Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium
Figures
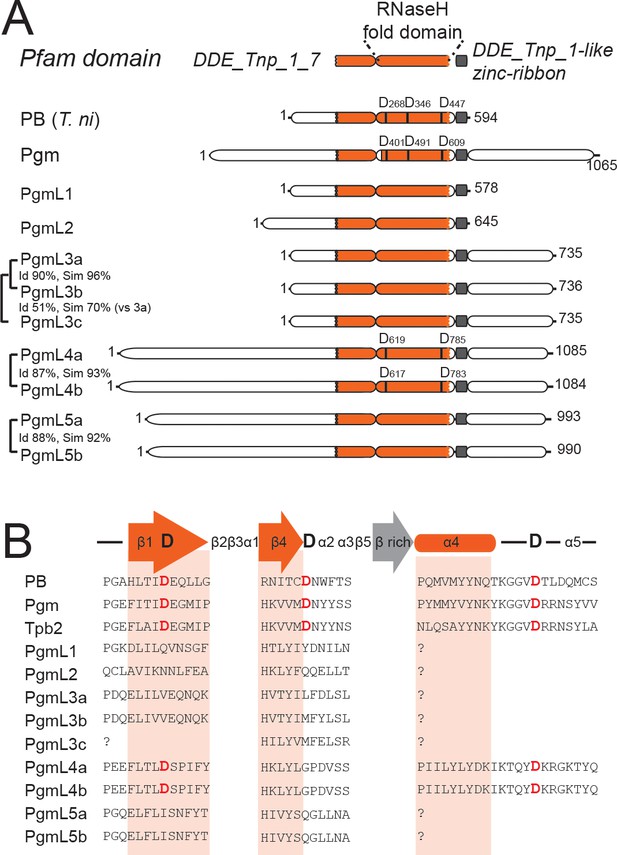
Novel domesticated PiggyBac transposases in Paramecium.
(A) Domain organization of the PiggyBac transposase (PB) from T. ni and of Paramecium PiggyBac-related proteins (Pgm and PgmLs). The Pfam domain DDE_Tnp_1_7 is shown as a bipartite orange domain, with the RNase H fold corresponding to its right part (conserved catalytic D residues are indicated by vertical bars). The DDE_Tnp_1-like zinc ribbon is in grey. Id: % of amino acid identity; sim: % of similarity. (B) Protein sequence alignment of the residues surrounding the three catalytic aspartic acids (DDD). Following secondary structure prediction, sequence alignments were adjusted manually, using the expected position of the three catalytic D residues in the first and fourth β strands and immediately downstream of the fourth α helix of the RNase H fold domain, respectively (Hickman et al., 2010). ‘?' indicates that the expected α4 helix could not be predicted using the PSIPRED secondary structure prediction software.

MUSCLE alignment of the cysteine-rich domains of ciliate domesticated PB transposases and other PB transposases.
The analysis involved 62 amino acid sequences of PB transposases and domesticated transposases from ciliates and other species. Amino acid sequences encompassing the cysteine-rich domain of each protein were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). All sequences used for the alignment are displayed in Supplementary file 3. Complete accession numbers can be found in Figure 1—figure supplement 2.
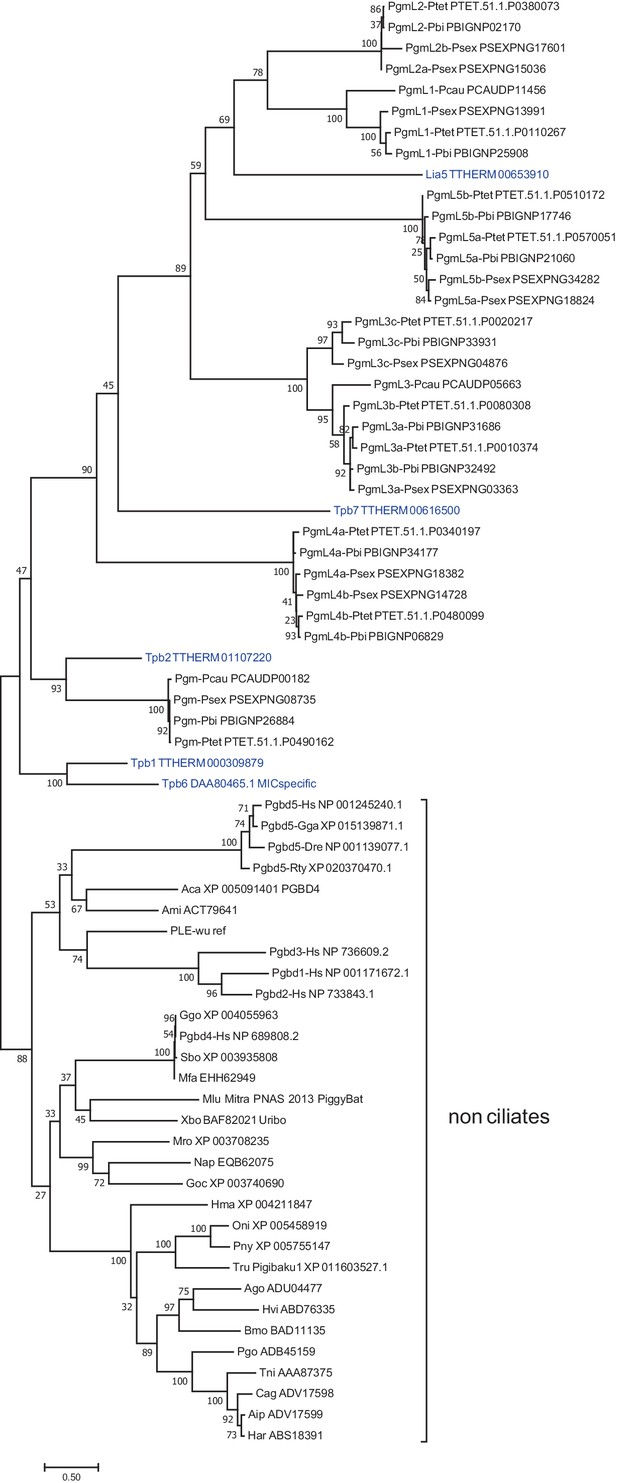
Maximum Likelihood tree of ciliate domesticated PB transposases and other PB transposases.
The tree includes 69 amino acid sequences of PB transposases and domesticated PB transposases from ciliates and other species. To construct the tree, the alignment of all transposase core domains (Supplementary file 1) was edited to remove specific insertions restricted to one particular PgmL family. All accession numbers are indicated, except for the PiggyBat transposase from Myotis lucifugus (Mitra et al., 2013). PB and domesticated PB proteins from Tetrahymena thermophila are in blue. The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al., 1992). The tree with the highest log likelihood (−35952.17) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (bootstrap = 100). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories (+G, parameter = 2.8098)). There were a total of 541 positions in the final dataset. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. Evolutionary analyses were conducted in MEGA7 (Kumar et al., 2016).
