Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism
Figures
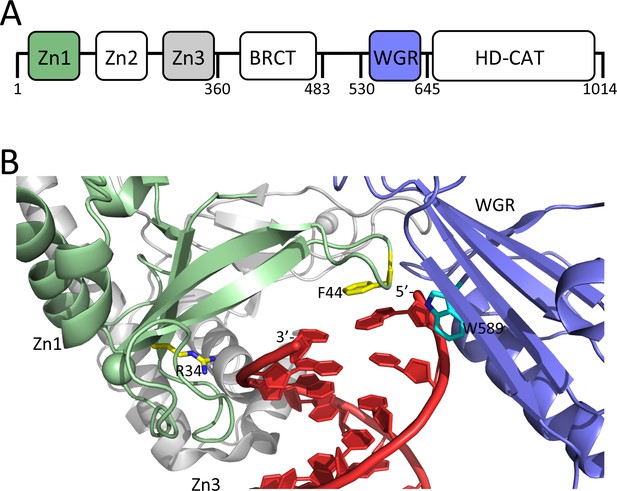
Domain organization of PARP1 and structural details of how PARP1 binds to a DSB.
(A) Schematic of the domains of PARP1; (B) DNA-binding domains (Zn1, green, Zn3 gray, and WGR, blue) of PARP1 engaging a DNA DSB (red). Residues R34 and F44 of the phosphate backbone grip and the base stacking loop in the Zn1 domain are shown in yellow and W589 in the WGR domain is shown in light blue. Coordinates were taken from 1dqy.
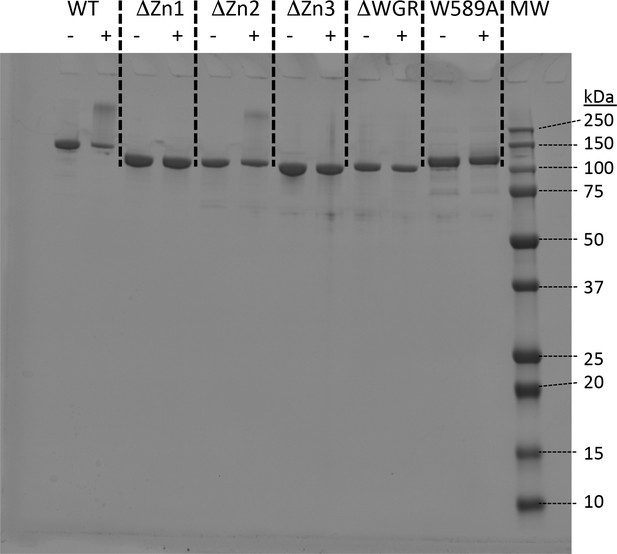
SDS-PAGE showing purified PARP1, its deletion constructs and the W589A point mutant.
Each construct was also assayed for activity in the presence (+) of NAD+ (200 µM) and one equivalent of p18mer DNA. Higher molecular weight smears are seen for WT and ΔZn2, indicating auto-PARylation.
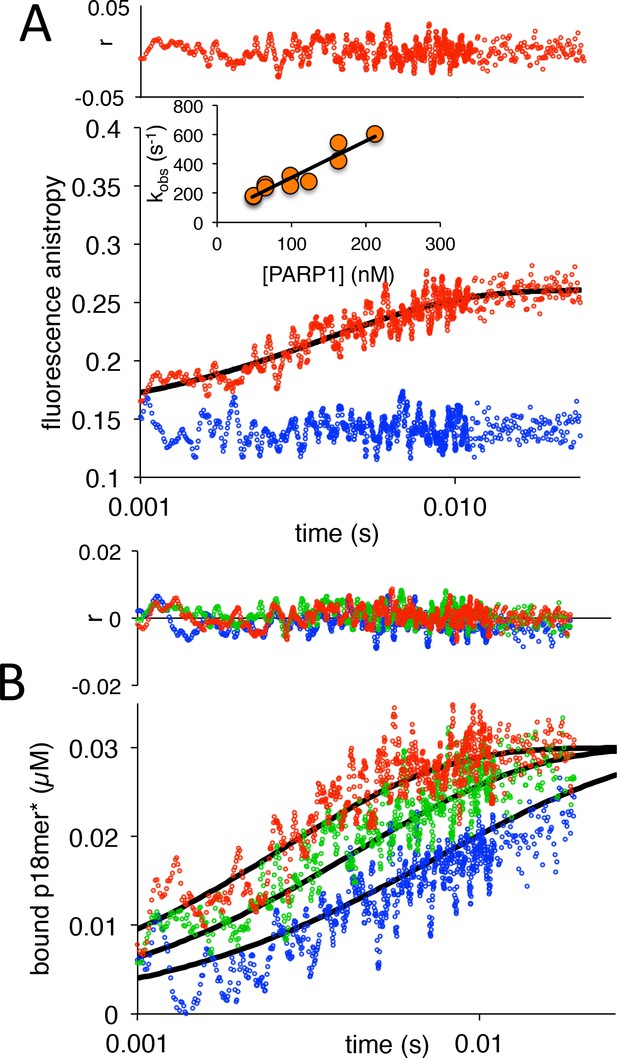
PARP1 association with DNA as monitored by fluorescence anistropy.
(A) Representative measurement of PARP1 association with DNA as monitored by fluorescence anisotropy. Shown are the data in the absence of PARP1 (blue) and in the presence of 83 nM PARP1 (red). The black line shows a first-order exponential fit to the data and the residuals (r) from this fit are shown above. The inset shows a replot of kobs vs. varying concentrations of PARP1. (B) Global fitting of three representative concentrations of PARP1 using the mechanism in Scheme 1: 50 nM in blue, 83 nM in green and 133 nM in red. Residuals (r) for the three concentrations are shown in the corresponding colors above.
-
Figure 2—source data 1
Raw data for representative measurement of PARP1 association with DNA as monitored by fluorescence anisotropy as shown in panel A.
The data shown in the inset are also included as labeled.
- https://doi.org/10.7554/eLife.37818.016
-
Figure 2—source data 2
Global fitting of three representative concentrations of PARP1 using the mechanism in Scheme 1 with raw data, best fits, and residuals as shown in panel B.
