Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies
Figures
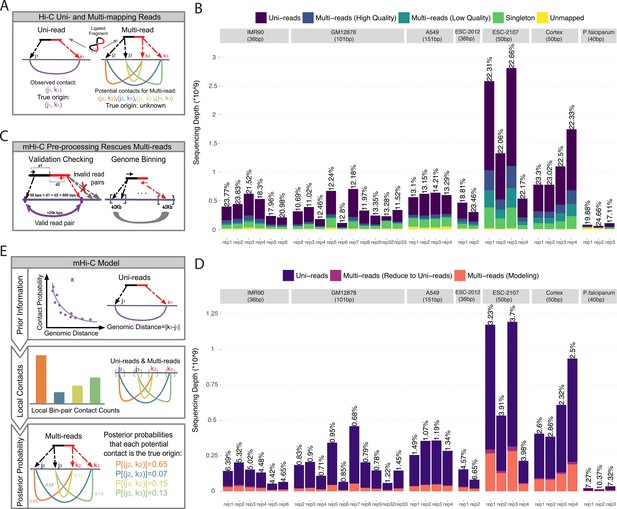
Overview of multi-reads and mHi-C pipeline.
(A) Standard Hi-C pipelines utilize uni-reads while discarding multi-mapping reads which give rise to multiple potential contacts. (B) The total number of reads in different categories as a result of alignment to reference genome across the study datasets. Percentages of high-quality multi-reads compared to uni-reads are depicted on top of each bar. (C) Multi-mapping reads can be reduced to uni-reads within validation checking and genome binning pre-processing steps. (D) Aligned reads after validation checking and binning. Percentage improvements in sequencing depths due to multi-reads becoming uni-reads are depicted on top of each bar. (E) mHi-C modeling starts from the prior built by only uni-reads to quantify the relationship between random contact probabilities and the genomic distance between the contacts. This prior is updated by leveraging local bin pair contacts including both uni- and multi-reads and results in posterior probabilities that quantify the evidence for each potential contact to be the true genomic origin.
-
Figure 1—source data 1
Detailed summary of study datasets.
- https://doi.org/10.7554/eLife.38070.009
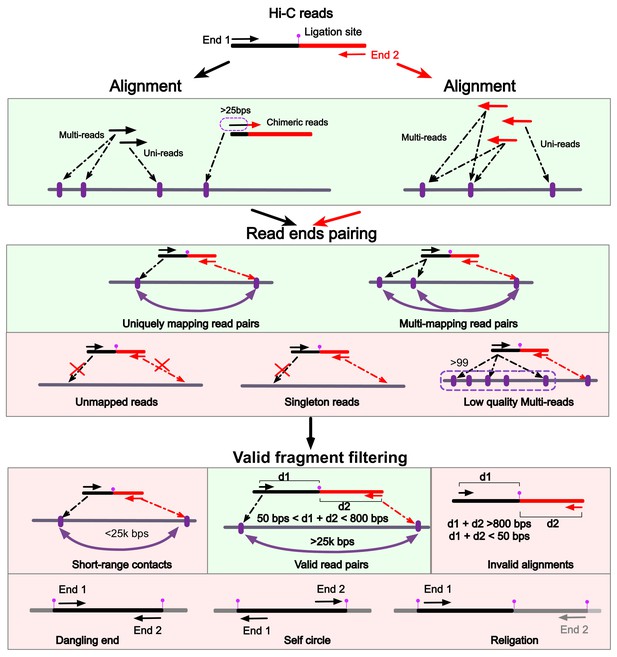
mHi-C pipeline (Alignment - Read end pairing - Valid fragment filtering).
1. Read ends are aligned to reference genome separately allowing multi-reads and chimeric reads to be rescued. 2. Read ends are paired by their read query names. Multi-reads form more than one read pair with the same read query name. Read ends that fail to align form either unmapped reads or singleton reads and are discarded. Multi-reads with ends aligning to more than 99 positions are regarded as low-quality multi-reads and are excluded from the downstream analysis. 3. Validation checking to filter short-range contacts and alignments far away from restriction enzyme recognition sites. Contacts residing within the same restriction fragment, that is dangling end or self-circle, as well as adjacent fragments (religation) are discarded. The above three processing steps are applied to each read independently enabling parallel implementation.
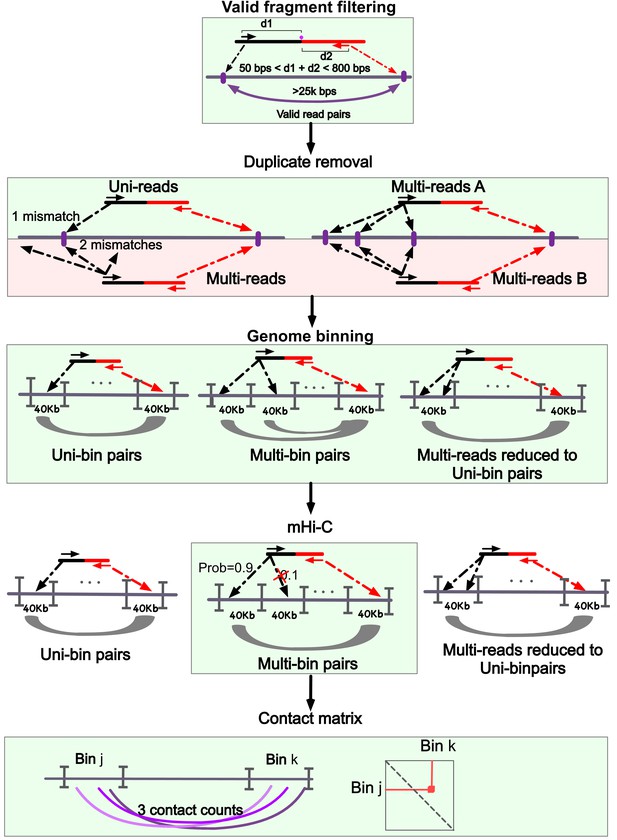
mHi-C pipeline (Duplicate removal - Genome binning - mHi-C).
4. PCR duplicates are removed to ensure that when a uni-read and a multi-read have the same alignment position and strand direction, the uni-read is kept. In the case of multi-reads that overlap with other multi-reads, the ones with alphabetically larger IDs are removed. 5. Genome is split into fix-sized non-overlapping intervals, that is bins or a fixed number of restriction fragments and, as a result, read alignment position pairs are reduced to bin pairs. Multi-reads, candidate alignment positions of which fall into the same bin, are reduced to uni-bin pairs. 6. mHi-C model estimates an allocation probability for each potential contact and enables filtering of contacts by thresholding this allocation probability.
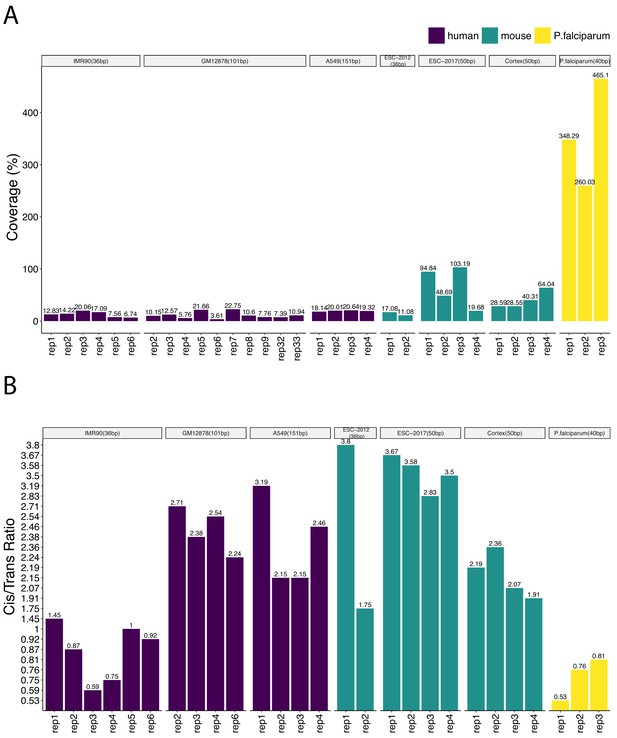
Coverage and cis-to-trans ratios across individual replicates of the study datasets as indicators of data quality.
(A) Coverage is approximated as the ratio of the sequencing depth to the genome size. (B) Cis-to-trans ratio is defined as the number of valid intra-chromosomal contacts divided by the number of valid inter-chromosomal contacts.
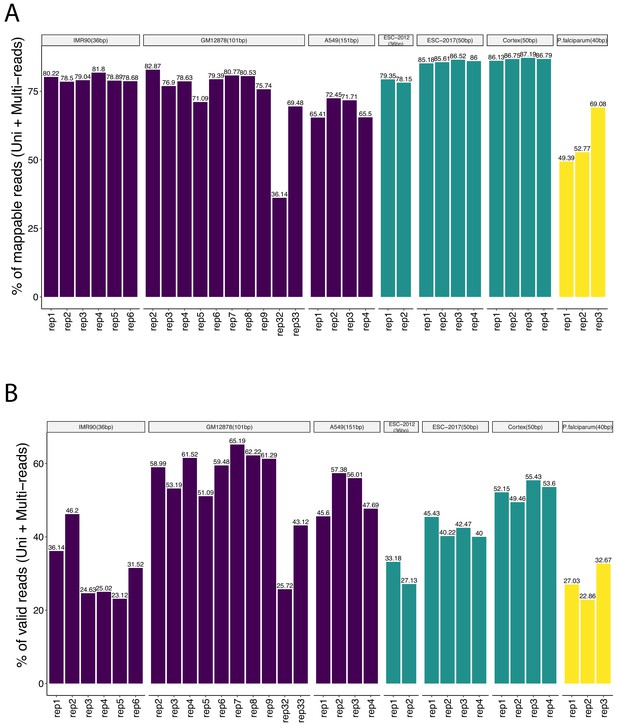
Percentages of (A) mappable and (B) valid reads over the set of all reads for individual replicates of the study datasets as an indicator of data quality.
Both the aligned uni-reads and multi-reads are taken into consideration.
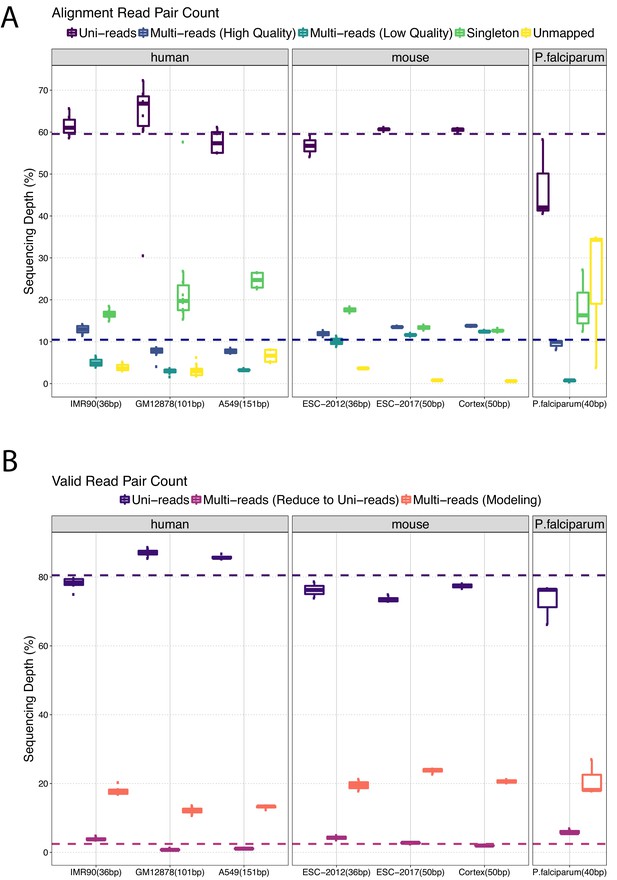
Categorization of reads after alignment across study datasets.