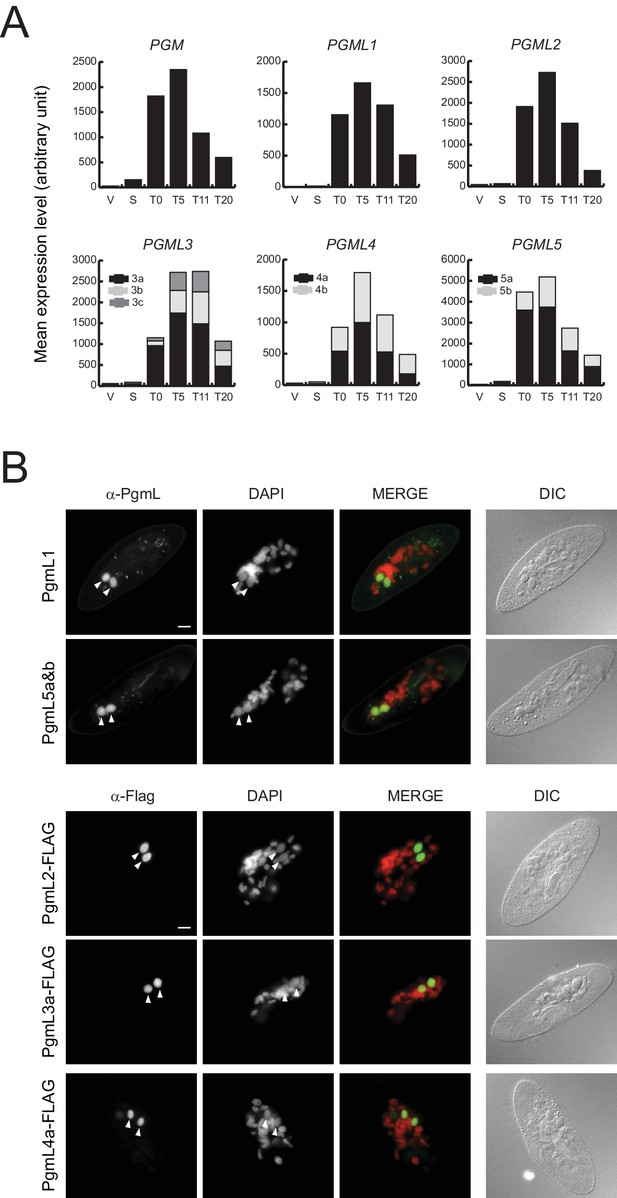
Expression and nuclear localization of PgmLs during autogamy.
(A) Normalized RNA-seq data were extracted from (Arnaiz et al., 2017) and used to calculate mean expression levels for each time-point. V: vegetative cells (V1.2); S: starved cells with meiotic micronuclei (S1.1 and S1.2); T0: T0.1 and T0.2; T5: T5.1 and T5.2; T11: T11.1 and T11.2; T20: T20.1 and T20.2. All time-points are in hours. (B) Immunofluorescence staining of PgmL proteins in autogamous cells. White arrowheads point to developing new MACs. Scale bar: 10 µm.
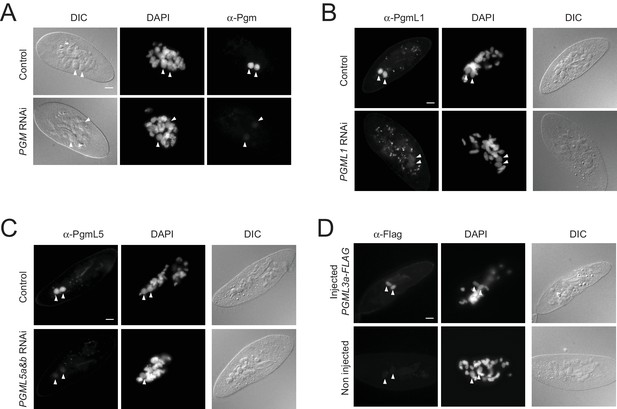
Validation of the specificity of antibodies directed against Pgm, PgmL1, PgmL5a, and the Flag peptide by immunofluorescence labelling of fixed cells.
(A) Immunostaining of Pgm in early autogamous cells subjected to control (ND7) or PGM RNAi. (B) Immunostaining of PgmL1 in early autogamous cells subjected to control (L4440) or PGML1 RNAi. (C) Immunostaining of PgmL5a and b in early autogamous cells subjected to control (L4440) or PGML5a and b RNAi. (D) Immunodetection of PgmL3a-FLAG in early autogamous cells injected with a PGML3a-FLAG fusion transgene (top panel). No FLAG signal was detected using the α-FLAG in non-injected cells (bottom panel). Developing MACs are indicated by white arrowheads. Scale bar is 10 μm.
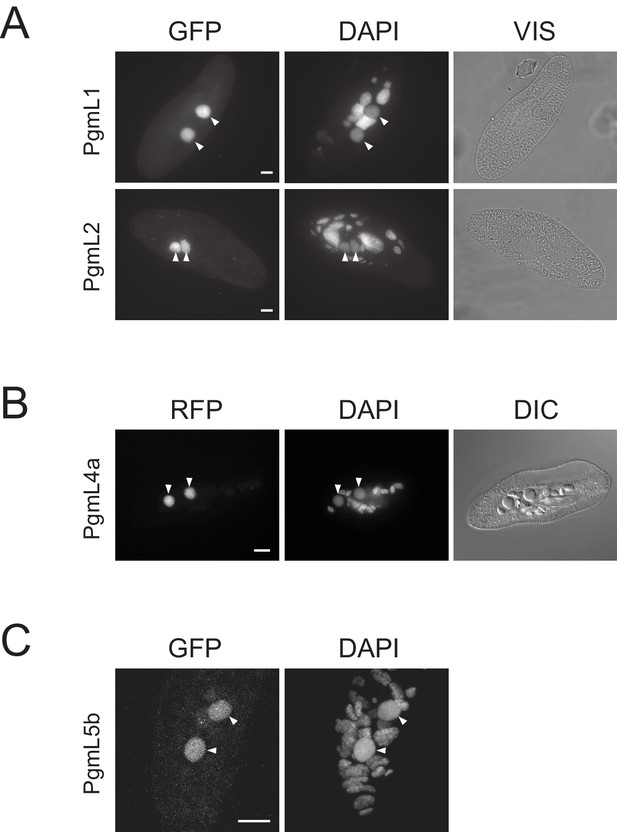
Localization of GFP and RFP fusions in developing new MACs.
Plasmids expressing GFP-PgmL1, GFP-PgmL2 and GFP-PgmL5b are derivatives of pUC19 and carry the EGFP coding sequence (Singh et al., 2014) fused to the 5’ end of each PGML coding sequence. They were linearized with ScaI (PGML1 and 2) or NdeI (PGML5b) prior to microinjection. Plasmid expressing PgmL4a-RFP is a derivative of pBL49g (Baudry et al., 2009), in which the PGM gene and its 5’ and 3’ UTRs were first replaced by the PGML4a gene and its upstream and downstream regulatory sequences: in this construct, a codon-optimized sequence encoding a TSSGGGSG linker followed by the mRFPmars protein was inserted between the last codon of PGML4a and the TGA stop codon. It was linearized with NgoMIV prior to microinjection. All plasmids carry the 5’ and 3’ transcription signals of the appropriate PGML gene (sequences available upon request). Following microinjection into the MAC of vegetative cells, transgenes are capped by addition of telomeric repeats at their ends (concatemers may form prior to telomere addition): they may then integrate into the somatic genome (through a mechanism that remains to be studied) or be maintained throughout vegetative growth as autonomously replicating mini-chromosomes (Gilley et al., 1988; Bourgain and Katinka, 1991; Katinka and Bourgain, 1992). Replication origins have not been characterized in Paramecium, but any injected DNA (including bacterial plasmids) may be replicated. Transgenes persist in old MAC fragments throughout autogamy and continue to be expressed if controlled by proper transcription signals, but are lost in the next sexual generation, after complete destruction of the old MACtransformed cells were grown for ~20 vegetative divisions and starved to induce autogamy. (A) Autogamous cells expressing GFP-PgmL1 or GFP-PgmL2. Cells were fixed for 10 min in 2% paraformaldehyde 1X PHEM, and stained with DAPI for 10 min before mounting them in Citifluor AF2 antifading solution (Biovalley). Epifluorescence microscopy imaging was performed using a Nikon Eclipse microscope, with a 63x oil objective. VIS: whole cells visualized under brightfield illumination. (B) Autogamous cell expressing PgmL4a-RFP. Cells were fixed using a previously published procedure (Marmignon et al., 2014) and epifluorescence microscopy imaging was performed as described (Dubois et al., 2017). DIC: whole cells visualized using differential interference contrast microscopy (Normarski). (C) Autogamous cell expressing GFP-PgmL5b. Cells were fixed and stained as mentioned in (Ignarski et al., 2014) for the immunostaining of histone modifications. Confocal imaging was carried out using an Olympus Fluoview Fv1000 confocal laser scanning microscope, with a 63X zoom four objective. Image analysis was performed using ImageJ 1.5 r. In all panels, the white bar represents 10 µm and arrowheads indicate the position of developing new MACs.