- https://doi.org/10.7554/eLife.37818.017
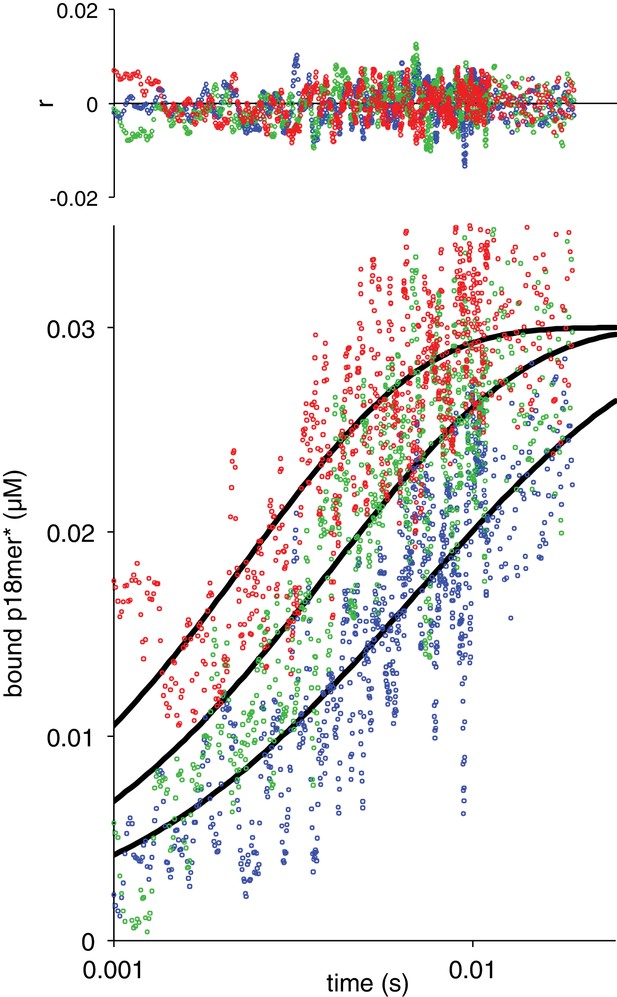
Association of ΔZn1 with p18mer*.
Global fits of the kinetic model in Scheme 1 for three representative protein concentrations: 42 nM in blue, 72 nM in green, 120 nM in red. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 2—figure supplement 1—source data 1
Association of ΔZn1 with p18mer*.
- https://doi.org/10.7554/eLife.37818.007
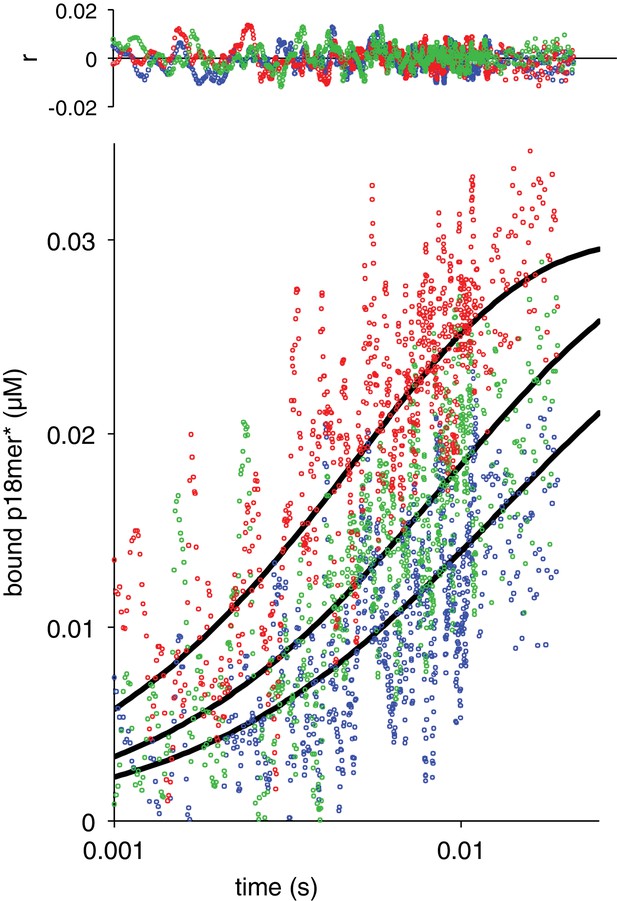
Association of ΔZn2 with p18mer*.
Global fits of the kinetic model in Scheme 1 for three representative protein concentrations: 35 nM in blue, 52 nM in green, 95 nM in red. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 2—figure supplement 2—source data 1
Association of ΔZn2 with p18mer*.
- https://doi.org/10.7554/eLife.37818.009
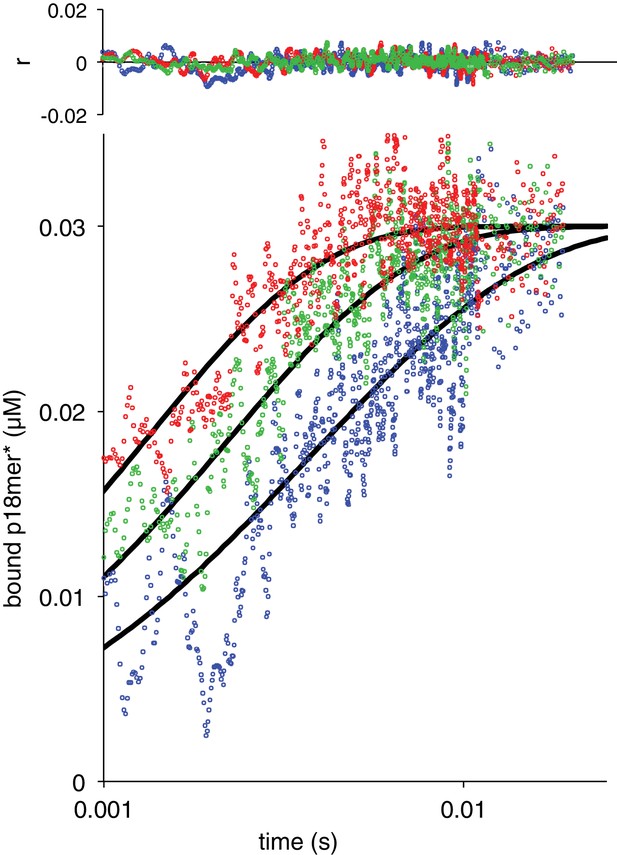
Association of ΔZn3 with p18mer*.
Global fits of the kinetic model in Scheme 1 for three representative protein concentrations 50 nM in blue, 83 nM in green, 133 nM in red. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 2—figure supplement 3—source data 1
Association of ΔZn3 with p18mer*.