(A) Percentages of mapped reads in different alignment categories. (B) Percentages of valid reads with both ends uniquely aligned to reference genome (Uni-reads), at least one end aligning to multiple positions and resulting in only one valid alignment after validation and binning (Multi-reads (Reduce to Uni-reads)), and reads with multiple potential valid alignment positions (Multi-reads (Modeling)).
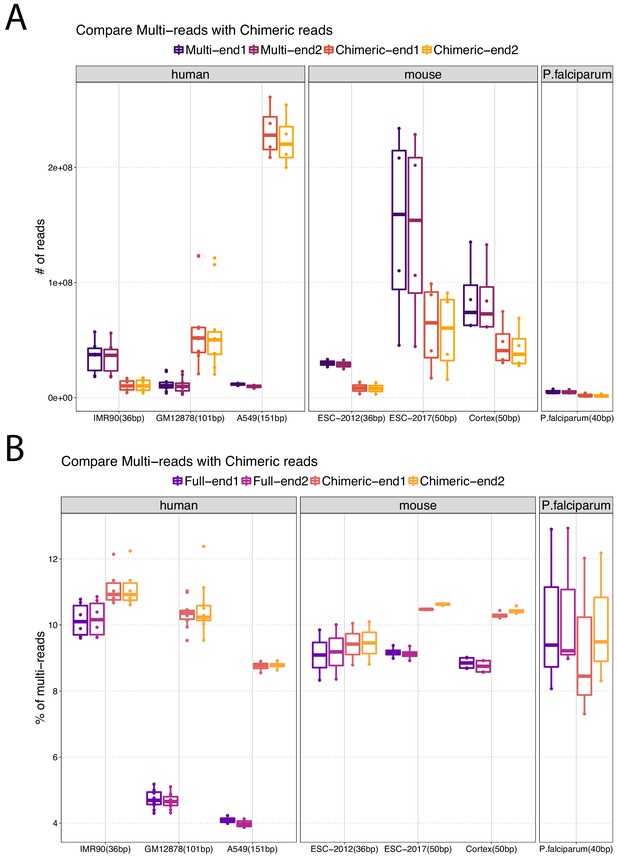
Comparison of the prevalence of multi-reads and chimeric reads, both of which require additional processing.
(A) Numbers of multi-reads compared to the numbers of chimeric reads at each read end level. For Hi-C datasets with shorter read lengths, multi-reads constitute a larger percentage of the usable reads compared to chimeric reads. (B) Proportion of multi-reads among full-length alignable reads compared with that among chimeric reads. As expected, chimeric reads lead to larger percentages of multi-reads.
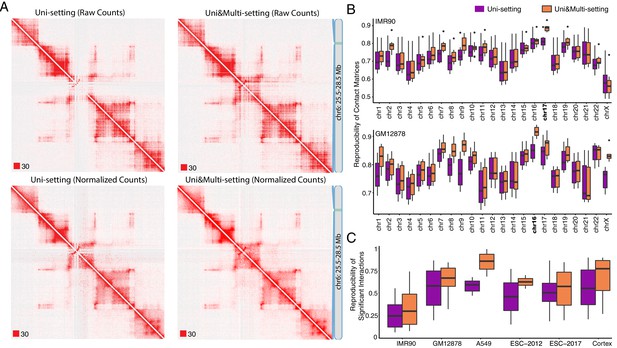
Global impact of multi-reads in Hi-C analysis.
(A) Contact matrices of GM12878 with combined reads from replicates 2–9 are compared under Uni-setting and Uni&Multi-setting using raw and normalized contact counts for chr6:25.5 Mb - 28.5 Mb. White gaps of Uni-reads contact matrix, due to lack of reads from repetitive regions, are filled in by multi-reads, hence resulting in a more complete contact matrix. Such gaps remain in the Uni-setting even after normalization. Red squares at the left bottom of the matrices indicate the color scale. (B) Reproducibility of Hi-C contact matrices by HiCRep across all pairwise comparisons between replicates under the Uni- and Uni&Multi-settings (IMR90 and GM12878 are displayed). (C) Reproducibility of the significant interactions across replicates of the study datasets. Reproducibility is assessed by overlapping interactions detected at FDR of 5% for pairs of replicates within each study dataset.
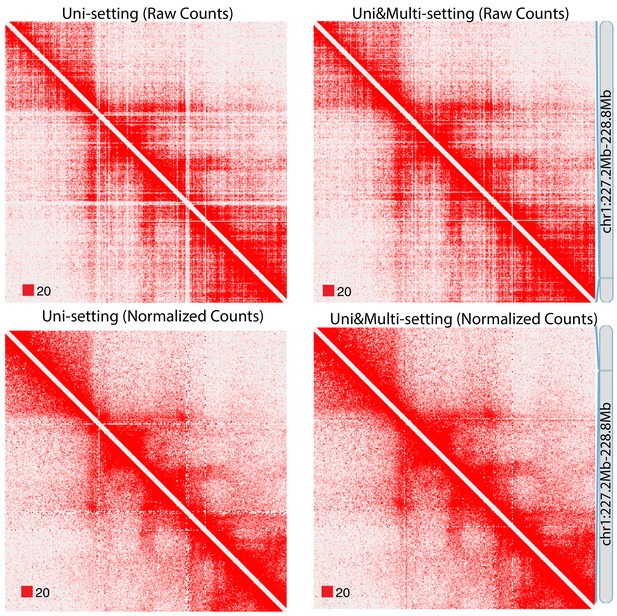
Raw and normalized contact matrices of GM12878 under Uni-setting and Uni&Multi-setting on chromosome 1.
https://doi.org/10.7554/eLife.38070.012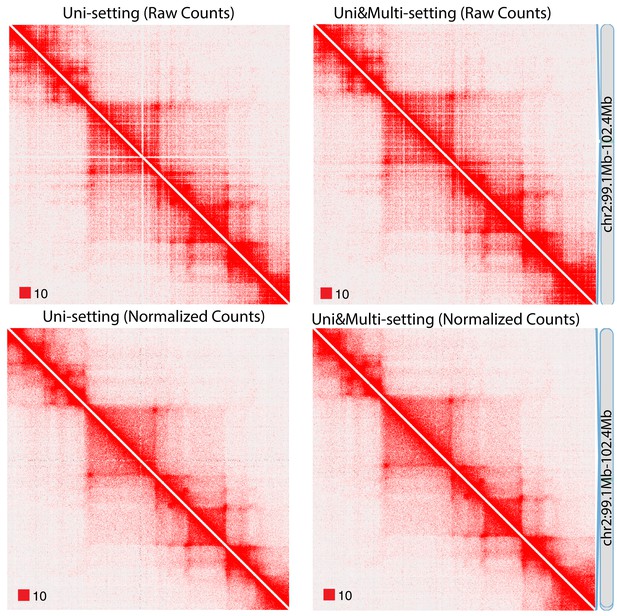
Raw and normalized contact matrices of GM12878 under Uni-setting and Uni&Multi-setting on chromosome 2.
https://doi.org/10.7554/eLife.38070.013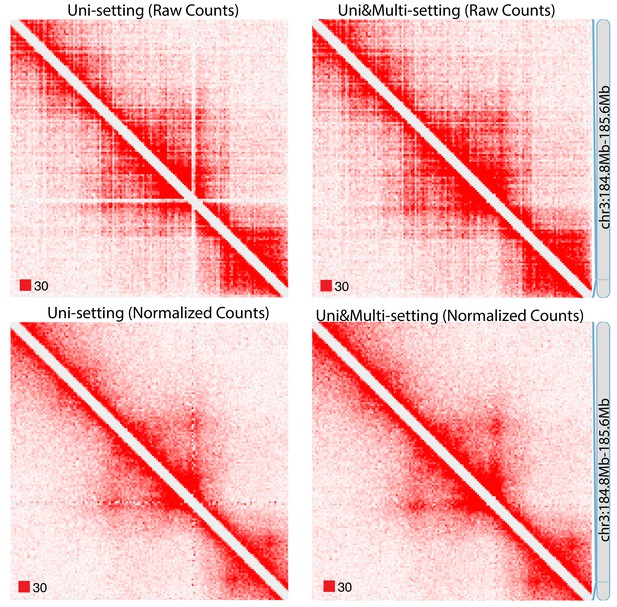
Raw and normalized contact matrices of GM12878 under Uni-setting and Uni&Multi-setting on chromosome 3.
https://doi.org/10.7554/eLife.38070.014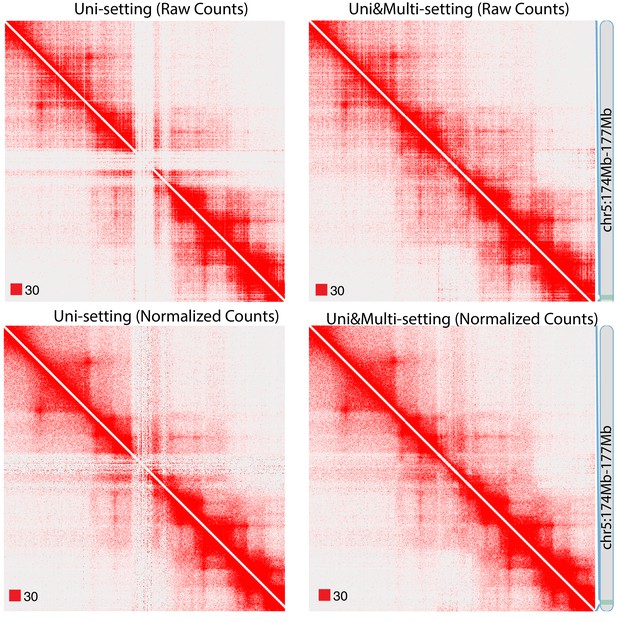
Raw and normalized contact matrices of GM12878 under Uni-setting and Uni&Multi-setting on chromosome 5.
https://doi.org/10.7554/eLife.38070.015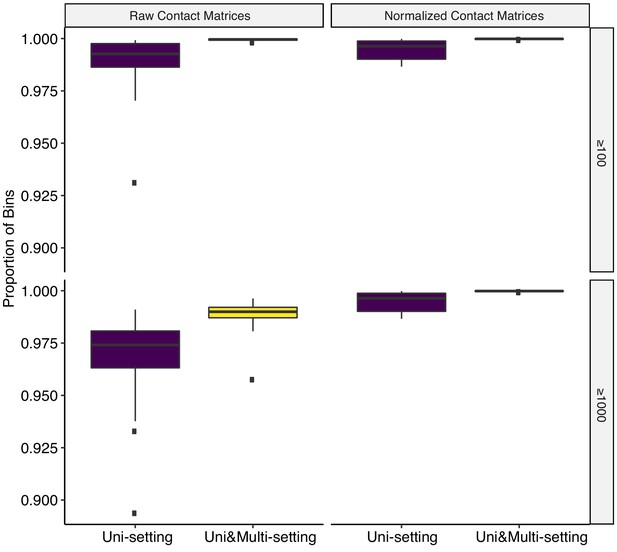
Proportion of bins that are covered by at least 100 (row 1) or 1000 (row 2) contacts for raw contact matrices (column 1) and normalized contact matrices (column2) under Uni- and Uni&Multi-settings for GM12878 with combined reads from replicates 2–9 at 5 kb resolution.