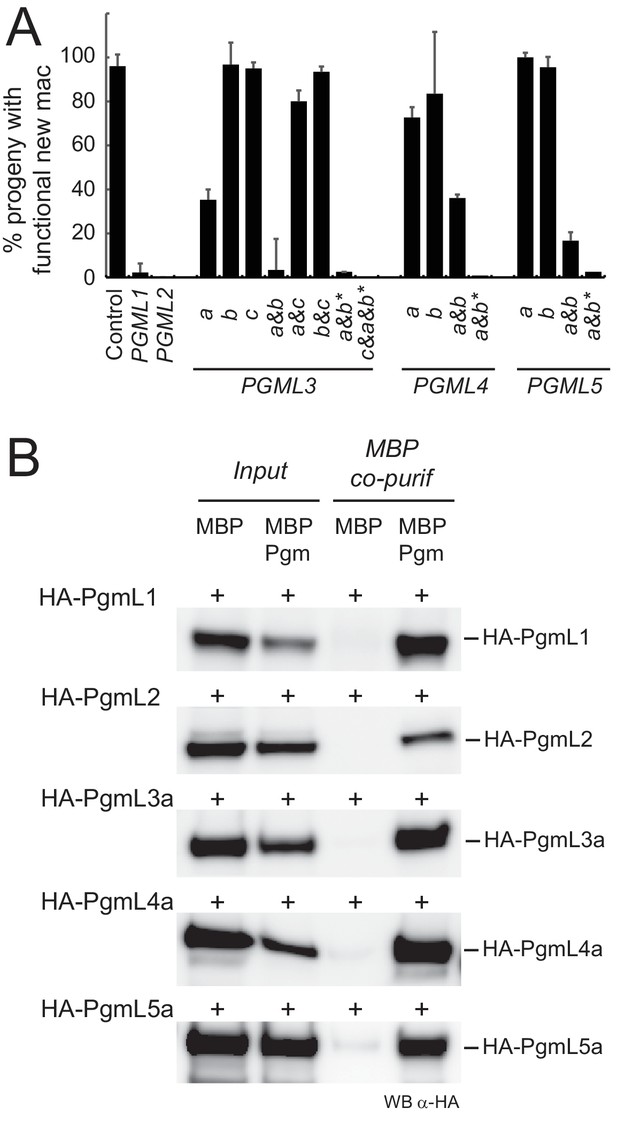
PgmLs are essential during autogamy and interact with Pgm in cell extracts.
(A) Effect of PGML KDs on the recovery of post-autogamous progeny with functional new MACs. For PGML1, PGML2 and PGML3c, only the results obtained using IF1 RNAi constructs (Figure 3—figure supplement 1) are shown. For groups of duplicated paralogs, individual gene KDs were performed using gene-specific IF2 constructs (Figure 3—figure supplement 1), while double KDs were performed using either IF2 or cross-hybridizing IF1 (*) constructs. Error bars represent standard deviations (n = 2 to 14, see Supplementary file 6) (B) Pull down of HA-PgmL fusions with MBP-Pgm using recombinant proteins expressed in insect cells. In each panel, the HA-tagged protein that was co-expressed with MBP or MBP-Pgm is indicated on the left and the band revealed on western blots (WB) using anti-HA antibodies is indicated on the right. The full-size blot with molecular weight marker is shown in Figure 3—figure supplement 2.
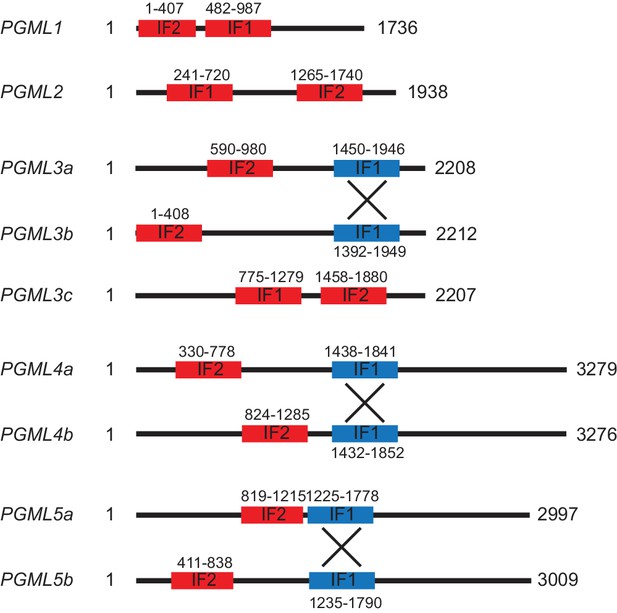
Map and coordinates of PGML feeding inserts.
Two feeding inserts (IF1 and IF2) were designed for each gene. Within multigenic PGML groups, gene-specific inserts are in red and inserts that are able to cross-silence other genes of the same family (* constructs in the main text) are in blue (the cross indicates that blue inserts can target the two paralogs simultaneously). The coordinates of each fragment refer to their 5’ and 3’ nucleotide positions, with +1 corresponding to the first base of the ATG start codon.
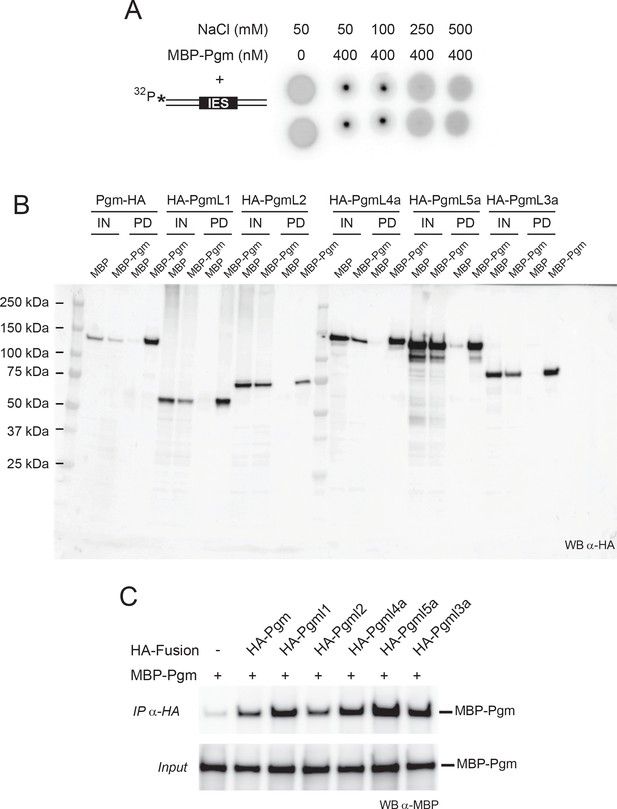
Co-precipitation of MBP-Pgm with HA-PgmL fusions.