- https://doi.org/10.7554/eLife.37818.011
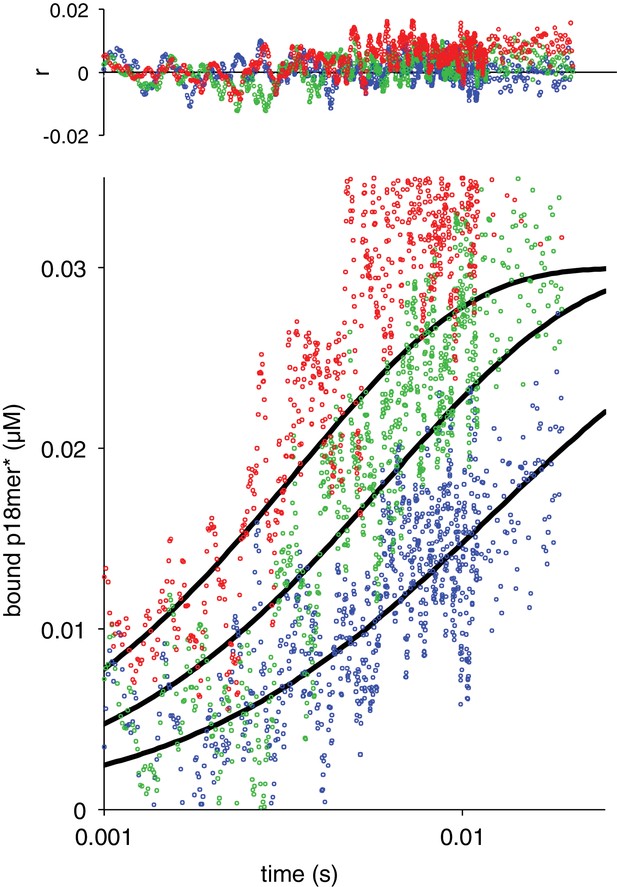
Association of ΔWGR with p18mer*.
Global fits of the kinetic model in Scheme 1 for three representative protein concentrations: 37 nM in blue, 74 nM in green, 129 nM in red. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 2—figure supplement 4—source data 1
Association of ΔWGR with p18mer*.
- https://doi.org/10.7554/eLife.37818.013
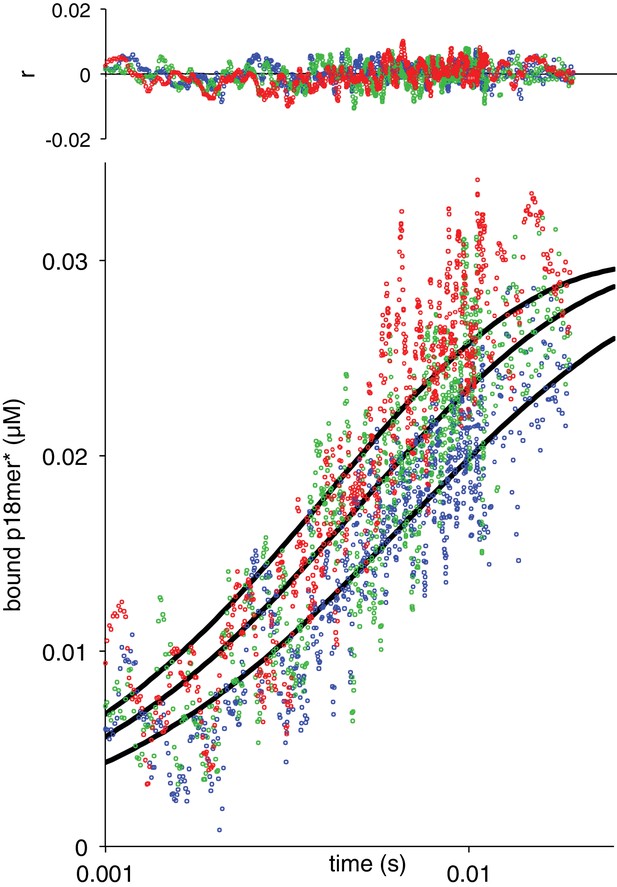
Association of W589A with p18mer*.
A global fit of the kinetic model in Scheme 1 to three representative protein concentrations is shown (38 nM in blue, 51 nM in green, and 63 nM in red).
-
Figure 2—figure supplement 5—source data 1
Association of W589A with p18mer*.
- https://doi.org/10.7554/eLife.37818.015
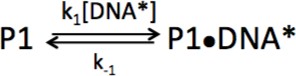
Kinetic model for association of PARP1 with DNA.
https://doi.org/10.7554/eLife.37818.018
Kinetic model for dissociation of PARP1 from labeled dDNA in the presence of competing unlabeled DNA.
https://doi.org/10.7554/eLife.37818.021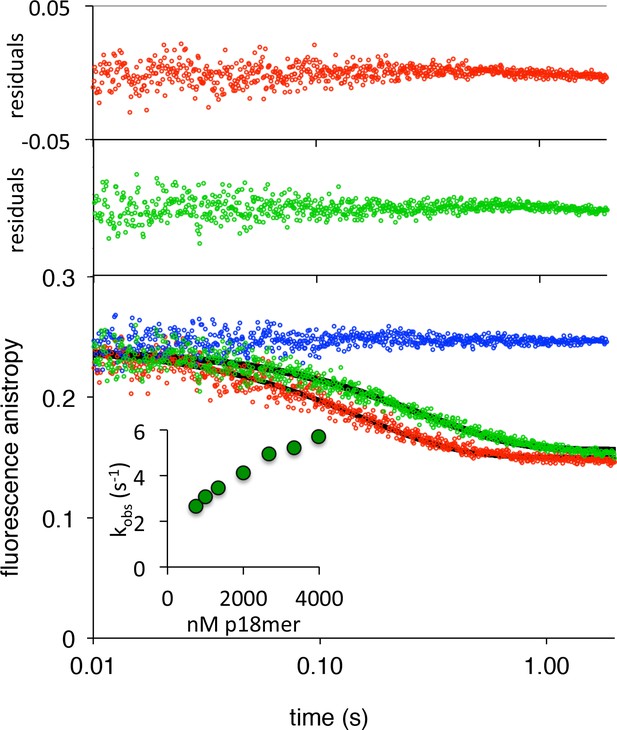
Representative measurement of PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Shown are the data in the absence of competitor DNA (in blue) and in the presence of 2.2 µM (green) and 4 µM DNA (red). The black line shows a first-order exponential fit to the data and the residuals from these fits are shown in the corresponding colors above. The inset shows a replot of kobs vs. varying concentrations of competitor DNA.
