the injection site (indicated by the white dashed line). (C) Representation of the area covered by human and chimpanzee migrating NPCs (D) Distance migrated by single NPCs from the injection site. (****p<0.0001, Mann-Whitney U test).

In vivo migration of human and chimpanzee NPCs two weeks after transplantation in mouse brain.
NPCs from human and chimpanzee were transduced with lentiviral vectors expressing either green fluorescent protein (green) or TdTomato (red). NPC lines from human and chimpanzee were mixed and grafted into the developing postnatal mouse cortex. (A) Low magnification image (4X) of grafted human (green) and chimpanzee (red) NPCs migrating away from the injection site (indicated by the white dashed line) (B) Higher magnification image (10X) from the box shown on (A). Scale bars: 10 µm.
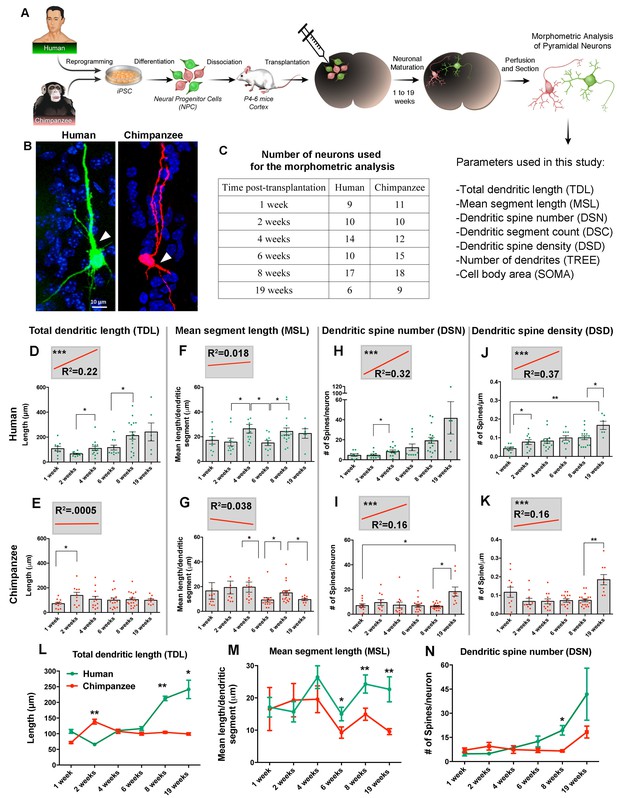
Comparative morphometric analyses on iPSC-derived transplanted neurons over time.
(A) Scheme of the transplantation experimental procedure. NPCs were obtained from both species and transduced with lentiviral vectors expressing either green fluorescent protein (GFP) or TdTomato (Red). The NPCs from both species were dissociated together and mixed before injection into brains of mice at postnatal day P4-6. The brains of the animals were harvested after 1, 2, 4, 6, 8 and 19 weeks post transplantation (PST). Each brain was cryosectioned and the anatomy of GFP- and TdTomato-positive pyramidal neurons in the cortex was analyzed. Parameters used for the morphometric analysis are listed. (B) Representative confocal images from human and chimpanzee pyramidal neurons eight weeks PST. Arrowheads indicate potential apical dendrites. Scale bar = 10 µm. (C) Number of pyramidal neurons used for the morphometric analysis at different time points following transplantation. (D–K) Graphs illustrating changes in morphometric variables in human and chimpanzee iPSC-derived pyramidal neurons over time after transplantation. (D, E) Total dendritic length; (F, G) mean segment length; (H, I) dendritic spine number; and (J, K) dendritic spine density. Each point in the bar graphs represents an individual neuron. Statistical significance is shown between adjacent time points and between the earliest (1 week) and the latest (19 weeks) time point after transplantation. The top left corner insets show regression lines (red) of respective variables correlated with weeks PST. Coefficient of determination (R2) and statistical significance are indicated for each regression analysis (***p<0.001, Pearson’s correlation test). (L–N) Comparative morphometric analyses on iPSC-derived transplanted neurons showed significant difference between species on specific time points. (L) Total dendritic length (TDL: 2 weeks, p=0.0100; 8 weeks, p=0.0039; 19 weeks, p=0.0316); (M) mean segment length (MSL: 6 weeks, p=0.0483; 8 weeks, p=0.0086; 19 weeks, p=0.0016); and (N) dendritic spine number (DSN: 8 weeks, p=0.0482). (*p<0.05; **p<0.01, Unpaired t test).