https://doi.org/10.7554/eLife.38070.016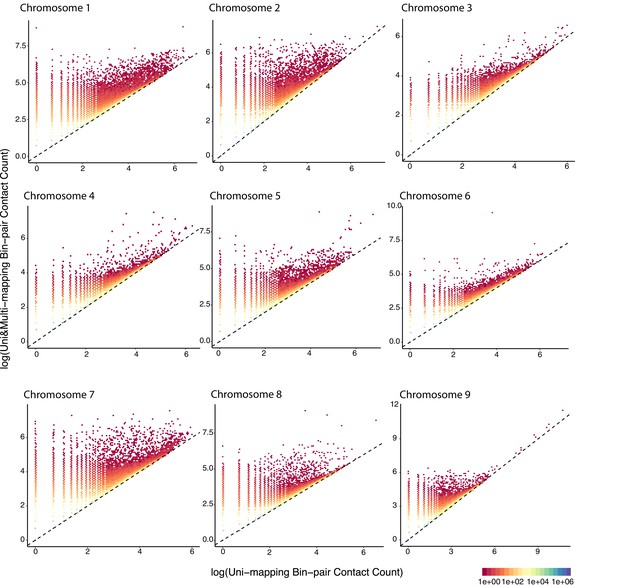
Bin coverage improvement of raw contact matrices under Uni&Multi-setting compared to Uni-setting for GM12878 with combined reads from replicates 2–9 at 5 kb.
Only chromosome 1–9 are shown and the pattern for the rest chromosomes are very similar. The dashed line is y = x.
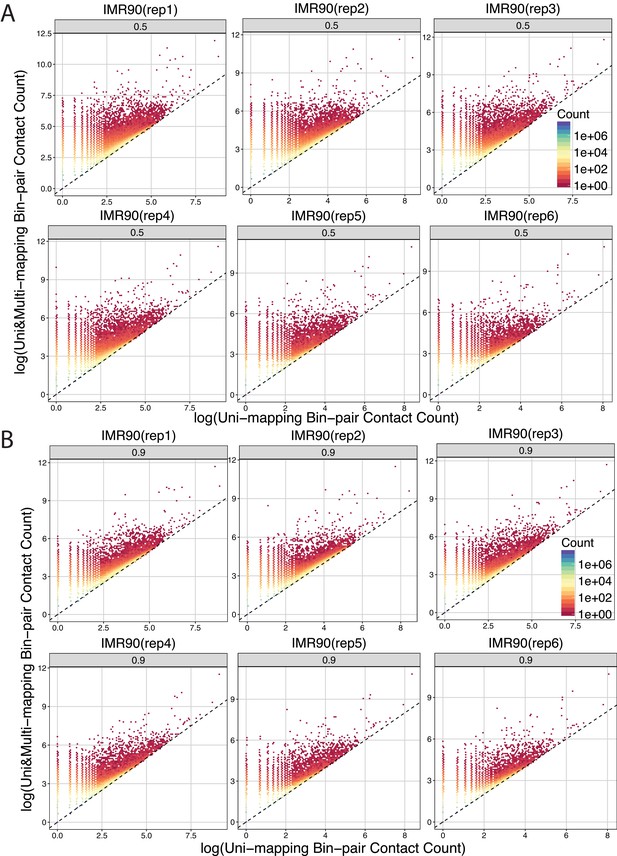
Bin coverage improvement of raw contact matrices under Uni&Multi-setting compared to Uni-setting for IMR90 at the individual replicate level for two different allocation probability thresholds.
Uni&Multi-setting for (A) includes multi-reads with a posterior probability > 0.5, whereas (B) depicts more strict filtering with an allocation probability > 0.9 The dashed line is y = x. mHi-C rescues multi-reads from valid ligation fragments, resulting in a significant increase in contact counts.
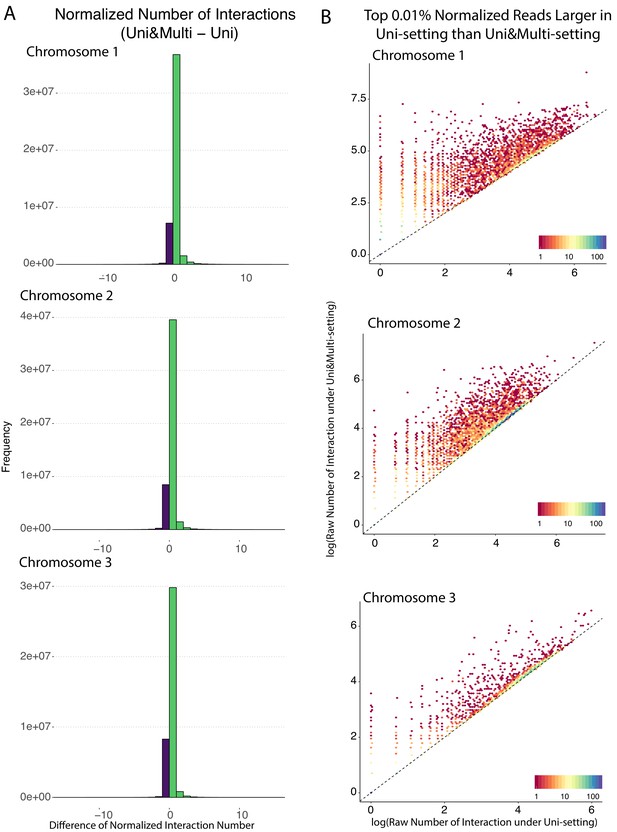
Bin coverage comparison of normalized contact matrices under Uni&Multi- and Uni-settings for GM12878 with combined reads from replicates 2–9 at 5 kb.
(A) Histogram of the bin-level differences between normalized contact counts of the Uni&Multi- and Uni-settings. Green bars represent increased counts under the Uni&Multi-setting, and purple ones indicate no change or decrease. Bins in the purple bar group are low coverage bins under the Uni-setting and have inflated normalized contact counts. Only chromosome 1–3 are shown, and the pattern for the rest of the chromosomes are very similar. (B) Raw contact count comparison of the top 0.01% bins of Panel A with drastically higher normalized contact counts under the Uni-setting compared to the Uni&Multi-setting (purple bars). The contact counts of these bins get inflated by normalization. The dashed lines are y = x.
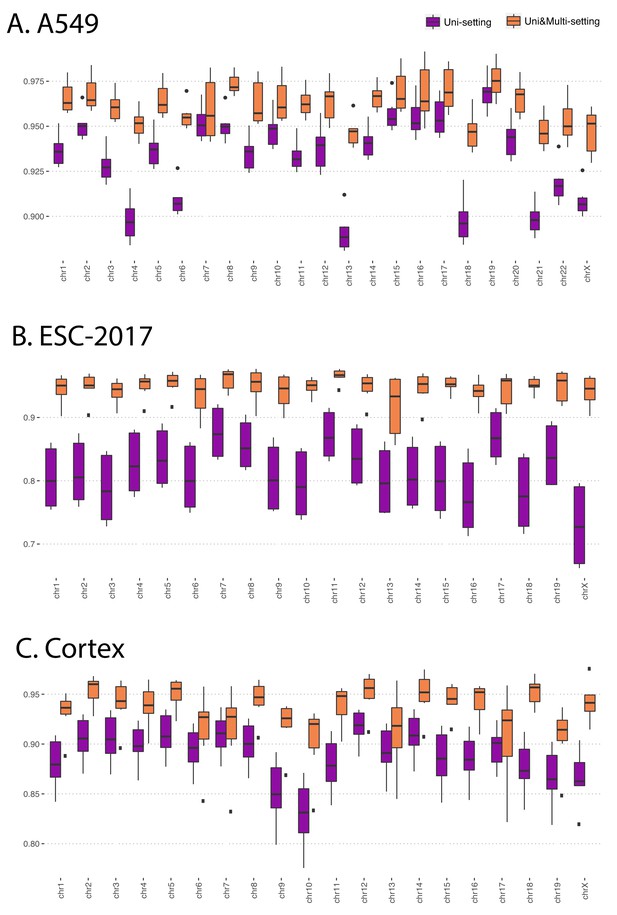
Reproducibility at the contact matrix level under the Uni- and Uni&Multi-settings in A549, ESC-2017 and Cortex cell lines.
Note that the box plots for the ESC-2012 cell line which only has two replicates are not displayed.
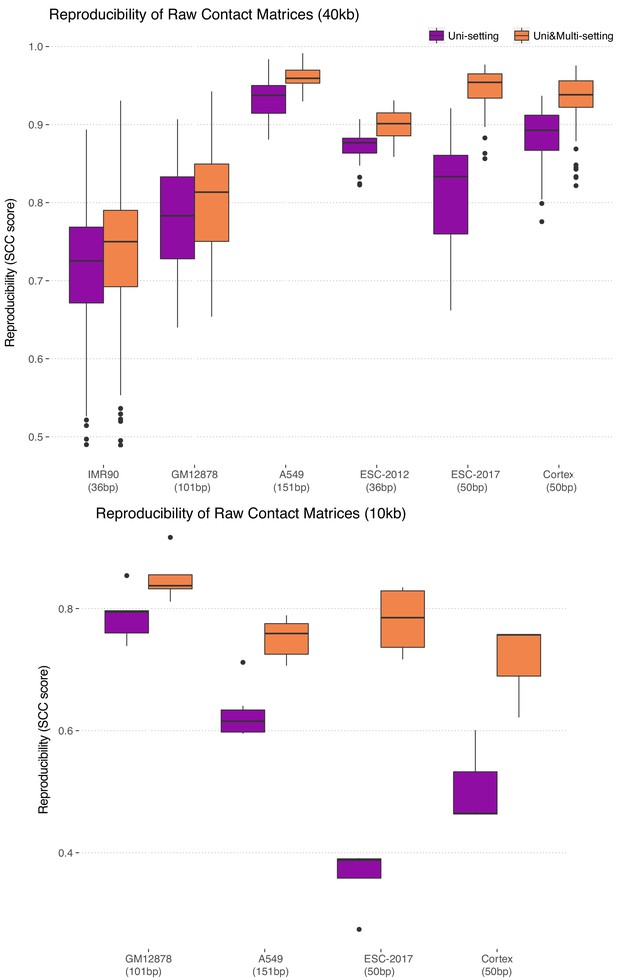
Reproducibility at the contact matrix level at resolutions 40 kb (low) and 10 kb (high) across study datasets.
https://doi.org/10.7554/eLife.38070.021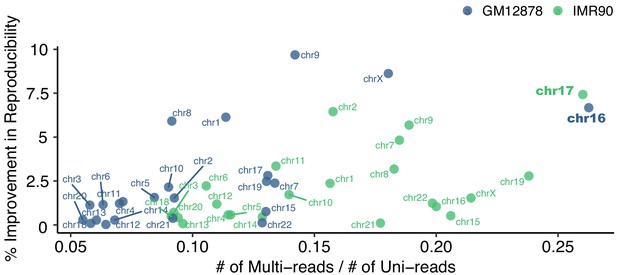
Percent improvement in reproducibility due to the Uni&Multi-setting versus the proportion of the number of valid multi-reads compared to the number of the uni-reads in the datasets.
https://doi.org/10.7554/eLife.38070.022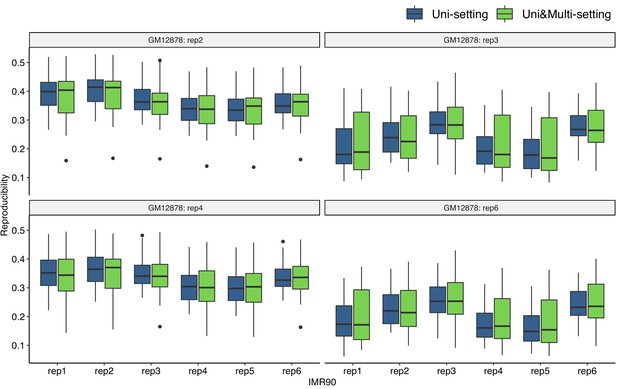
Reproducibility at the contact matrix level under the Uni- and Uni&Multi-settings between GM12878 and IMR90 at 40 kb resolution.