-
Figure 3—source data 1
Representative measurement of PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
The data shown in the inset are also included as labeled.
- https://doi.org/10.7554/eLife.37818.025
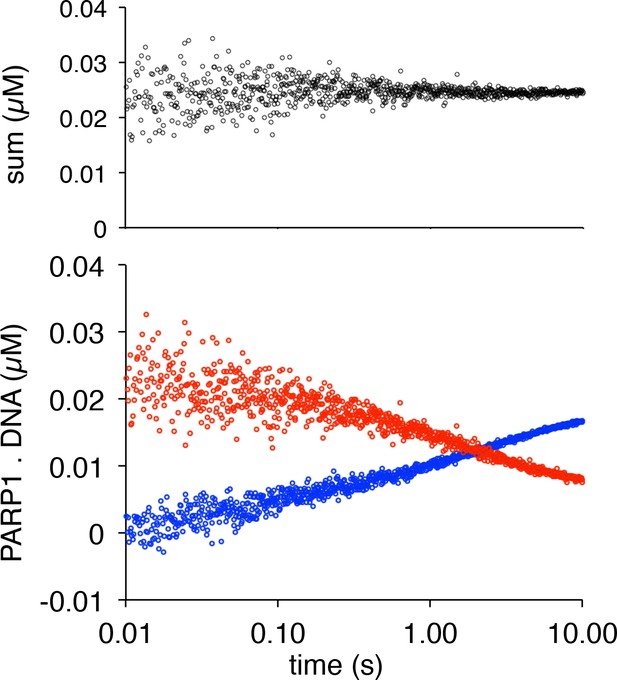
Label-swap experiment demonstrating that monitoring p18mer* and p18mer release are kinetically identical.
The pseudo-residuals generated by addition of the two signals are centered at the total concentration of the probe DNA (25 nM).
-
Figure 3—figure supplement 1—source data 1
Label-swap experiment demonstrating that monitoring p18mer* and p18mer release are kinetically identical.
The pseudo-residuals generated by addition of the two signals are centered at the total concentration of the probe DNA (25 nM).
- https://doi.org/10.7554/eLife.37818.024

Kinetic model for the dissociation of PARP1 from labeled DNA that depends on formation of a ternary complex with the unlabeled DNA.
https://doi.org/10.7554/eLife.37818.026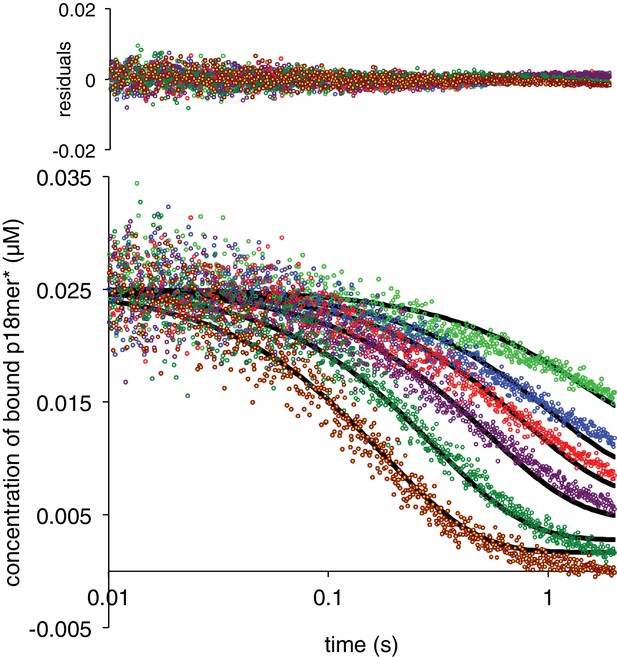
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Global fitting of six representative concentrations of competitor p18mer DNA using the mechanism in Scheme 3: 76 nM (light green), 149 nM (blue), 225 nM (red), 398 nM (violet), 1 µM (dark green), and 4 µM (brown). Residuals for the seven concentrations are shown overlaid in the corresponding colors above.
-
Figure 4—source data 1
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Global fitting of six representative concentrations of competitor p18mer DNA using the mechanism in Scheme 3: 76 nM (light green), 149 nM (blue), 225 nM (red), 398 nM (violet), 1 µM (dark green), and 4 µM (brown). Residuals for the seven concentrations are shown overlaid in the corresponding colors above.
- https://doi.org/10.7554/eLife.37818.038
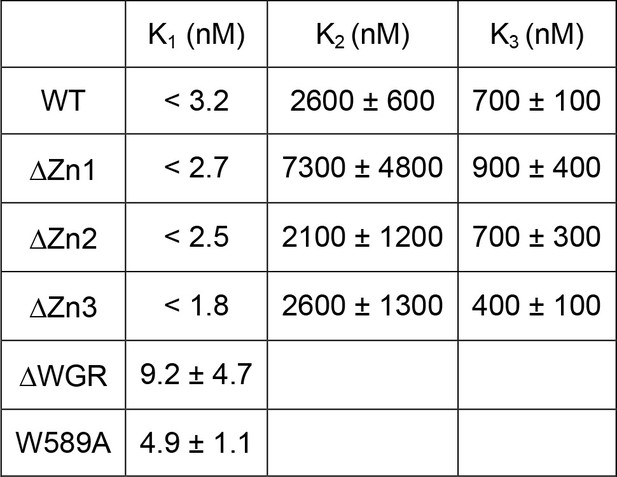
Thermodynamic parameters for DNA binding to PARP1.
All values (mean and standard deviation) were derived from the kinetic parameters in Table 1.
-
Figure 4—figure supplement 1—source data 1
Thermodynamic parameters for DNA binding to PARP1.