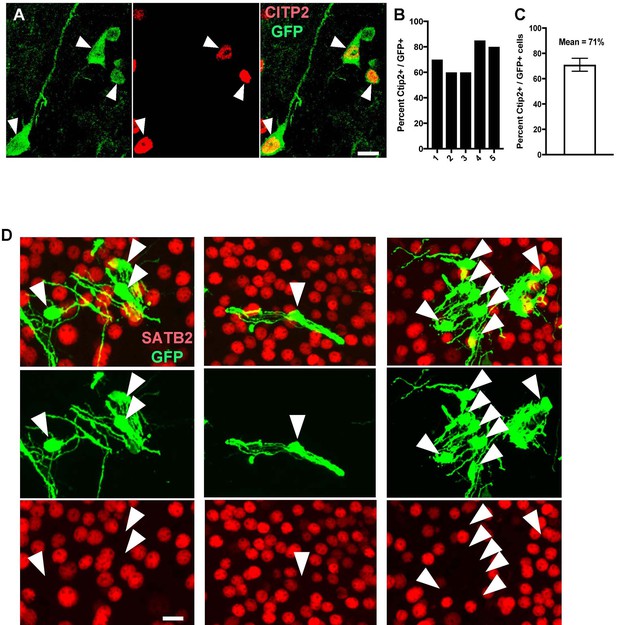
In vivo differentiation of human and chimpanzee neurons post-transplantation.
(A) Images of pyramidal neurons (arrowheads) transplanted in mouse cortex showing co-localization with deep layer (layer V-VI) cortical marker CTIP2. (B–C) The percentage of CTIP2-positive cells was similar in different transplanted animals (example showing five different animals) and between human and chimpanzee transplanted cells (mean of 71% of co-localization). (D) Images of neurons transplanted in mouse cortex (arrowheads) showing absence of co-localization with upper cortical layer marker (SATB2).
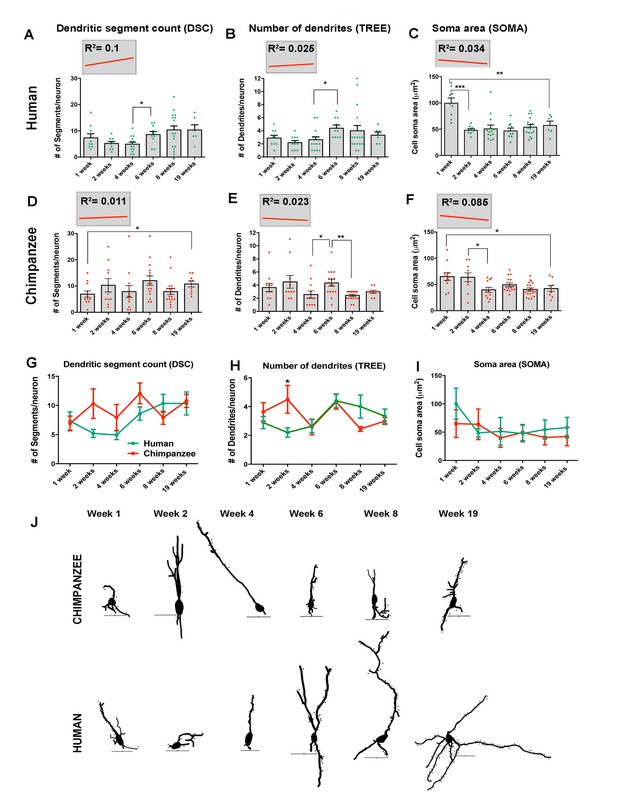
Morphometric analysis of human and chimpanzee neurons post- transplantation.