(A) Control DRaCALA DNA binding assay (Differential Radial Capillary Action of Ligand Assay, see [Donaldson et al., 2012]). MBP-Pgm was purified from insect cells. Purified MBP-Pgm (400 nM final concentration) was mixed with a 32P-radiolabeled 80 bp double-strand DNA fragment carrying IES 51A1835 from the surface antigen A gene (25 nM final concentration) in 25 mM HEPES pH7.5, 0.1 mg/ml BSA, 0.5 mM DTT and 50 mM NaCl-containing buffer. NaCl was then added to reach 50, 100, 250 or 500 mM final concentration and complexes were loaded in duplicate onto a Nitrocellulose Hybond ECL membrane. DNA binding is detected at 50 and 100 mM NaCl, while complexes are destabilized at higher salt concentrations (250 mM NaCl and above). (B) Pull down of each HA-PgmL fusion with MBP-Pgm. The full-size western blot is identical to the one used for Figure 3. Recombinant MBP-Pgm was co-expressed in insect cells with each indicated HA-tagged protein, cell extracts were prepared as indicated in Materials and methods and MBP-Pgm was pulled down using amylose beads. HA-tagged proteins were revealed using monoclonal α-HA antibodies (HA-7 from Sigma Aldrich). Expected sizes: 128 kDa (Pgm-HA), 69 kDa (HA-PgmL1), 76 kDa (HA-PgmL2), 88 kDa (HA-PgmL3a), 127 kDa (HA-PgmL4a) and 116 k Da (HA-PgmL5a). ‘Precision Plus Protein Standards’ from Bio-Rad were used as molecular weight markers. (C) Co-immunoprecipitation of MBP-Pgm with each HA-PgmL fusion. For each lane, 1.5 μg of monoclonal α-HA antibodies were incubated overnight at 4°C on a rotating wheel with 10 μL of protein A sepharose beads (GE Healthcare). The coated beads were incubated with the same cell extracts as in (B) for 2 hr at 4°C, then washed 3 times with 1 mL of lysis buffer A and re-suspended in Laemmli buffer (Laemmli, 1970) before electrophoresis in SDS-polyacrylamide gels. MBP-Pgm was detected using HRP-coupled α-MBP antibodies according to the manufacturer’s instructions (New England Biolabs).
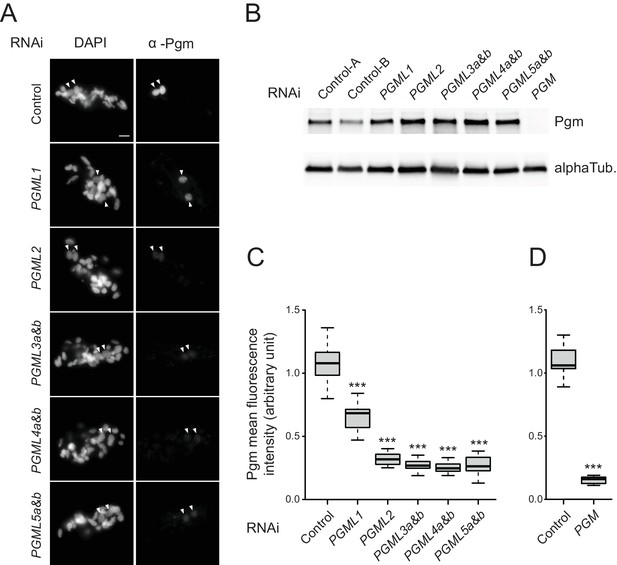
Expression and localization of Pgm in PGML KDs.
(A) Immunostaining of Pgm in early autogamous cells subjected to control (L4440) or PGML RNAi. Developing MACs are indicated by white arrowheads. Scale bar is 10 μm. (B) Western blot analysis of Pgm expression in early autogamous cells subjected to control (L4440: two independent controls A and B are shown), PGML or PGM RNAi. (C) Boxplot representation of the distribution of Pgm fluorescence intensities quantified in 30–55 μm2 developing MACs subjected to the different RNAi shown in (A). This size window corresponds to the maximal Pgm signal in the control (Figure 4—figure supplement 1) and was chosen to quantify nuclear Pgm immunofluorescence for all KDs, since no significant size difference was noticed for developing MACs relative to the control. For each condition, 19 to 35 developing MACs were analyzed. (D) Independent set of experiments showing the quantification of Pgm fluorescence intensity in 30–55 μm2 developing MACs following control (ND7) or PGM RNAi. 11 and 12 MACs were analyzed, respectively. In (C) and (D): *** for p<0.001 in a Mann-Whitney-Wilcoxon statistical test (see Materials and methods for details).
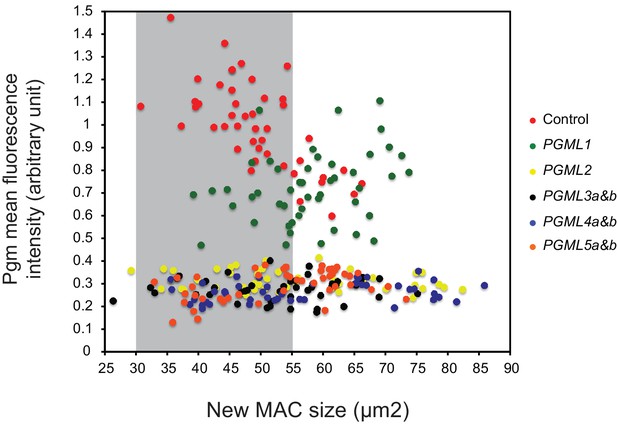
Plot of Pgm mean immunofluorescence intensity vs developing MAC size in cells subjected to control or PGML RNAi.
For each RNAi condition, 44 to 48 developing MACs were analyzed on slides carrying whole cells immunostained with α-Pgm antibodies (Figure 4). The Pgm mean fluorescence intensity was plotted against the size of the developing MACs. As previously reported, the intensity of the Pgm signal varies during MAC development (Dubois et al., 2017). In control experiments (here using L4440), maximal level of Pgm expression was observed for MAC sizes ranging between 30 and 55 μm2: this size window is highlighted in grey.
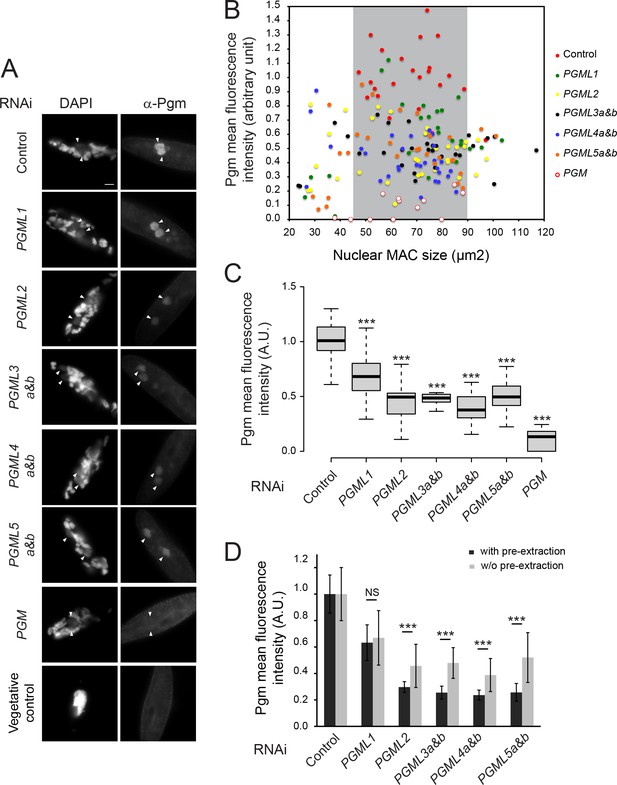
Immunolocalization of Pgm without Triton extraction in PGML knockdowns.