Each box contains reproducibility measurements on 23 chromosomes between every two pairs of replicates from GM12878 and IMR90. Within each panel, one GM12878 replicate contact matrix is compared with each of the six IMR90 replicates respectively for Uni- and Uni&Multi-settings.
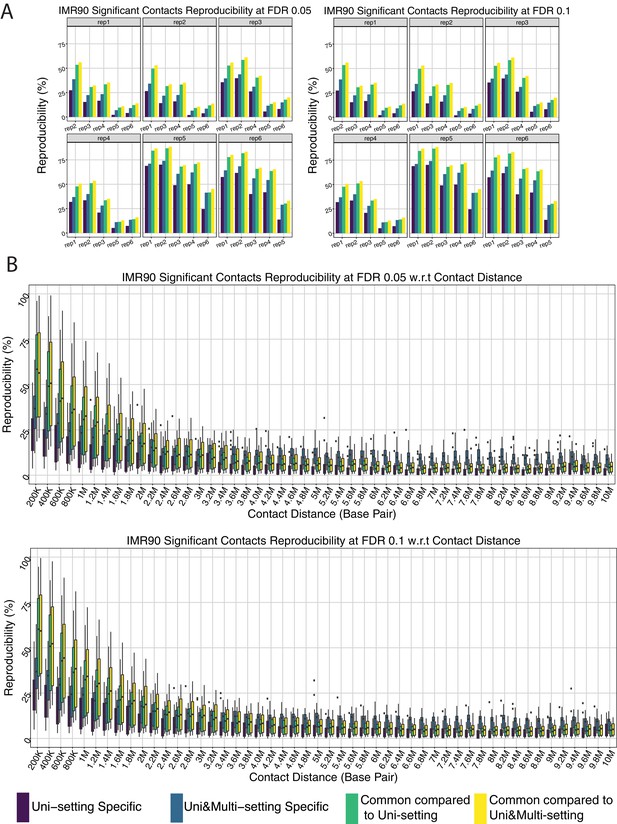
Reproducibility of significant interactions for IMR90.
(A) Significant interactions are classified into three categories: Uni-setting specific or Uni&Multi-setting specific or common to both. Reproducibility is evaluated by the percentage of significant interactions reproduced in another replicate within the same category. (B) Reproducibility of significant interactions stratified by genomic distance. Reproducibility is evaluated by the percentage of significant interactions reproduced in another replicate within the same category and genomic distance range.
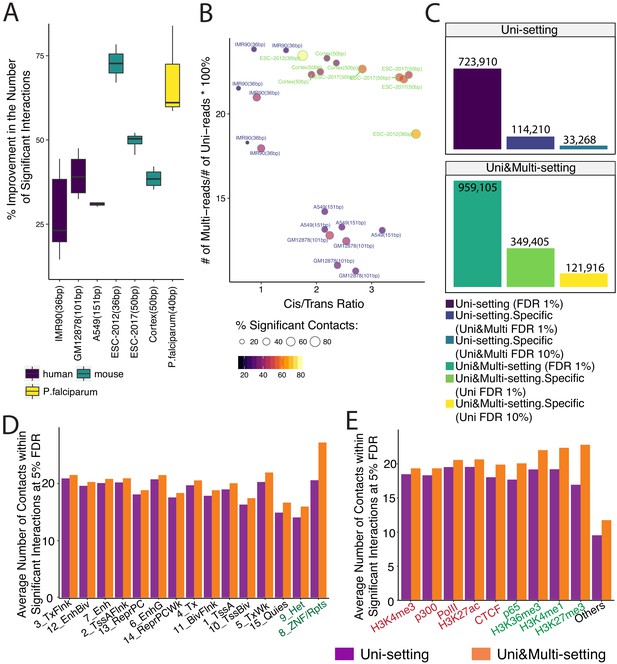
Gain in the numbers of novel significant interactions by mHi-C and their characterization by chromatin marks.
(A) Percentage increase in detected significant interactions (FDR 5%) by comparing contacts identified in Uni&Multi-setting with those of Uni-setting across study datasets at 40 kb resolution. (B) Percentage change in the numbers of significant interactions (FDR 5%) as a function of the percentage of mHi-C rescued multi-reads in comparison to uni-read and cis-to-trans ratios of individual datasets at 40 kb resolution. (C) Recovery of significant interactions identified at 1% FDR by analysis at 10% FDR, aggregated over the replicates of GM12878 at 40 kb resolution. Detailed descriptions of the groups are provided in Figure 3—figure supplement 6. (D) Average number of contacts falling within the significant interactions (5% FDR) that overlapped with each chromHMM annotation category across six replicates of IMR90 identified by Uni- and Uni&Multi-settings. (E) Average number of contacts (5% FDR) that overlapped with significant interactions and different types of ChIP-seq peaks associated with different genomic functions (IMR90 six replicates). Red/Green labels denote smaller/larger differences between the two settings compared to the differences observed in the ”Others’ category that depict non-peak regions.
-
Figure 3—source data 1
Percentage of improvement in the number of significant interactions across six studies at resolution 40 kb.
- https://doi.org/10.7554/eLife.38070.043
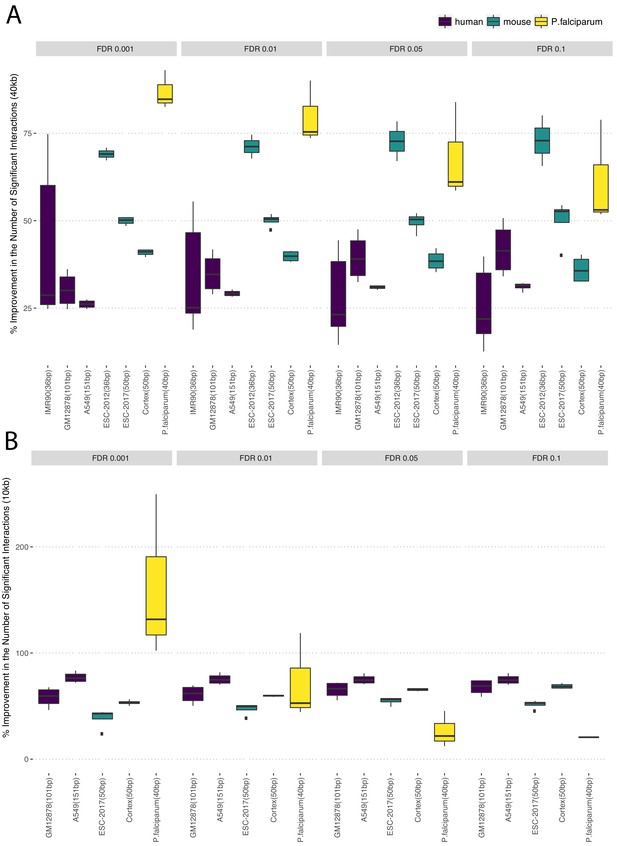
Percentage change in the numbers of significant interactions under the Uni&Multi-setting compared to Uni-setting at 0.1%, 1%, 5% and 10% FDR thresholds and resolutions (A) 40 kb and (B) 10 kb.
https://doi.org/10.7554/eLife.38070.026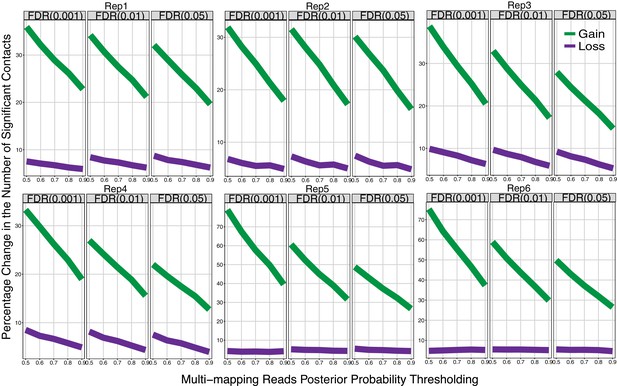
Comparison of significant interactions as a function of posterior probabilities of multi-read assignment (IMR90 40 kb).
Percentage change in the numbers of significant interactions gained (Green) and lost (Purple) by the Uni&Multi-setting compared to the Uni-setting across individual IMR90 replicates for varying FDR and allocation probability thresholds.
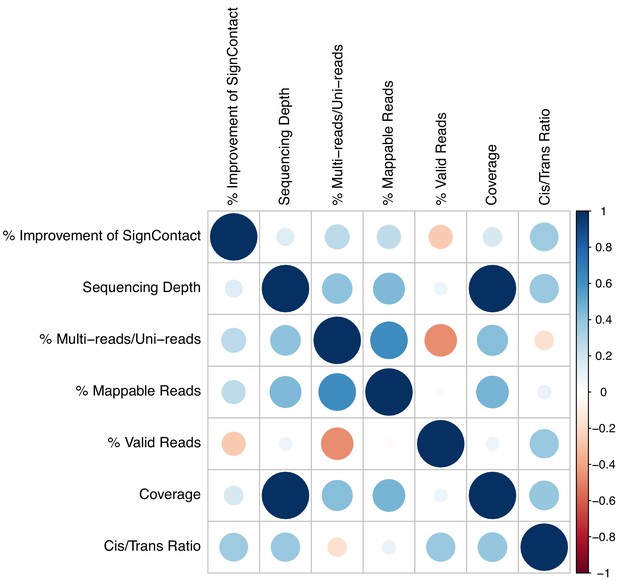
Heatmap for marginal correlations of percentage increase in the number of identified significant interactions (FDR 5%) with indicators of data quality across study datasets excluding P. falciparum at 40 kb.
The relative percentage of multi-reads added to uni-reads as well as cis-to-trans ratio is leading impact factors (with p-values 0.005) followed by the percentage of mappable reads and valid reads.