(A–F) Graphs illustrating changes in morphometric variables in human and chimpanzee iPSC-derived pyramidal neurons over time post-transplantation. (A, B) Dendritic segment count (DSC); (C, D) number of dendrites (TREE); (E, F) soma area (SOMA). Each point in the graph represents an individual neuron. Statistical significance is shown between adjacent time points and between the earliest (1 week) and the latest (19 weeks) time point post-transplantation. The top left corner insets show regression lines (red) of respective variables correlated with weeks post transplantation. Coefficient of determination (R2) is indicated for each regression analysis. No statistical significance was observed for the regression lines obtained. (G–I) Comparative morphometric analyses in human and chimpanzee iPSC-derived transplanted neurons. (G) DSC; (H) TREE and (I) SOMA. (*p<0.05; **p<0.01, Unpaired t test). (J) Sample tracings of neurons included in the analysis examples show chimpanzee (top) and human (bottom) iPSC-derived neurons for each time point after transplantation (Week 1 – Week 19). Dots represent dendritic spines. Scale bar: 25 µm. *Includes neurons for 8 and 7.5 weeks after transplantation.
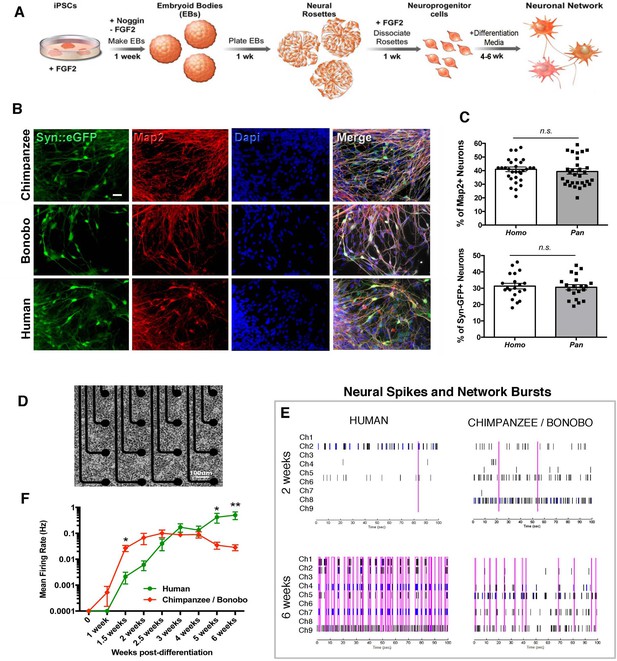
Neuronal functional analysis of human, chimpanzee and bonobo over time.
(A) Differentiation scheme using EBs, neural rosettes and NPCs as intermediate cell lines. (B) iPSC-derived neurons in culture can be visualized by infection with a lentiviral vector carrying the EGFP reporter gene under the activity of the neuron-specific Synapsin promoter (Syn::eGFP). Most of the eGFP-positive neurons expressed the mature neuronal marker Synapsin. Scale bar: 20 µm. (C) No significant change was observed in the percentage of neurons expressing the neuronal markers Map2AB and Synapsin::eGFP between human, chimpanzee and bonobo lines. (D–E) Multielectrode array (MEA) analysis was performed in differentiating neurons from human, chimpanzee and bonobo lines. (D) Example of chimpanzee neurons growing on top of electrodes on a multiwell plate. (E) Representative MEA traces on two- and six-week-old neurons from humans, chimpanzees and bonobos showing neural spikes (black dashes), neural bursts (blue dashes) and network bursts among channels (pink lines). (F) Comparative analysis between species over differentiation time showing that while chimpanzee and bonobo neurons have accelerated functional maturation (1.5 weeks, p=0.0147), human neural cultures show increased firing rates at later time points (5 weeks, p=0.0239 and 6 weeks, p=0.0018). (*p<0.05; **p<0.01, Unpaired t test).