- https://doi.org/10.7554/eLife.37818.029
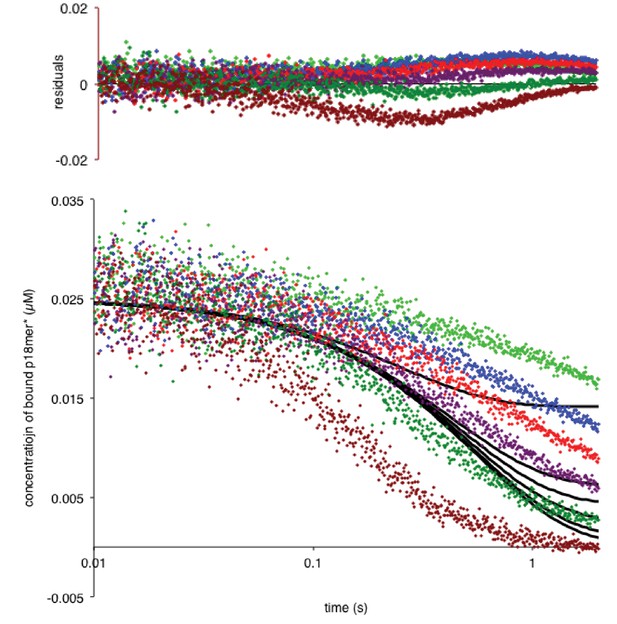
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Global fitting of six representative concentrations of competitor DNA using the mechanism in Scheme 2: 76 nM (light green), 149 nM (blue), 225 nM (red), 398 nM (violet), 1 µM (dark green), and 4 µM (brown). Residuals for the seven concentrations are shown overlaid in the corresponding colors above.
-
Figure 4—figure supplement 2—source data 1
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Poor global fitting of six representative concentrations of competitor DNA using the mechanism in Scheme 2.
- https://doi.org/10.7554/eLife.37818.031
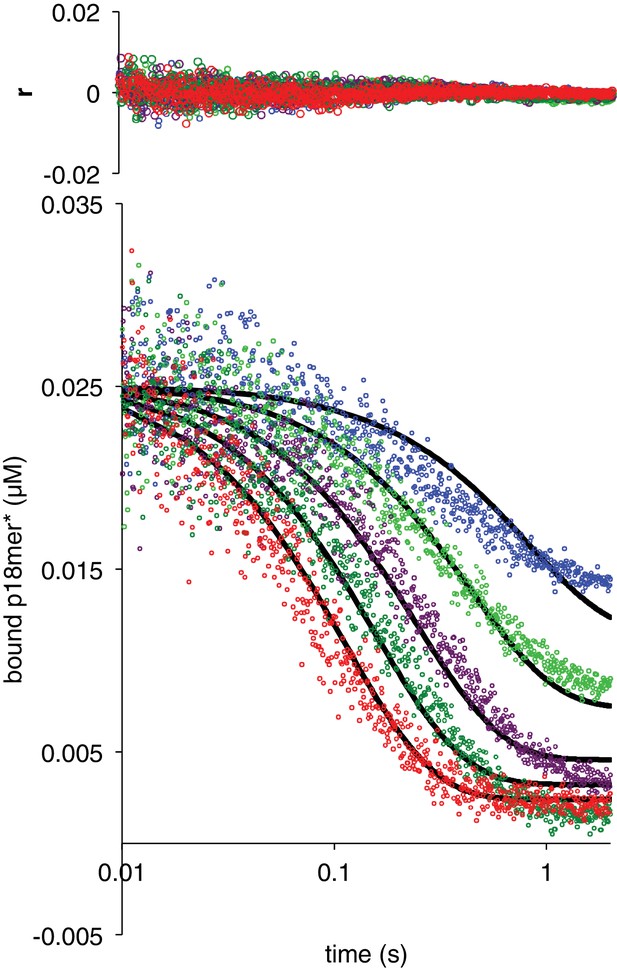
Dissociation of ΔZn1 from p18mer*.
Global fits of five representative concentrations of competitor DNA are shown: 0.15 µM (blue), 0.4 µM (light green), 1 µM (violet), 2 µM (dark green), and 4 µM (red) to the kinetic mechanism in Scheme 3. Residuals for the five concentrations are shown overlaid in the corresponding colors above each plot.
-
Figure 4—figure supplement 3—source data 1
Dissociation of ΔZn1 from p18mer*.
- https://doi.org/10.7554/eLife.37818.033
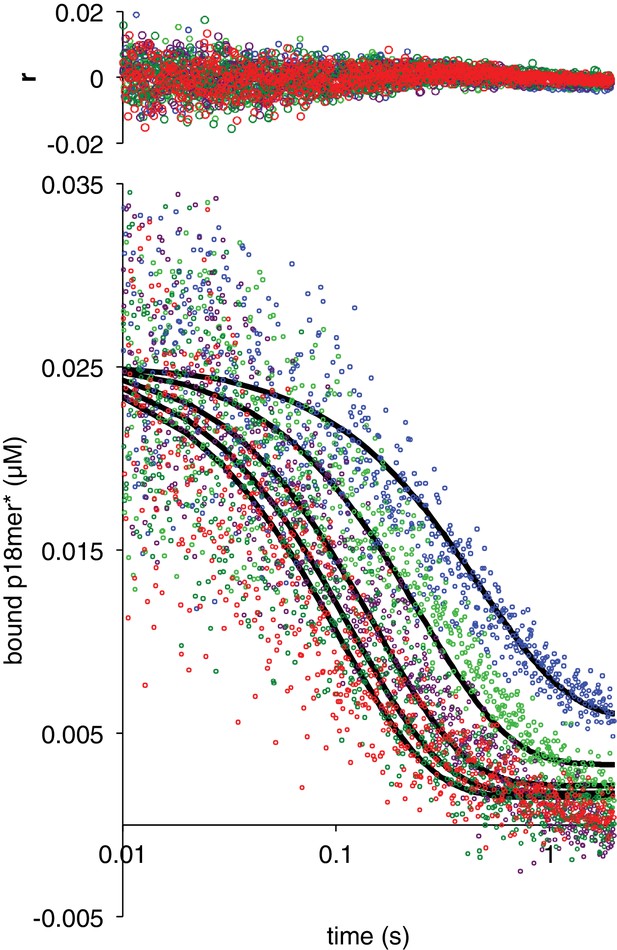
Dissociation of ΔZn2 from p18mer*.
Global fits of five representative concentrations of competitor DNA are shown: 0.15 µM (blue), 0.4 µM (light green), 1 µM (violet), 2 µM (dark green), and 4 µM (red) to the kinetic mechanism in Scheme 3. Residuals for the five concentrations are shown overlaid in the corresponding colors above each plot.
-
Figure 4—figure supplement 4—source data 1
Dissociation of ΔZn2 from p18mer*.