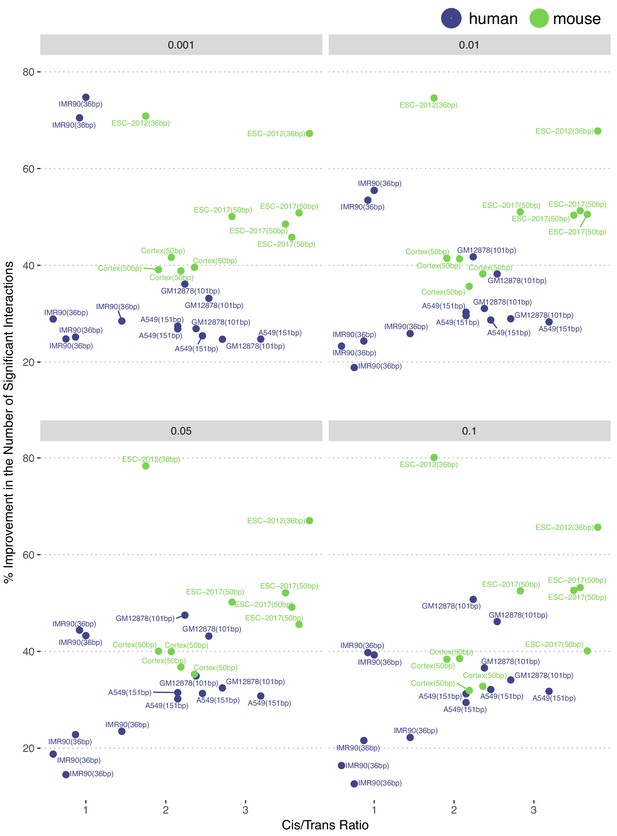
Percentage change in the numbers of significant interactions with respect to cis-to-trans ratio excluding P. falciparum at 40 kb.
Cis-to-trans ratio is defined as the number of valid intra-chromosomal contact counts divided by the number of valid inter-chromosomal contact counts.
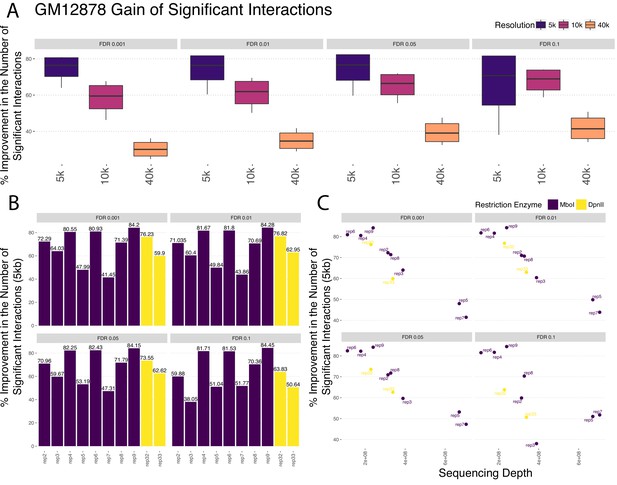
Percentage change in the numbers of significant interactions of GM12878 datasets at different resolutions.
(A) Percentage change in the numbers of significant interactions of GM12878 datasets at 5 kb, 10 kb, 40 kb resolutions, respectively, across varying FDR thresholds. (B) Percentage change in the numbers of significant interactions of 8 replicates that are based on MboI as the restriction enzyme and two replicates with DpnII as restriction enzyme at 5 kb resolution across different FDR thresholds. (C) Percentage change in the numbers of significant interactions with respect to the replicate sequencing depth of all 10 replicates of GM12878 at 5 kb resolution across different FDR thresholds.
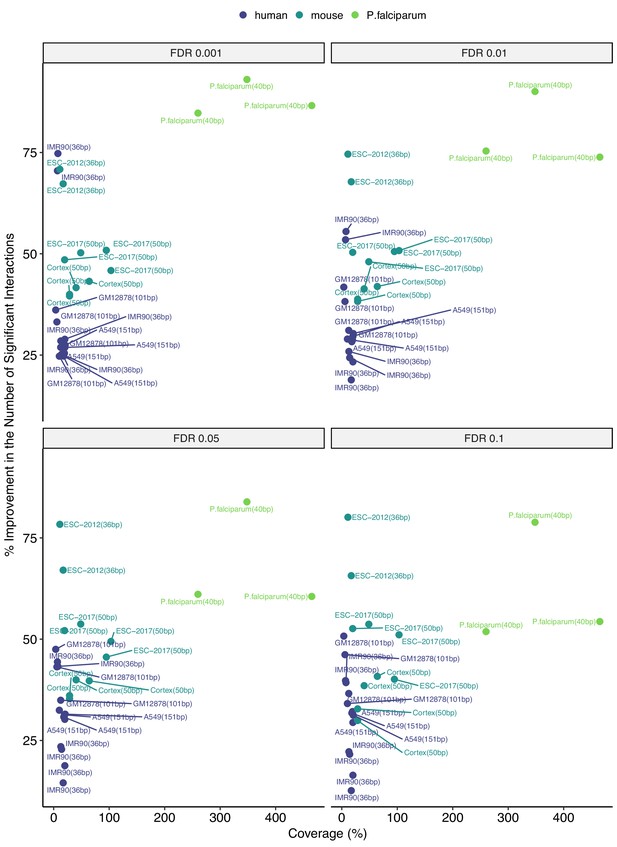
Percentage change in the numbers of significant interactions with respect to coverage at 40 kb.
Coverage is approximated as the ratio of the sequencing depth to the genome size.
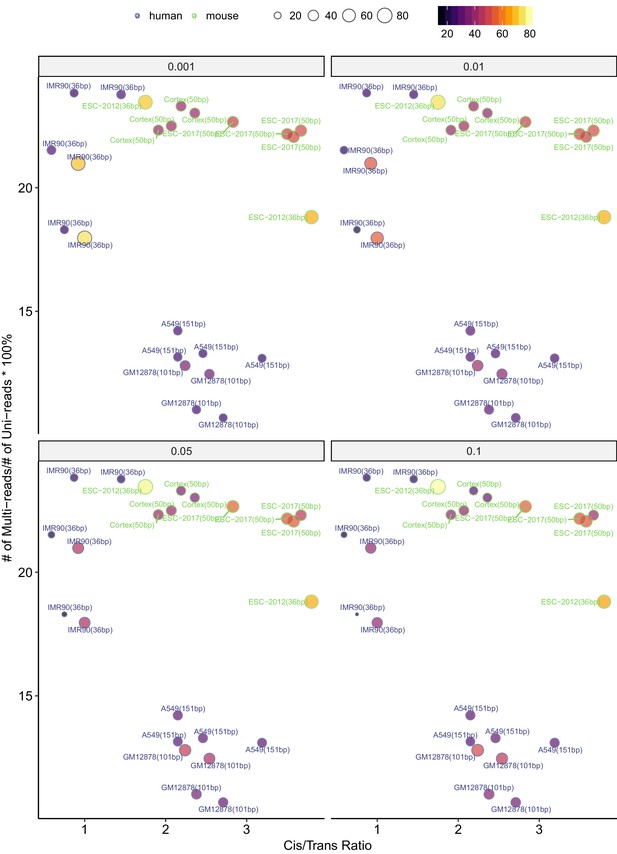
Percentage change in the numbers of significant interactions as a function of the percentage of mHi-C rescued multi-reads in comparison to uni-reads and cis-to-trans ratios at 40 kb.
https://doi.org/10.7554/eLife.38070.030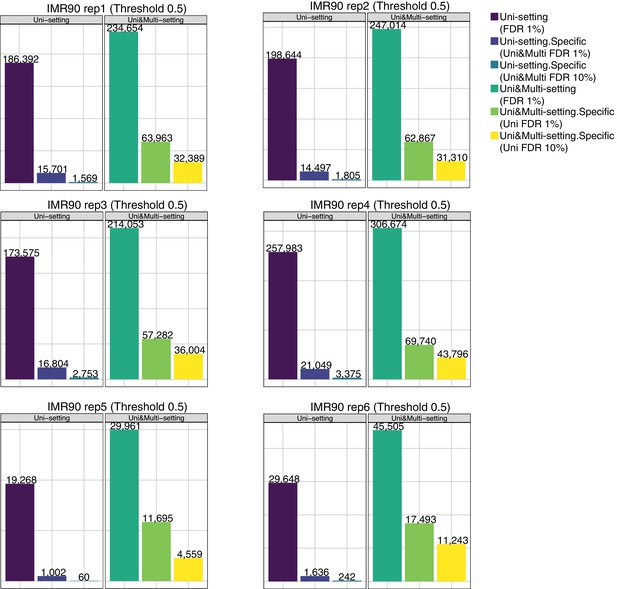
Recovery of significant interactions identified at FDR 1% by analysis at FDR 10% for each of six replicates of IMR90 at 40 kb.
Uni&Multi-setting. Specific (Uni FDR 10%) is the set of significant interactions identified at 1% FDR by the Uni&Multi-setting but are still unrecoverable by the Uni-setting even with a liberal FDR of 10%. The detailed descriptions of the groups are as follows: Uni-setting (FDR 1%): # of significant interactions identified by the Uni-setting at 1% FDR. Uni&Multi-setting (FDR 1%): # of significant interactions identified by the Uni&Multi-setting at 1% FDR. Uni-setting.Specific (Uni&Multi FDR 1%): # of significant interactions identified by Uni-setting (FDR 1%) but not by Uni&Multi-setting at 1% FDR. Uni-setting.Specific (Uni&Multi FDR 10%): # of significant interactions identified by Uni-setting (FDR 1%) but not by Uni&Multi-setting at 10% FDR. Uni&Multi-setting.Specific (Uni FDR 1%): # of significant interactions identified by Uni&Multi-setting (FDR 1%) but not by Uni-setting at 1% FDR. Uni&Multi-setting.Specific (Uni FDR 10%): # of significant interactions identified by Uni&Multi-setting (FDR 1%) but not by Uni-setting at 10% FDR.
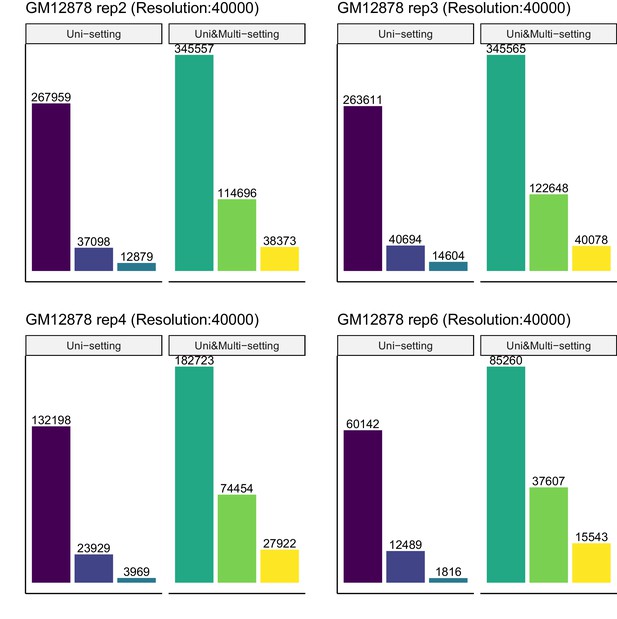
Recovery of significant interactions identified at FDR 1% by analysis at FDR 10% for each of four replicates of GM12878 at 40 kb resolution.