(A) Immunostaining of Pgm in cells subjected to control (L4440), PGML or PGM RNAi. Cells were fixed for 10 min in PHEM +2% formaldehyde, permeabilized for 15 min in PHEM +1% Triton before TBST +3% BSA washes. The following steps of the immunostaining were performed as described in the supplementary Materials and methods. Developing MACs are indicated by white arrowheads. The last panel (Vegetative control) shows the background immunostaining of a vegetative cell in the control RNAi culture. Scale bar is 10 μm. (B) Plot of Pgm mean fluorescence intensity vs size in developing MACs. Since background immunostaining is relatively high under these experimental conditions, the mean Pgm fluorescence intensity was calculated by measuring the mean fluorescence intensity in developing MACs minus the mean fluorescence intensity for vegetative cells on the slide. Under these experimental conditions, the size of the developing MACs is larger than in standard immunostaining conditions including the pre-extraction step (Figure 4—figure supplement 1), and the maximum level of Pgm in control cells is observed for MAC sizes falling within the 45–90 μm2 range (highlighted in grey). 12 to 33 developing MACs were quantified for each condition. (C) Boxplot representation of Pgm mean fluorescence intensity (in arbitrary units: A.U.) for developing MACs ranging between 45–90 μm2 in size. 10 to 25 MACs were quantified for each RNAi. (D) Mean Pgm fluorescence intensity in PGML KDs after immunostaining with or without the pre-extraction step. Data plotted in (C) were compared to those plotted in Figure 4C, after normalization by the mean control value obtained in each experimental condition (with or without pre-extraction). Error bars represent the standard deviation for each dataset. *** for p<0.001 (Mann-Whitney-Wilcoxon test).
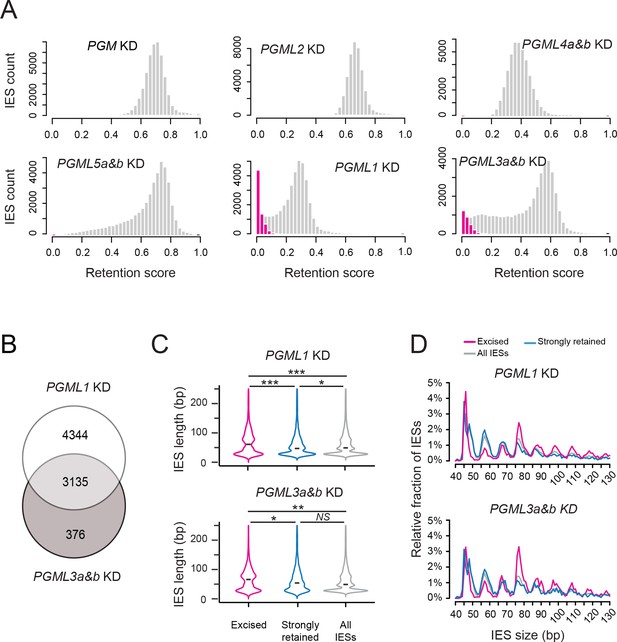
Analysis of IES retention in PGML KDs.
(A) Distribution of IES retention scores (IRS) in PGML KDs. Grey bars represent the distribution of all IESs over IRS ranging from 0 to 1 (by bins of 0.025). The distribution obtained in a previously published PGM KD (Arnaiz et al., 2012) is shown as a control. Absolute values of IRS should not be compared from one KD to the other, due to variable contamination by old MAC fragments. For PGML1 and PGML3a and b KDs, the distribution of statistically non-significantly retained IESs (i.e. excised IESs) is superimposed in magenta. (B) Venn diagram representing the overlap between the sets of excised IESs in PGML1 or PGML3a and b KDs (C) Violin plots of IES length distributions for the population of non-significantly retained IESs (magenta; n = 7479 in PGML1 KD and n = 3511 in PGML3a and b KD), the same number of IESs with the highest retention scores (blue) and all IESs (grey). The black dash shows the median of each distribution. Plots were drawn using the ggplot2 R-package (Wickham, 2009). Size distributions were compared using a Mann-Whitney-Wilcoxon statistical test and p values are indicated for each comparison (***: p<2.2 10−16; **: 2.2 10−16<p<10−10; *: 10−10<p<5 10−2; NS: p>5.5 10−2) (D) Comparative analysis of the relative fraction of IESs in each size peak among the populations of excised (magenta) and strongly retained (blue) IESs in PGML1 and PGML3a and b KDs, and the whole IES population (grey). Only IESs between 40 and 130 bp are represented.
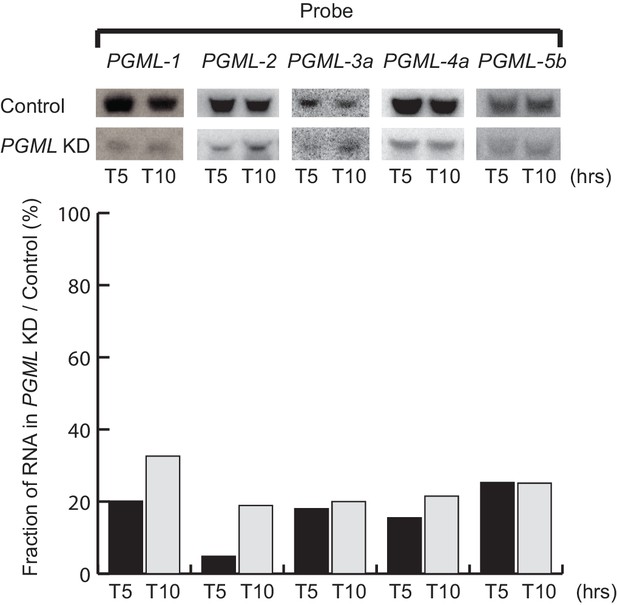
Northern blot analysis of PGML mRNA during autogamy in PGML knockdowns.
Top panels: Detection of PGML mRNAs by northern blot hybridization. RNAi against the ND7 gene was used as a control. PGML1 and PGML2 genes were knocked down individually using their respective IF1 RNAi-inducing plasmids (Figure 3—figure supplement 1). Each of PGML4 or PGML5 groups of paralogs was knocked down as a whole by feeding Paramecium cells with a 1:1 mixture of induced bacteria harboring IF1 plasmids targeting RNAi against both genes a and b. For PGML3 genes, the most highly expressed a and b genes were knocked down through the same double feeding procedure using cross-reacting IF1 plasmids. PGML hybridization probes (indicated on top) were the gene-specific IF2 inserts (Figure 3—figure supplement 1). Time-points are in hours after T0, the time at which 50% of cells had a fragmented MAC. Bottom panel: Quantification of mRNA levels in PGML KDs, as the percentage of steady state mRNA amounts observed in the control RNAi. Acquisition of PGML hybridization signals was performed using a Typhoon phosphorimager and quantified using the ImageQuant TL software (GE Healthcare Life Sciences). For each lane, 17S rRNA was used as a loading control for normalization.
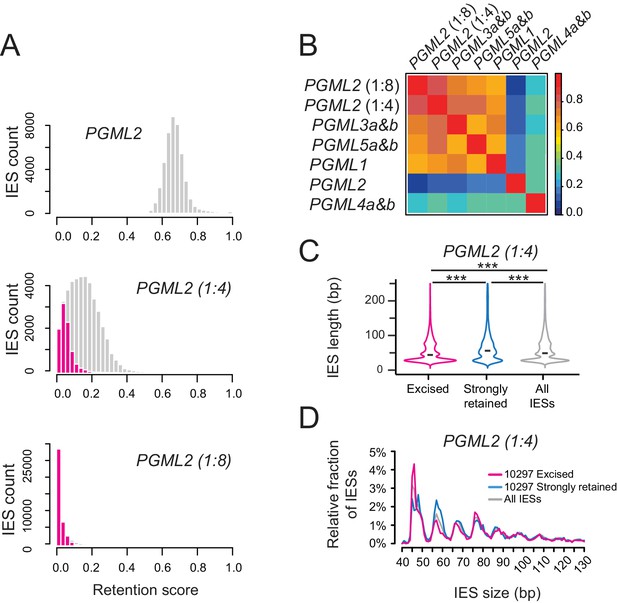
Analysis of IES retention scores in partial PGML2 KDs.