- https://doi.org/10.7554/eLife.37818.035
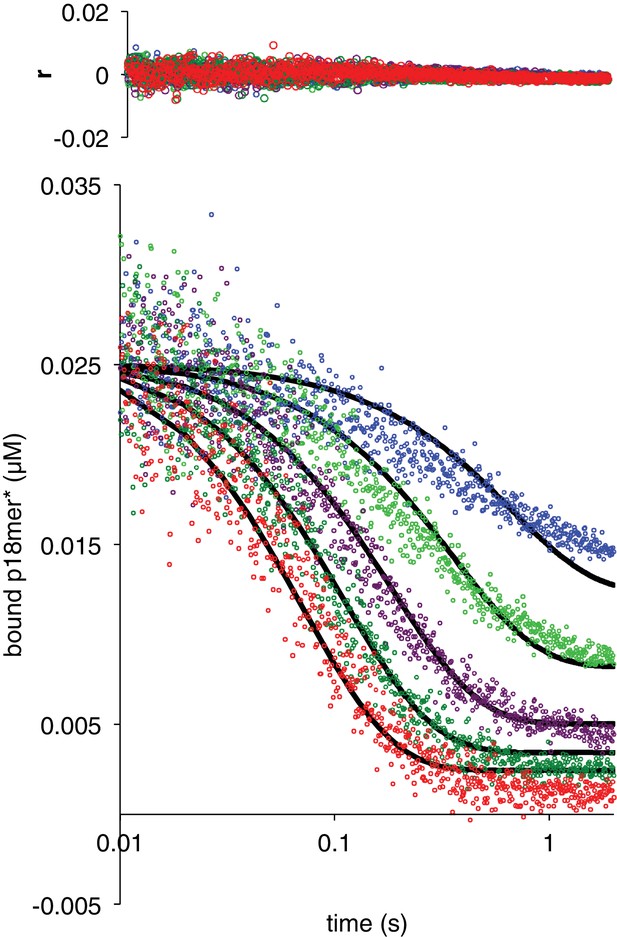
Dissociation of ΔZn3 from p18mer*.
Global fits of five representative concentrations of competitor DNA are shown: 0.15 µM (blue), 0.4 µM (light green), 1 µM (violet), 2 µM (dark green), and 4 µM (red) to the kinetic mechanism in Scheme 3. Residuals for the five concentrations are shown overlaid in the corresponding colors above each plot.
-
Figure 4—figure supplement 5—source data 1
Dissociation of ΔZn3 from p18mer*.
- https://doi.org/10.7554/eLife.37818.037
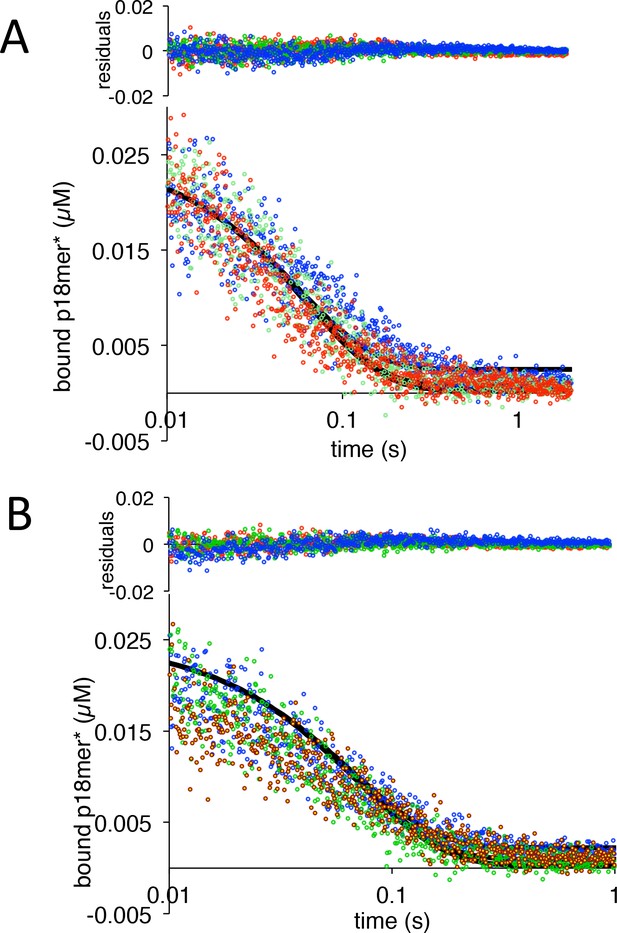
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
Global fitting of three representative concentrations of competitor p18mer DNA using the mechanism in Scheme 2: 0.4 µM in blue, 1.3 µM in green, and 4 µM in red for (A) ΔWGR and (B) W589A. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 5—source data 1
PARP1 dissociation from DNA as monitored by fluorescence anisotropy for ΔWGR from panel A.
- https://doi.org/10.7554/eLife.37818.040
-
Figure 5—source data 2
PARP1 dissociation from DNA as monitored by fluorescence anisotropy for W589A from panel B.
- https://doi.org/10.7554/eLife.37818.041
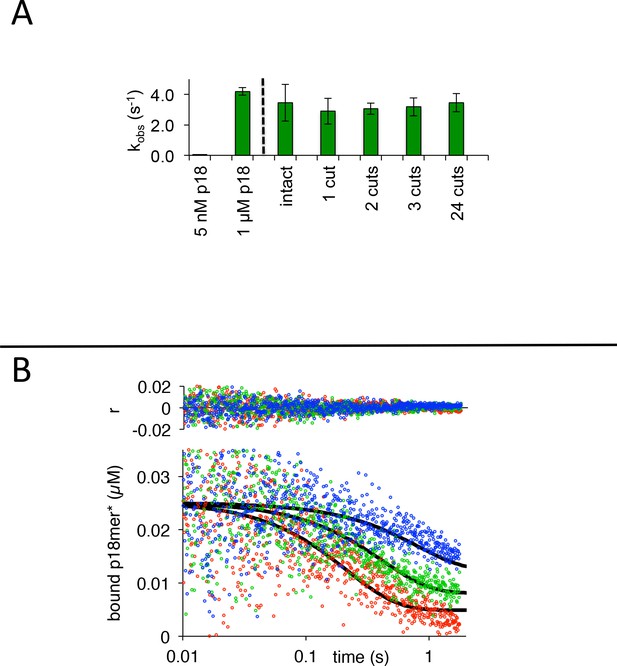
PARP1 dissociation from DNA as monitored by fluorescence anisotropy.