Color labels are the same as Figure 3—figure supplement 8.
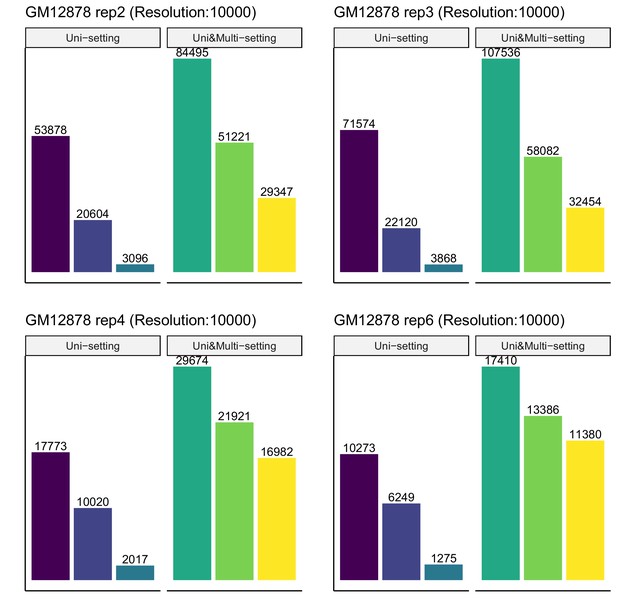
Recovery of significant interactions identified at FDR 1% by analysis at FDR 10% for each of four replicates of GM12878 at 10 kb resolution.
Color labels are the same as Figure 3—figure supplement 8.
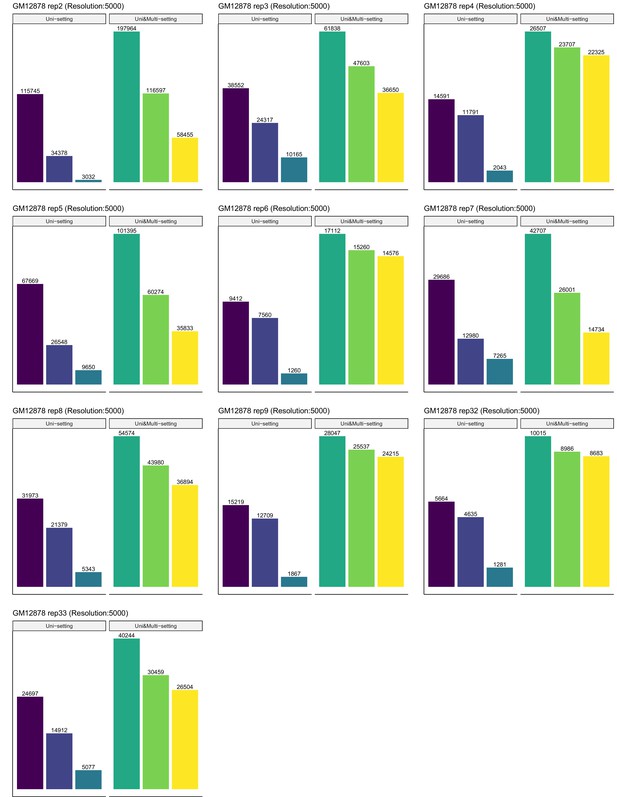
Recovery of significant interactions identified at FDR 1% by analysis at FDR 10% for each of ten replicates of GM12878 at 5 kb resolution.
Color label is the same as Figure 3—figure supplement 8.
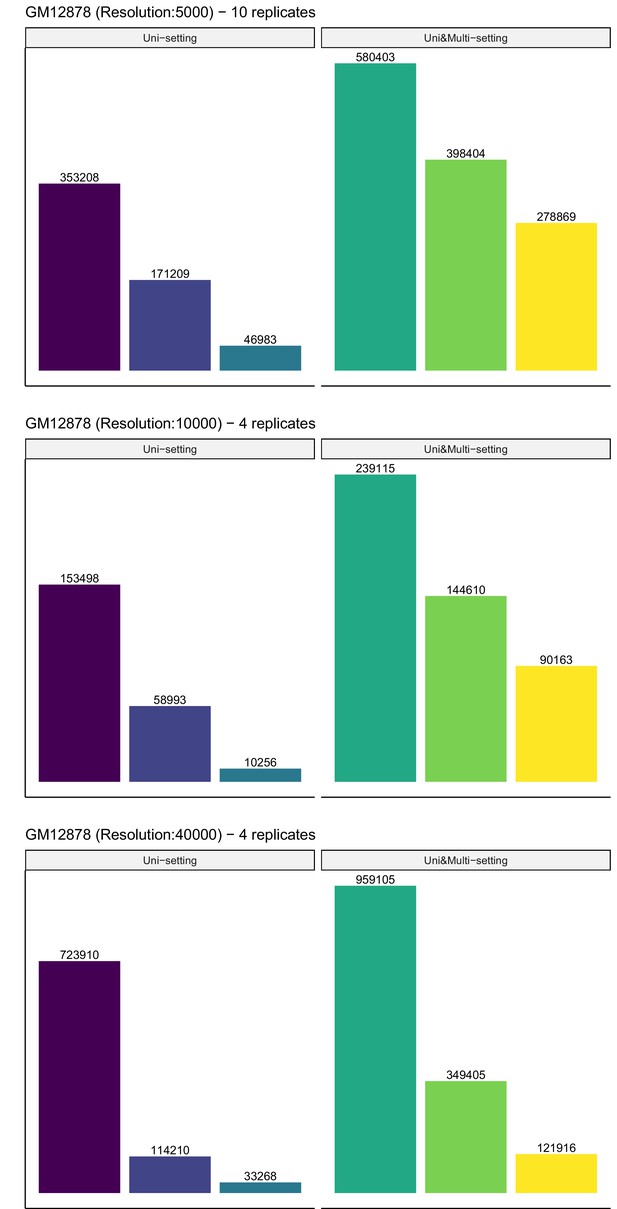
Recovery of significant interactions identified at FDR 1% by analysis at FDR 10% for GM12878 summed across replicates at 5 kb, 10 kb, and 40 kb resolutions.
Color label is the same as Figure 3—figure supplement 8.
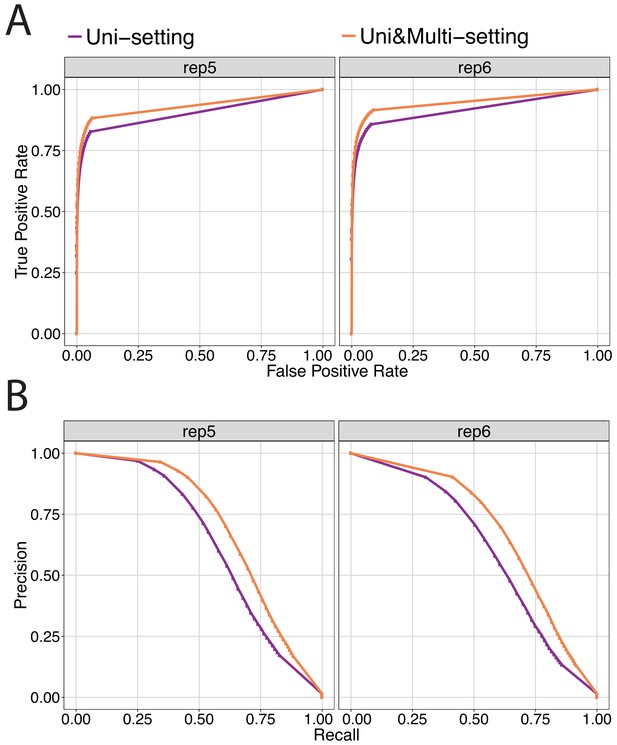
ROC and PR curves for replicates 5 and 6 of IMR90.
Sets of ‘True’ interactions and ‘True’ non-interactions are defined by reproducible significant/insignificant interactions across replicate 1–4 of both Uni-setting and Uni&Multi-setting (See Materials and methods). Significant interactions of replicates 5 and 6 are utilized to compare ROC (A) and PR (B) curves among the Uni- and Uni&Multi-settings.
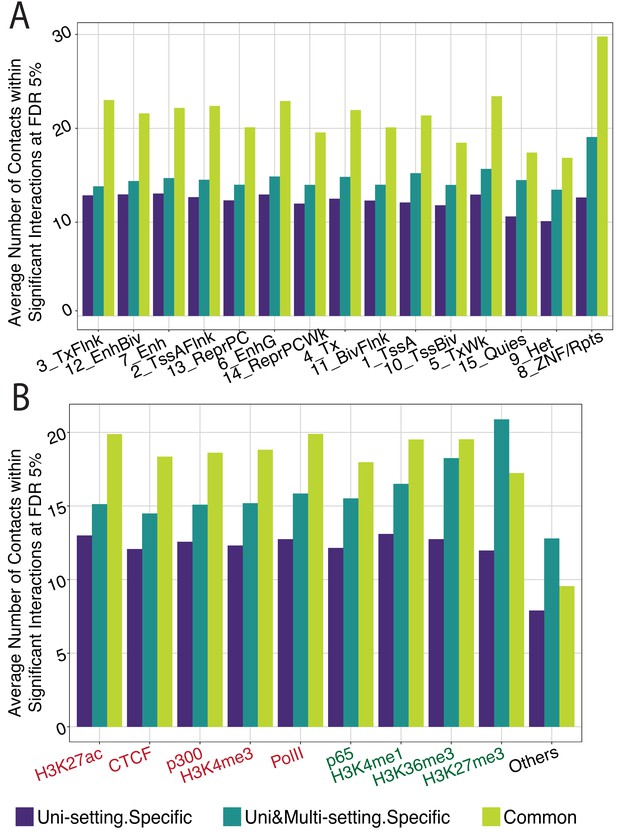
Quantification of significant interactions for chromHMM states and ChIP-seq peak regions (IMR90).
(A) Grouping the significant interactions into three groups (Uni-setting specific, Uni&Multi-setting specific, Common to both settings) reveals the largest enrichment differences in chromHMM annotation categories related to repetitive regions, such as Zinc Finger Genes & Repeats as well as Heterochromatin. (B) Average number of significant interactions across regions with a variety of ChIP-seq signals. Red/green labels denote smaller/larger differences between Uni-setting specific and Uni&Multi-setting specific compared to the differences observed in the ‘Others’ category that depicts non-peak regions.