(A) Distribution of IES retention scores in complete and partial PGML2 KDs. Dilutions of PGML2 RNAi-inducing bacteria are indicated in each panel. For each condition, the distribution of statistically non-significantly retained IESs (i.e. efficiently excised IESs) is plotted in magenta.(B) Spearman’s rank correlation coefficients between IES retention scores in all PGML KDs, including PGML2 partial KDs. The graph was drawn using the ‘levelplot’ function from the ‘lattice’ package (Sarkar, 2008) (see Figure 5—figure supplement 3 for details). (C) Violin plots of the distributions of IES length for the population of 10297 non-significantly retained IESs in the partial PGML2 KD (1:4 dilution) (magenta), the same number of IESs with the highest retention scores (blue) and all IESs (grey). The black dash shows the median of each distribution. Violin plots were drawn using the ggplot2 R-package (Wickham, 2009). Size distributions were compared using a Mann-Whitney-Wilcoxon statistical test and p values are indicated for each comparison (***: p<2.2 10−16; **: 2.2 10−16<p<10−10; *: 10−10<p<5 10−2; NS: p>5.5 10−2). (D) Comparative analysis of the relative fraction of IESs in each size peak among the populations of efficiently excised (magenta) and strongly retained (blue) IESs in the partial PGML2 KD (1:4), and the whole IES population (grey). Only IESs between 40 and 130 bp are represented.
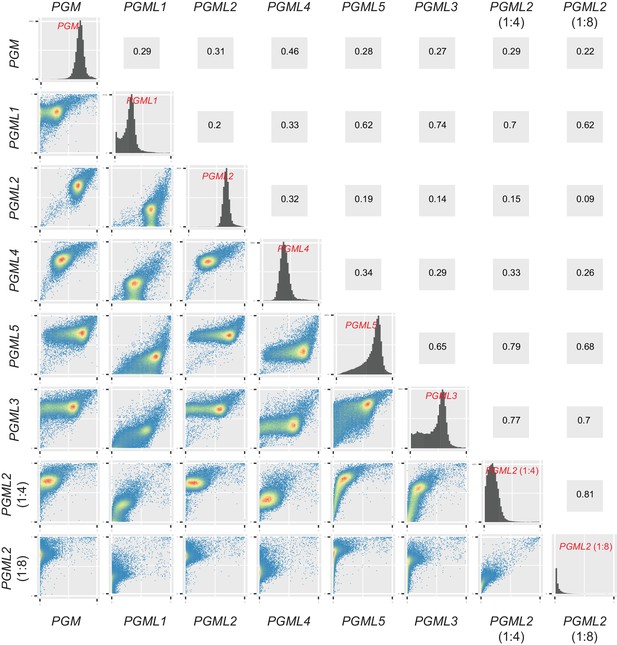
Correlation between IES retention scores in PGM, PGML and partial PGML2 knockdowns.
The distributions of IES boundary scores are displayed in the diagonal for each knockdown. For each pair of KDs, level plots of IES retention scores were drawn using the ‘ggplot2’ package (Wickham, 2009) and are shown below the diagonal. Spearman’s rank correlation coefficients are displayed at the symmetrical position above the diagonal.
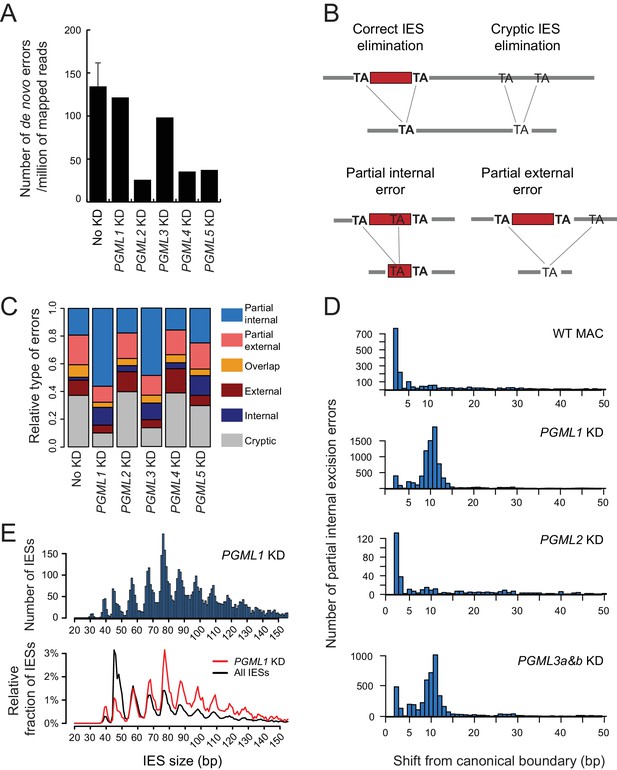
IES excision errors in PGML KDs.
(A) Number of IES excision errors in the MAC of vegetative cells before autogamy (No KD) and in the new MAC of autogamous cells upon each PGML KD (de novo errors). For the No KD sample, the error bar represents the standard deviation for five replicates (V samples in Supplementary file 9). (B) Major classes of IES excision errors found in the different samples. In external or internal errors, the two alternative TAs are misplaced (one on each flanking side or both inside of a reference IES, respectively). Overlapping errors use one TA inside and the other outside of reference IESs. (C) Distribution of the different classes of de novo excision errors in PGML KDs. As a control, the distribution of pre-existing errors found in the old MAC is shown for a vegetative culture (see Supplementary file 9). (D) Position of alternative excision boundaries used in partial internal excision errors, relative to the canonical boundary of the reference IES. WT: vegetative MAC; for PGML KDs, only de novo errors were considered. (E) Size distribution of IESs exhibiting partial internal errors in a PGML1 KD. Upper panel: size distribution of all IESs with partial internal errors. Lower panel: the black curve shows the fraction of IESs of each size relative to the total number of IESs in the genome; the red curve shows the fraction of IESs of each size among the population of IESs showing at least one partial internal error in a PGML1 KD. In both panels, only IESs with an alternative boundary at >2 bp from the canonical one were counted. In the bottom panel, IESs shorter than 35 bp were not considered (see Figure 6—figure supplement 4).
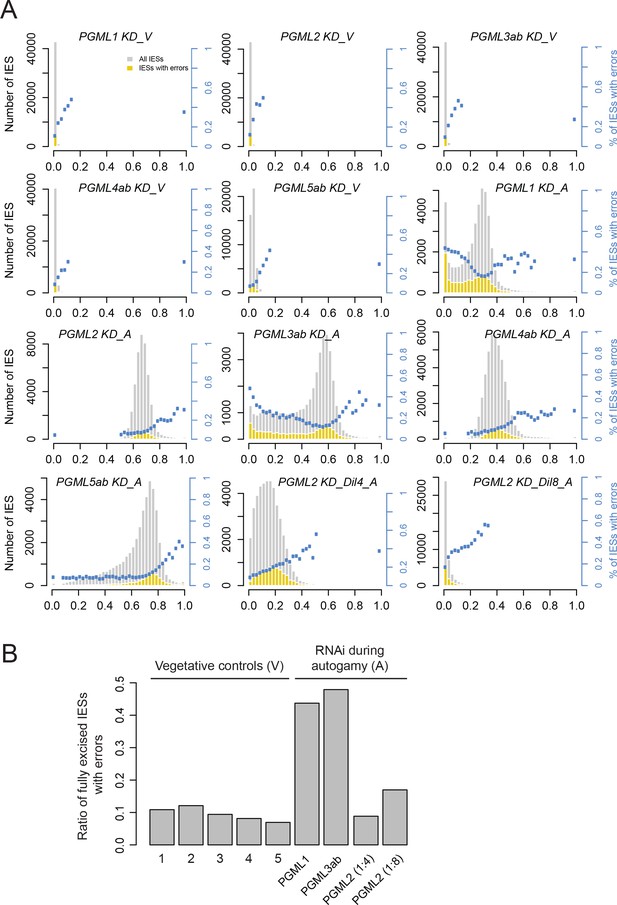
The number of excision errors increases for IESs with the lowest retention scores in PGML1 or PGML3a and b knockdowns.