(A) The observed rate of dissociation triggered by 5 nM of intact or variably cut plasmid is compared to 5 nM and 1 µM p18mer. (B) Global fitting of three representative concentrations of competitor intact plasmid DNA using the mechanism in Scheme 3: 0.7 nM in blue, 2.0 nM in green, and 5.8 µM in red. Residuals for the three concentrations are shown overlaid in the corresponding colors above.
-
Figure 6—source data 1
The observed rate of dissociation triggered by 5 nM of intact or variably cut plasmid is compared to 5 nM and 1 µM p18mer.
- https://doi.org/10.7554/eLife.37818.044
-
Figure 6—source data 2
Global fitting of three representative concentrations of competitor intact plasmid DNA using the mechanism in Scheme 3.
- https://doi.org/10.7554/eLife.37818.045
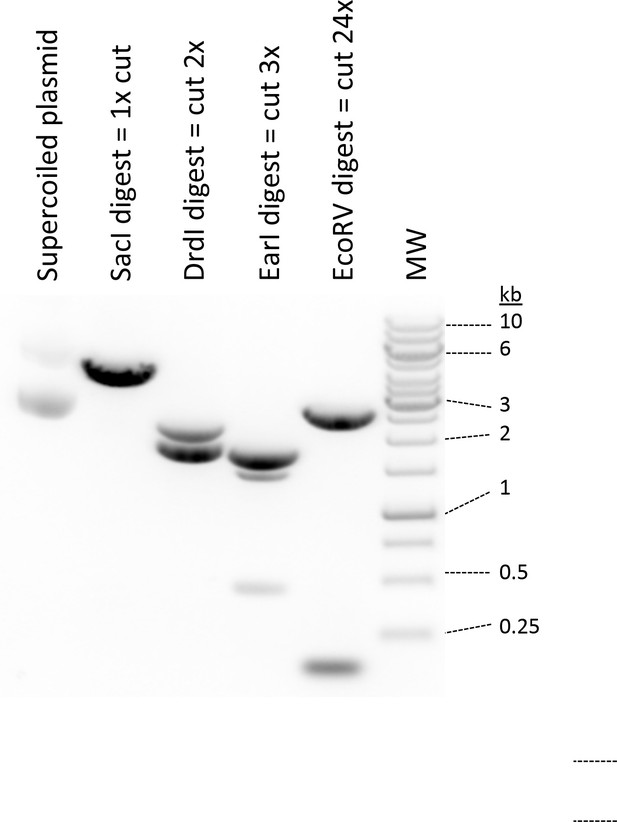
DNA 1% agarose gel demonstrating the various forms of plasmid used to demonstrate that undamaged DNA can effectively promote the monkey-bar mechanism for PARP1 to move from one segment of DNA to another.
Note the characteristic faster migration of supercoiled DNA compared to singly-cut plasmid. Also note that digest with EcoRV yields the parent plasmid and 12 copies of the 147 bp inserts.
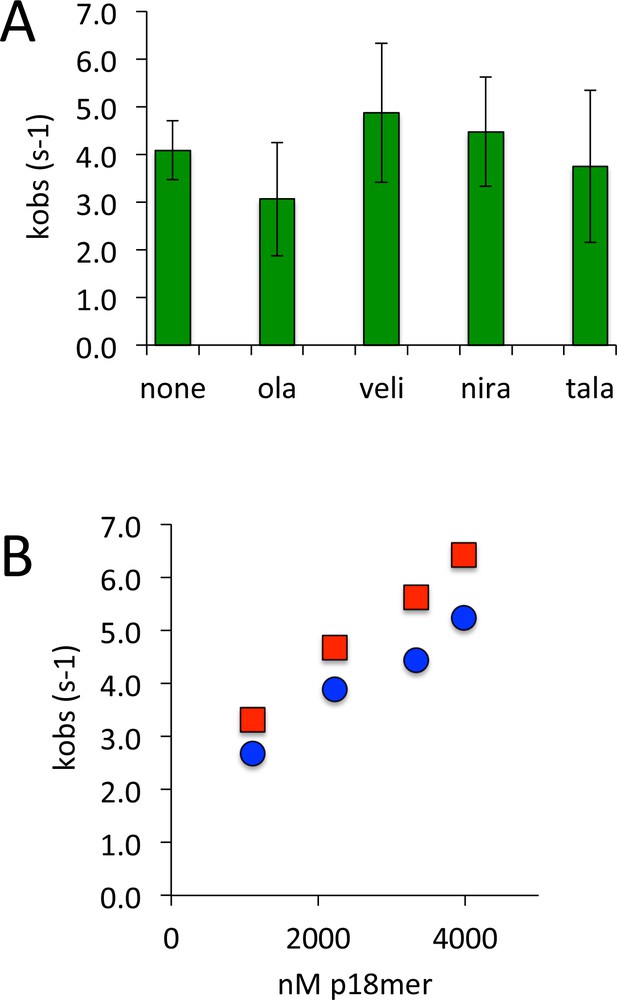
PARP1 dissociation from DNA as monitored by fluorescence anisotropy in the presence of various inhibitors of PARP1.
(A) Apparent kobs at 1 µM competitor DNA using four different inhibitors (50 nM) (ola = olaparib, veli = veliparib, nira = niraparib, tala = talazoparib). (B) Apparent kobs at 1–4000 nM competitor DNA for PARP1 alone (red) and PARP1 the presence of 50 nM talazoparib (blue).
-
Figure 7—source data 1
PARP1 dissociation from DNA as monitored by fluorescence anisotropy in the presence of various inhibitors of PARP1.
- https://doi.org/10.7554/eLife.37818.052
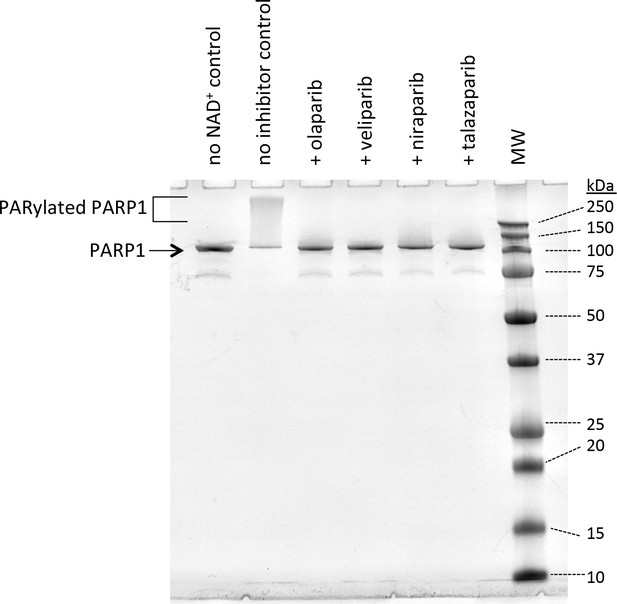
Smear assay of PARP1 demonstrating the effectiveness of four different inhibitors in blocking the activity of PARP1.