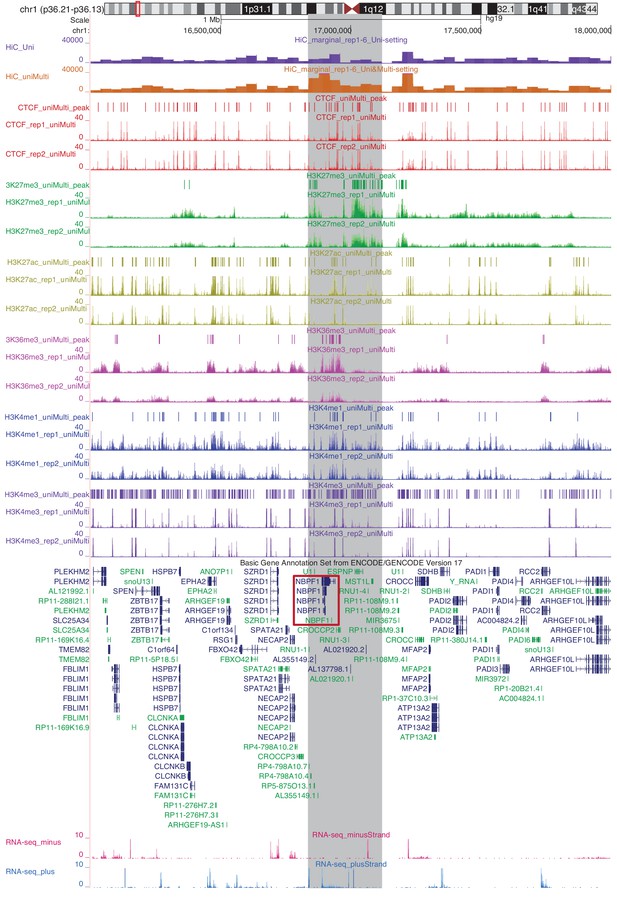
Marginalized Hi-C signal (contact counts aggregated across the genomic coordinates for six replicates of IMR90), ChIP-seq coverage and peaks and gene expression for chr1:16,000,000–18,000,000.
Highlighted in grey is a region with significantly different marginal Hi-C signal between Uni-setting and Uni&Multi-setting.
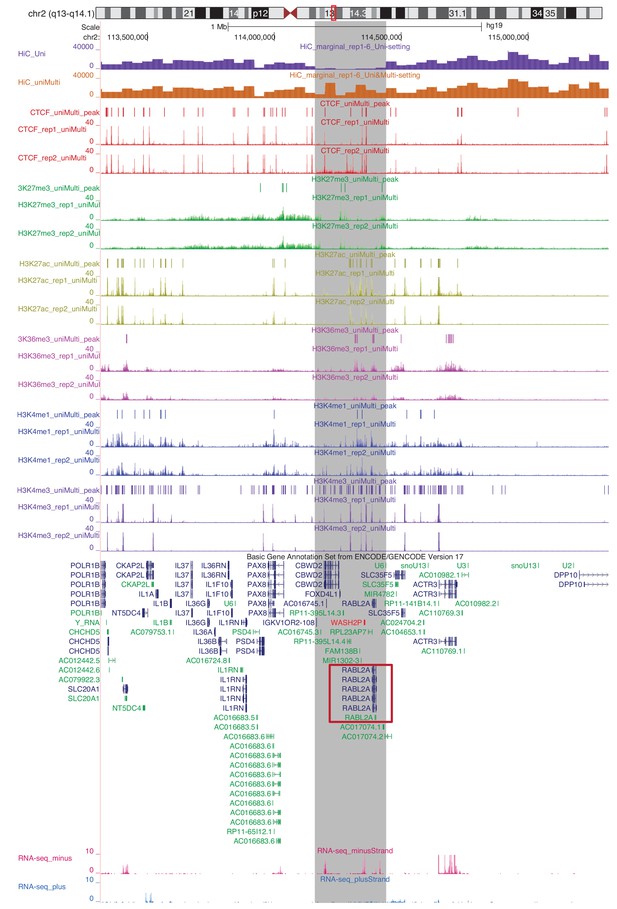
Marginalized Hi-C signal (contact counts aggregated across the genomic coordinates for six replicates of IMR90), ChIP-seq coverage and peaks and gene expression for chr2:113460,000–116,000,000.
Highlighted in grey is a region with significantly different marginal Hi-C signal between Uni-setting and Uni&Multi-setting.
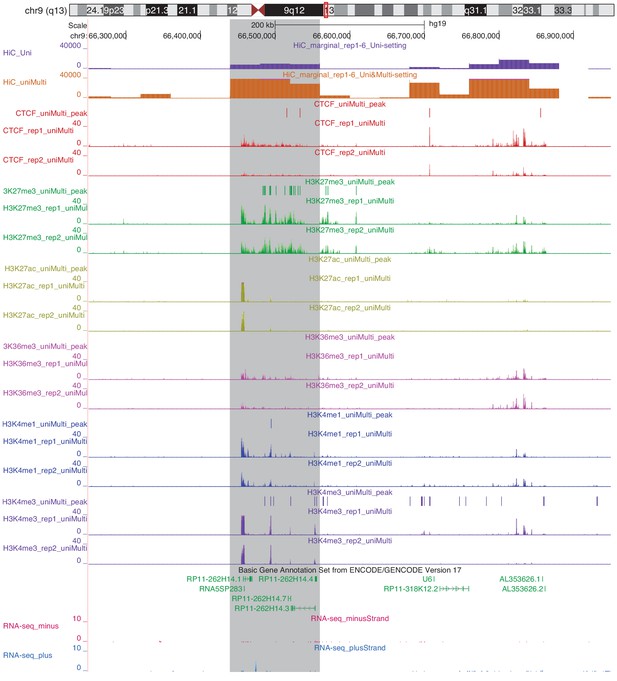
Marginalized Hi-C signal (contact counts aggregated across the genomic coordinates for six replicates of IMR90), ChIP-seq coverage and peaks and gene expression for chr9:66,250,000–66,950,000.
Highlighted in grey is a region with significantly different marginal Hi-C signal between Uni-setting and Uni&Multi-setting.
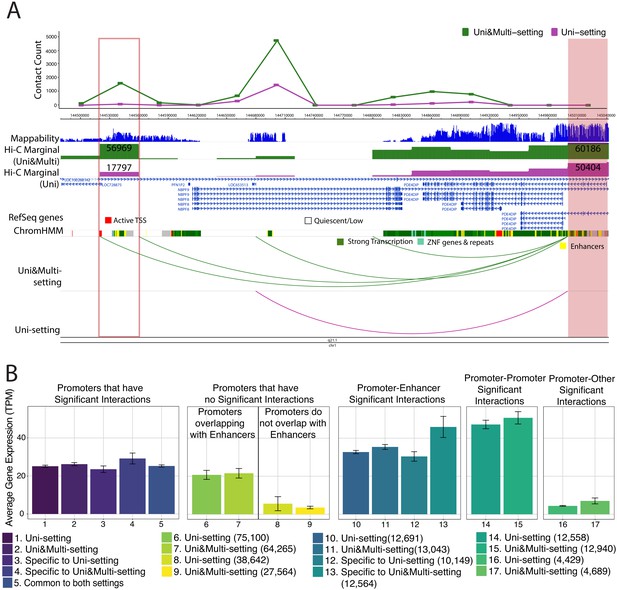
Novel promoter-enhancer interactions are reproducible and associated with actively expressed genes.
(A) mHi-C identifies novel significant promoter-enhancer interactions (green arcs) that are reproducible among at least two replicates in addition to those reproducible under the Uni-setting (purple arcs). Shaded and the boxed regions correspond to the anchor and target bins, respectively. The top track displays the contact counts associated with the anchor bin under Uni- and Uni&Multi-settings. Related chromHMM annotation color labels are added around the track. The complete color labels are consistent with ChromHMM 15-state model at https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html. (B) Average gene expression with standard errors for five different scenarios of interactions that group promoters into six different categories. In the first panel, significant interactions involving promoters are classified into five settings, and the average gene expressions across genes with the corresponding promoters are depicted. The second panel involves two alignment settings and genes without any promoter interactions at 5% FDR. This panel is further separated into two categories: promoters that overlap with enhancer annotated regions and those that do not. The latter one serves as the baseline for average expression. Genes contributing to the third and fourth panel have promoter-enhancer, promoter-promoter interactions at 5% FDR. The fifth panel considers genes promoters of which have significant interactions with non-enhancer and non-promoter regions. Numbers in the parenthesis correspond to the number of transcripts in each category.
-
Figure 4—source data 1
The number of significant promoter-enhancer Hi-C interactions at FDR 5% under Uni-setting and Uni&Multi-setting, respectively, for six replicates of IMR90.
- https://doi.org/10.7554/eLife.38070.049
-
Figure 4—source data 2
Significant promoter-enhancer interactions at FDR 5% under Uni-setting and Uni&Multi-setting for six replicates of IMR90 with the number of contacts.
- https://doi.org/10.7554/eLife.38070.050
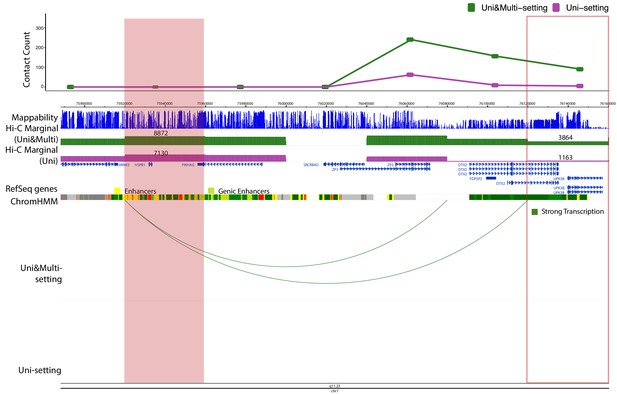
Examples of significant promoter-enhancer interactions reproducible among six replicates under Uni- and Uni&Multi-settings (IMR90) on chromosome 7.
https://doi.org/10.7554/eLife.38070.046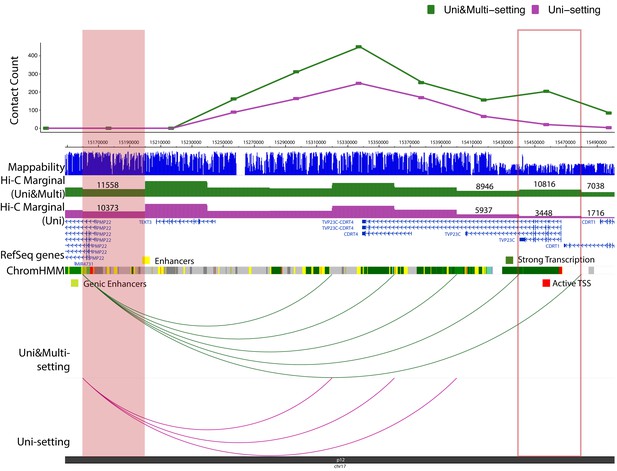
Examples of significant promoter-enhancer interactions reproducible among 6 replicates under Uni- and Uni&Multi-settings (IMR90) on chromosome 17.
https://doi.org/10.7554/eLife.38070.051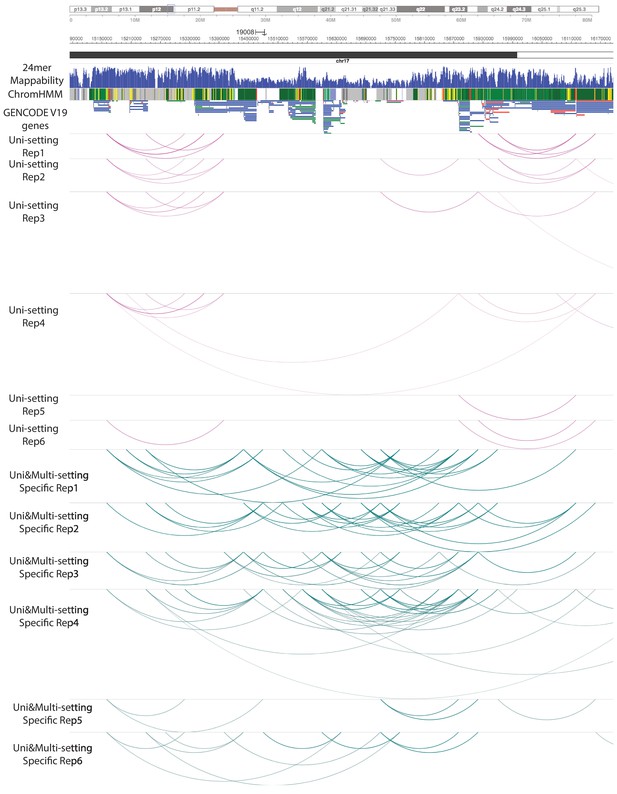
Significant promoter-emhancer interactions under Uni- and Uni&Multi-settings across 6 IMR90 replicates (Chromosome 17).