(A) Fraction of IESs with errors as a function of boundary scores. For each dataset (see Supplementary file 9), the distribution of IES boundary scores is shown in grey (interval width: 0.025). For each interval, the number of IESs showing at least one excision error is superimposed in yellow and blue dots represent the ratio of IESs with at least one error (for intervals containing at least 50 IESs). All classes of errors were considered in this analysis. To allow comparison between PGML1, PGML3 and partial PGML2 KDs, raw error counts were used for all plots. (B) Plot of the fraction of IESs showing at least one excision error among fully excised IESs (boundary score <0.025). Vegetative controls are five independent cultures before autogamy in each RNAi-inducing medium (numbers refer to PGML groups).
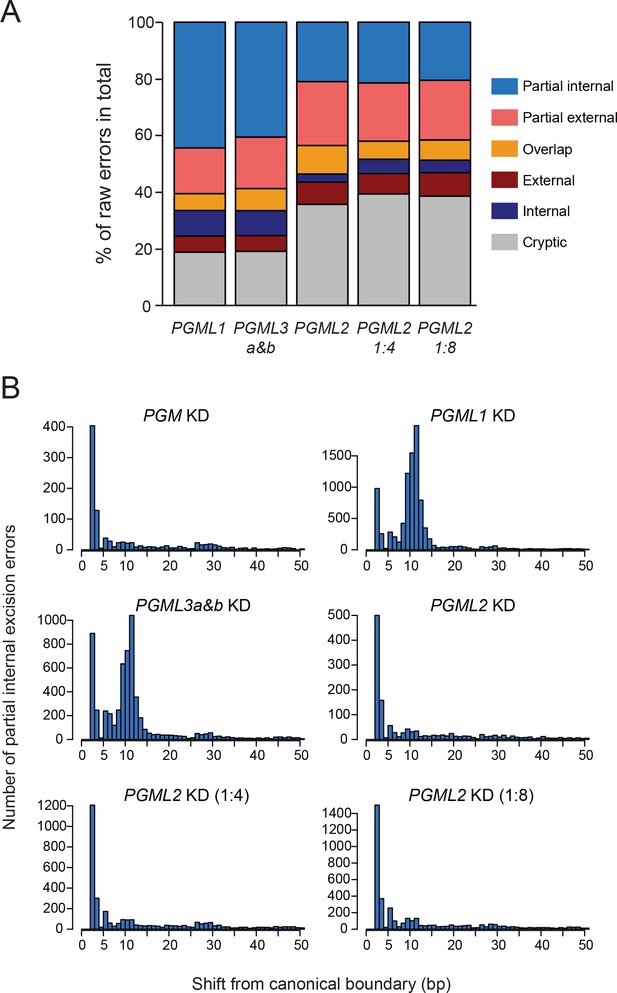
Raw counts of IES excision errors in PGML1, PGML3a and b and partial PGML2 knockdowns.
(A) Raw counts of IES excision errors in PGML1, PGML3a and b and partial PGML2 KDs. Here, and in contrast to Figure 6C, the distribution of different classes of IES excision errors for each condition was plotted without subtracting the contribution of the old MAC from raw error counts. Over-representation of partial internal excision errors was still detected in PGML1 and PGML3a and b KDs. See Supplementary file 9 for details. (B) Position of alternative excision boundaries used in partial internal excision errors in PGM and PGML KDs, relative to the canonical boundary of the reference IES. As in panel A, all partial internal errors were considered for each sample without subtracting the contribution of the old MAC, leading to higher error counts for each position than shown in Figure 6D.
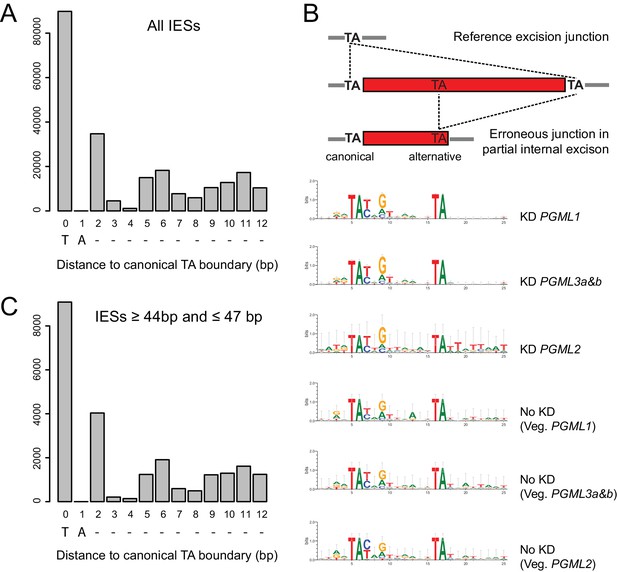
Alternative excision boundaries used in partial internal IES excision errors.
(A) Fraction of IES ends with a TA dinucleotide localized at each indicated distance from the reference TA boundary. Position 0 corresponds to the T of the reference TA at each IES end. (B) SeqLogos of canonical (left) and alternative internal (11–12 bp distant) TAs erroneously used in PGML KDs were determined using the ‘weblogo’ software, version 3.3 (Schneider and Stephens, 1990; Crooks et al., 2004). (C) Same analysis as in panel A, restricted to IESs ranging from 44 to 47 bp in length.
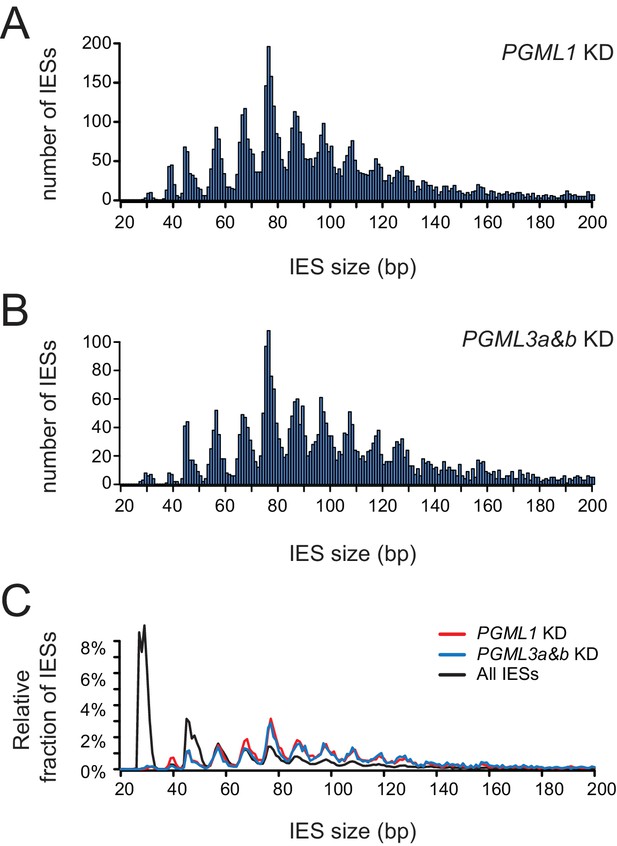
Size distribution of IESs with partial internal excision errors in PGML1 or PGML3a and b KDs.