Each inhibitor was added at 1 µM final concentration in the presence of 0.5 µM PARP1, 1 µM p18mer, and 500 µM NAD+, and the reactions were allowed to proceed for 5 min at room temperature.
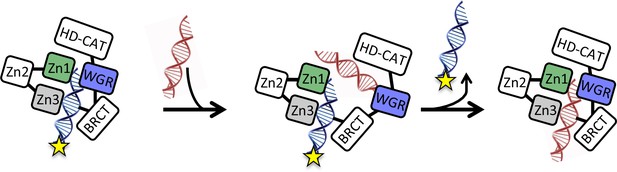
Model for the monkey bar mechanism for PARP1 depicting the proposed role of the WGR domain in capturing the second DNA strand prior to release of the originally bound DNA strand and subsequent re-arrangement around the second DNA.
https://doi.org/10.7554/eLife.37818.053Tables
Kinetic parameters for DNA association and dissociation
All values (mean and standard deviation) were derived from global fitting in Kintek Explorer of data from at least three independent experiments performed on different days and using at least two independent preparations of protein. All k1 rate constants were derived using the kinetic model in Scheme 1. All other rate constants came from using the kinetic model in Scheme 3, except for the k-1 values for ΔWGR and W589A, which were derived using Scheme 2. *The value of k2 assumes the full range of 5–450 possible binding sites per 4.5 kb plasmid (see Results).
| k1 (nM−1s−1) | k-1 (s−1) | k2 (nM−1s−1) | k-2 (s−1) | k3 (s−1) | k-3 (nM−1s−1) | |
|---|---|---|---|---|---|---|
| WT | 3.1 ± 0.2 | <10 | 0.043 ± 0.019 | 102 ± 22 | 9.7 ± 0.8 | 0.013 ± 0.002 |
| ΔZn1 | 3.7 ± 0.4 | <10 | 0.018 ± 0.012 | 97 ± 19 | 21 ± 5 | 0.027 ± 0.009 |
| ΔZn2 | 4.0 ± 1.3 | <10 | 0.073 ± 0.015 | 145 ± 66 | 15 ± 4 | 0.023 ± 0.007 |
| ΔZn3 | 5.5 ± 0.3 | <10 | 0.034 ± 0.013 | 76 ± 66 | 19 ± 7 | 0.050 ± 0.016 |
| ΔWGR | 2.4 ± 1.1 | 18.7 ± 2.7 | ||||
| W589A | 4.2 ± 0.5 | 20.2 ± 2.4 | ||||
| WT (with intact plasmid) | 0.041–3.7 | 161 ± 8 | 9 ± 3 | 0.032 ± 0.006 |
-
Table 1–source data 1
Kinetic parameters for DNA association and dissociation
- https://doi.org/10.7554/eLife.37818.020
Fluorescence accumulation after DNA damage.
The differences in values of Deff and F (mean and standard error of the mean) were determined to be statistically significant using an unpaired t-test with p=0.0094 and p=0.016, respectively. The raw data for all the nuclei are provided as source data.
| Wild-type | W589A | |
|---|---|---|
| Deff (µm2/s) | 3.7 ± 0.6 | 2.1 ± 0.2 |
| F | 44 ± 3 | 33 ± 3 |
| n | 28 | 30 |
-
Table 2–source data 1
Fluorescence accumulation after DNA damage; values from the table.
- https://doi.org/10.7554/eLife.37818.047
-
Table 2–source data 2
Normalized values for amount of fluorescence detected after DNA damage for each of 28 nuclei of WT PARP1.
- https://doi.org/10.7554/eLife.37818.048
-
Table 2–source data 3
Normalized values for amount of fluorescence detected after DNA damage for each of 30 nuclei of W589A mutant.
- https://doi.org/10.7554/eLife.37818.049
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Mus musculus) | Mouse embryo fibroblasts (MEF) | Dantzer Lab, University of Strasbourg, France | cells routinely tested negative for Mycoplasma contamination by Cell Culture Facilities | |
| Transfected construct (Homo sapiens) | pEGFP-C3-PARP1 | Dantzer Lab, University of Strasbourg, France | ||
| Peptide, recombinant protein | PARP1 | Pascal Lab, University of Montreal | ||
| Chemical compound, drug | NAD+ | Sigma | N1511 | 34 mM stock solution; pH'd to ~ 7 |
| Chemical compound, drug | olaparib | Selleck | S1060 | 10 mM stock solution in DMSO |
| Chemical compound, drug | veliparib | Selleck | S1004 | 10 mM stock solution in DMSO |
| Chemical compound, drug | niraparib | Selleck | S2741 | 10 mM stock solution in DMSO |
| Chemical compound, drug | talazoparib | Selleck | S7048 | 10 mM stock solution in DMSO |
| Chemical compound, drug | JetPei | Polyplus Transfection | 101–40N | |
| Chemical compound, drug | Hoechst 33342 | InVitrogen | H1399 | |
| Software, algorithm | MatLab code | doi.org/10.1101/373043 | DNA Repair Analysis Toolbox.mltbx | for conversion of microscope data to text files |
| Software, algorithm | MatLab code | doi.org/10.1101/373043 | Bioformats Toolbox (v1.0.4b).mltbx | need as an accessory for the DNA Repair Analysis Toolbox |
| Software, algorithm | Mathematica code | doi.org/10.1101/373043 | for analysis of microscope text files by free diffusion model | |
| Software, algorithm | Kintek Explorer | http://kintek-corp.com/ | for analysis of kinetic data |
Additional files
-
Source code 1
DNA repair analysis toolbox
- https://doi.org/10.7554/eLife.37818.054
-
Source code 2
Bioformats image toolbox
- https://doi.org/10.7554/eLife.37818.055
-
Source code 3
Q-FADD 0.13.nb
- https://doi.org/10.7554/eLife.37818.056
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37818.057