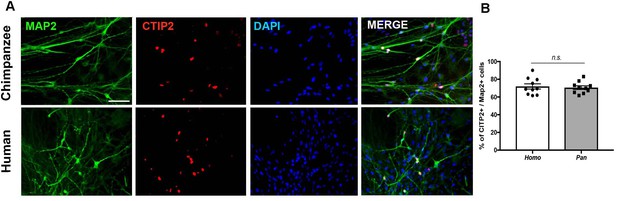
Immunocytochemistry of iPSC-derived neurons from humans and chimpanzees.
(A) Representative images from neurons stained with pan neuronal marker (Map2) and cortical marker (CTIP2). Scale bar = 50 µm. (B) Quantification of the percentage of Map2-positive cells that express CITP2. No significant change was observed in the percentage of neurons expressing CTIP2 in vitro between human and chimpanzee cells.
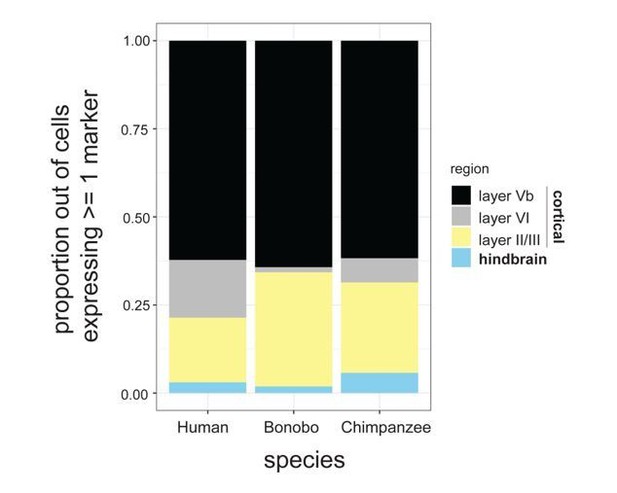
Proportion of cells expressing known markers for different cortical layers as well as hindbrain markers.
Proportions are expressed out of the total number of NPCs expressing at least 1 marker.
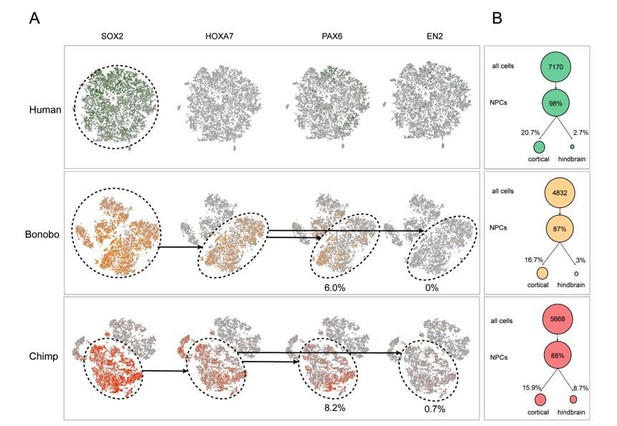
Single cell analysis on human and Pan NPCs.
(A) T-SNE of NPCs from human (top; green), bonobo (middle; orange), and chimpanzee (bottom; red). Populations that expressed SOX2, SOX2 and HOXA7, or all four markers, are noted in dashed circles. Percentage of PAX6+ or EN2+ cells out of all HOXA7-expressing NPCs are noted below the respective ovals. (B) Percentage of NPCs expressing known cortical or hindbrain markers for each species (human = green, bonobo = orange, chimpanzee = red). The existence of NPCs that were double positive for HOXA7 and PAX6 in bonobos and chimpanzees and the striking absence of HOXA7 expression in any human NPCs further supported that HOXA7 expression was an intrinsic characteristic of Pan NPCs, independent of regional identity.
Additional files
-
Supplementary file 1
Table showing the full list of genes differentially expressed in nonhuman primates versus human.
- https://doi.org/10.7554/eLife.37527.015
-
Supplementary file 2
Table showing the list of genes in the Gene Ontology (GO) category of migration that are differentially expressed in nonhuman primates versus human.
- https://doi.org/10.7554/eLife.37527.016
-
Supplementary file 3
Summary of the subjects and clones utilized for each experiment and additional information on RNA sequencing (total counts, raw counts and normalized counts).
- https://doi.org/10.7554/eLife.37527.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37527.018