(A) Size distribution of IESs exhibiting partial internal errors in a PGML1 KD. (B) Same as panel A for a PGML3a and b KD. (C) For each IES size, the black curve shows the fraction of IESs relative to total number of IESs in the genome, the red (PGML1 KD) and blue (PGML3a and b KD) curves show the fraction of IESs of each size among the population of IESs showing at least one partial internal error. In all panels, only IESs with an alternative boundary located >2 bp from the canonical one were counted.
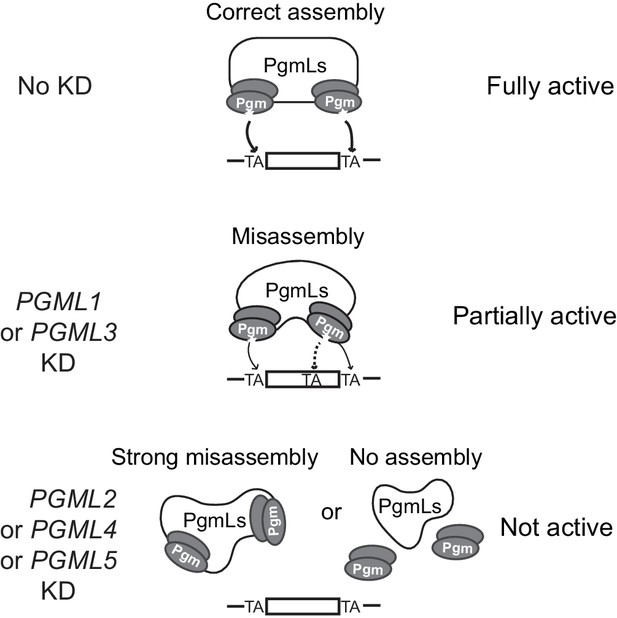
Model for IES excision mediated by a multicomponent Pgm/PgmLs complex.
This figure summarizes the observed effects of PGML KDs on Pgm-mediated IES excision. In line with previously published data (Dubois et al., 2017) and known properties of the T. ni PiggyBac transposase (Jin et al., 2017), the catalytically active form of Pgm is assumed to be a dimer. In the absence of information about the stoichiometry of the complex, one Pgm homodimer (active catalytic site drawn as a star) is represented at each IES boundary, with a large bridging structure formed by all PgmLs. In a fully assembled complex, PgmL subunits are proposed to drive the correct positioning of the Pgm catalytic site onto both TA cleavage sites (indicated by arrows). Following PGML KDs, we propose two distinct situations. In PGML1 or PGML3 KDs, the depleted complexes can exist but are misassembled. As a consequence, Pgm nuclear stability is reduced (the phenotype is more pronounced in a PGML3 KD than in a PGML1 KD) and Pgm activity is altered, because incorrect positioning of catalytic subunits generates specific partial internal excision errors at low frequency (a dotted arrow points to the erroneously targeted alternative TA). In the other three KDs, IES excision complexes depleted for PgmL2, PgmL4 or PgmL5 are totally inactive. This might result either from strong misassembly of the complex or its dissociation (or non-assembly).
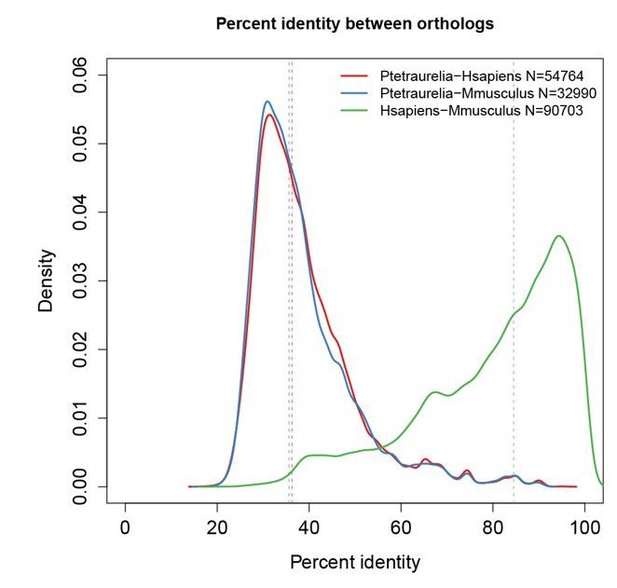
Paramecium proteins are highly divergent relative to their mammalian orthologs.
Percent identity between orthologs was calculated using the InParanoid tool from Cildb (Arnaiz et al., 2009, Database 2000 bap022; Arnaiz et al., 2014, Cilia 3:9; http://cildb.cgm.cnrs-gif.fr/). Dotted lines indicate the median of each distribution. Overall, Paramecium proteins share only 35% identity (median) with mammalian proteins, while mouse and human proteins share 95% identity with each other.
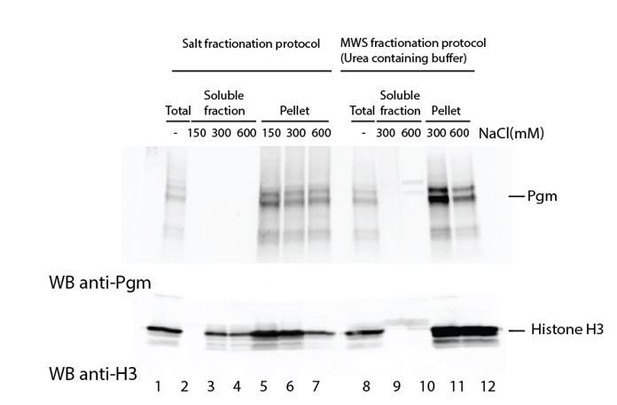
Attempts to extract endogenous Pgm from chromatin in control cells.
Paramecium nuclei were purified by low-speed centrifugation of autogamous cell lysates, as described (Arnaiz et al., 2012). Immunoblotting was performed using α-Pgm 2659-GP antibodies for Pgm detection (Dubois et al., 2017) and commercial antibodies for the detection of histone H3 (Merck Millipore # 07-690).
Additional files
-
Supplementary file 1
MUSCLE alignment of the transposase core domains of ciliate domesticated PB transposases and other PB transposases
- https://doi.org/10.7554/eLife.37927.024
-
Supplementary file 2
Table of Pgm and PgmL proteins encoded by published Paramecium genomes and their ParameciumDB accession numbers
- https://doi.org/10.7554/eLife.37927.025
-
Supplementary file 3
Sequences of the cysteine-rich domains used for the alignment shown in Figure 1—figure supplement 1
- https://doi.org/10.7554/eLife.37927.026
-
Supplementary file 4
Sequences of the transposase core domains used for the alignement shown in Supplementary file 1.
- https://doi.org/10.7554/eLife.37927.027
-
Supplementary file 5
Sequence of the synthetic PGML genes used for protein production in insect cells
- https://doi.org/10.7554/eLife.37927.028
-
Supplementary file 6
Analysis of post-autogamous progeny in small-scale PGML knockdowns,
- https://doi.org/10.7554/eLife.37927.029
-
Supplementary file 7
Analysis of post-autogamous progeny in middle- and large-scale PGML knockdowns
- https://doi.org/10.7554/eLife.37927.030
-
Supplementary file 8
DNA-seq datasets from ENA project PRJEB24171 (this study)
- https://doi.org/10.7554/eLife.37927.031
-
Supplementary file 9
Analysis of IES excision reads in PGM and PGML knockdowns
- https://doi.org/10.7554/eLife.37927.032
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37927.033