This is the individual replicate level data for Figure 4—figure supplement 2 in a large genome region.
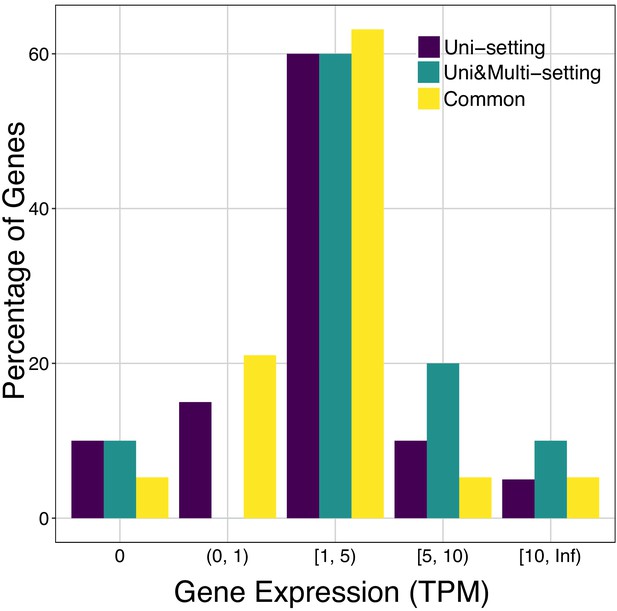
Expression distribution of genes promoters of which have significant promoter interactions (IMR90).
Genes harboring significant interactions (at 5% FDR) within their promoters are grouped into different gene expression categories.
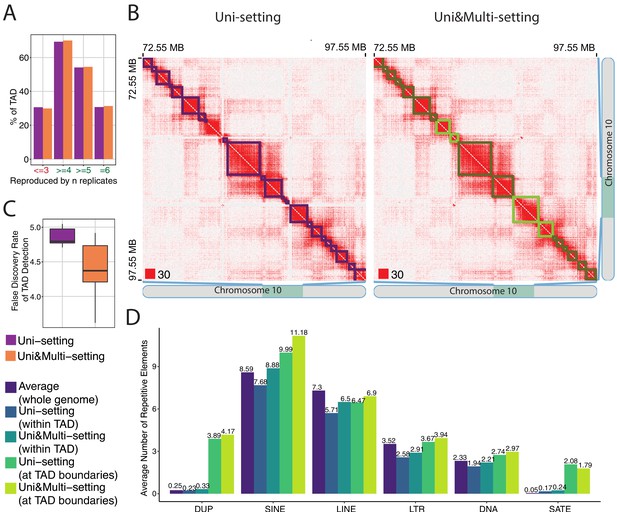
mHi-C rescued multi-reads refine detected topologically associating domains.
(A) Percentage of topologically associating domains (TADs) that are reproducibly detected under Uni-setting and Uni&Multi-setting. TADs that are not detected in at least 4 of the six replicates are considered as non-reproducible. (B) Comparison of the contact matrices with superimposed TADs between Uni- and Uni&Multi-setting for chr10:72,550,000–97,550,000. Red squares at the left bottom of the matrices indicate the color scale. TADs affected by white gaps involving repetitive regions are highlighted in light green. Light green outlined areas correspond to new TAD boundaries. (C) False discovery rate of TADs detected under two settings. TADs that are not reproducible and lack CTCF peaks at the TAD boundaries are labeled as false positives. (D) Average number of repetitive elements at the boundaries of reproducible TADs compared to those within TADs and genomewide intervals of the same size for GM12878 at 5 kb resolution.
-
Figure 5—source data 1
Topologically associating domains detected by DomainCaller (Dixon et al., 2012) under Uni&Multi-setting for six replicates of IMR90.
- https://doi.org/10.7554/eLife.38070.064
-
Figure 5—source data 2
Topologically associating domains detected by Arrowhead (Rao et al., 2014) under Uni&Multi-setting for ten replicates of GM12878.
- https://doi.org/10.7554/eLife.38070.065
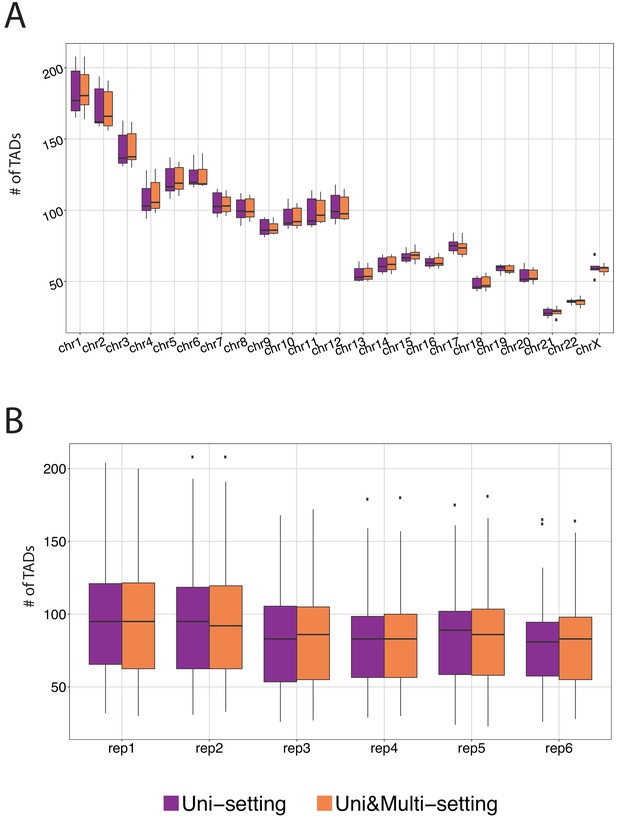
The number of topologically associating domains (TADs) detected in each chromosome under Uni-setting and Uni&Multi-setting (IMR90).
(A) Total number of TADs identified across six replicates for each chromosome. (B) Total number of TADs identified across 23 chromosomes for each replicate.
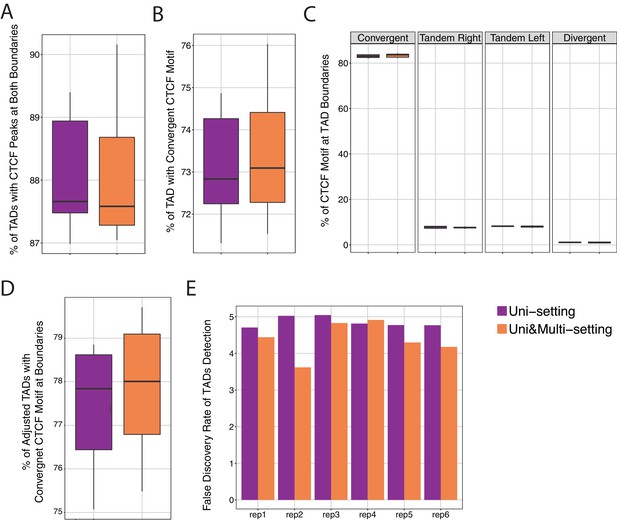
Comparison of CTCF peaks at the boundaries of topologically associating domains (TADs) under Uni-setting and Uni&Multi-setting across six replicates of IMR90.
(A) Percentages of TADs that have CTCF peaks at boundaries. (B) Percentages of TADs that have both CTCF peaks and convergent CTCF motifs at the boundaries. (C) Percentages of four types of CTCF motifs orientations at TAD boundaries. Convergent motif pairs are those with a forward strand motif upstream of TAD boundaries and a reverse strand motif downstream of TAD boundaries. Tandem Right, similarly, represents forward-forward CTCF motif pairs. Tandem Left refers to reverse-reverse motif pairs. Divergent is reverse-forward motif pairs (D) Some TAD boundaries are adjusted under Uni&Multi-setting. Box plots depict the percentage of adjusted TADs that have convergent CTCF motifs at the boundaries. (E). False discovery rate of TADs detected under the two settings. TADs that are not reproducible and lack CTCF convergent motifs at the TAD boundaries are considered as false positives.
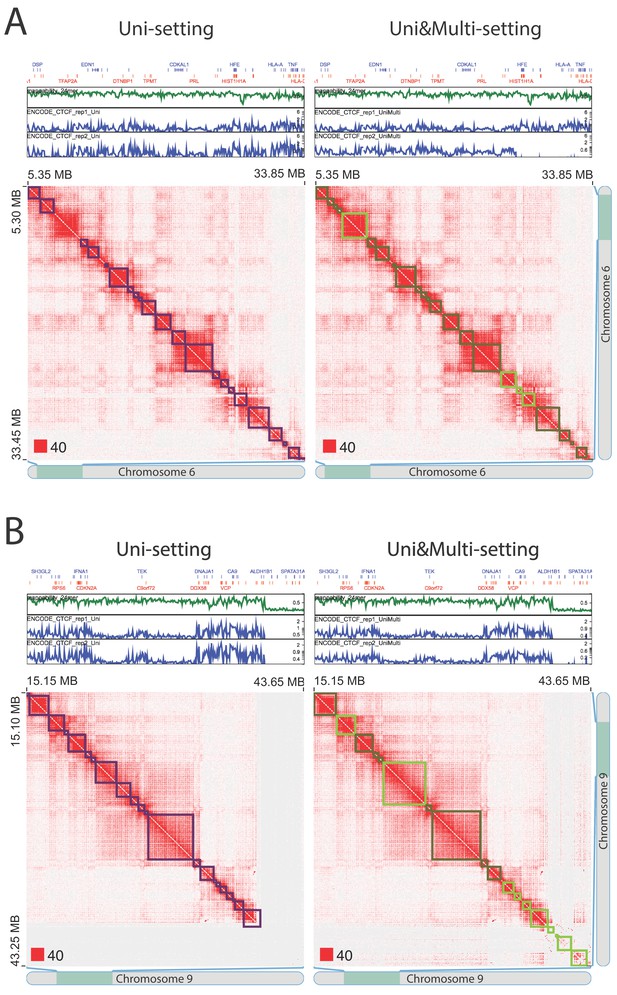
Novel topologically associating domains (TADs) with CTCF peaks at TAD boundaries (IMR90).
Gene tracks, 24mer mappability tracks as well as CTCF peaks are displayed above the contact matrices. (A) Example at chr6:5,350,000–33,850,000. Even in the lack of obviously low mappable contact gaps, multi-reads can enhance the existing interaction signal and reveal detectable TAD structures supported by CTCF peaks. (B) Example on chr9:15,150,000–43,650,000. TAD structure, supported by CTCF peaks at the TAD boundaries, becomes detectable as multi-reads fill in the gap in the contact matrix. Red squares at the left bottom of the matrices indicate the color scale.
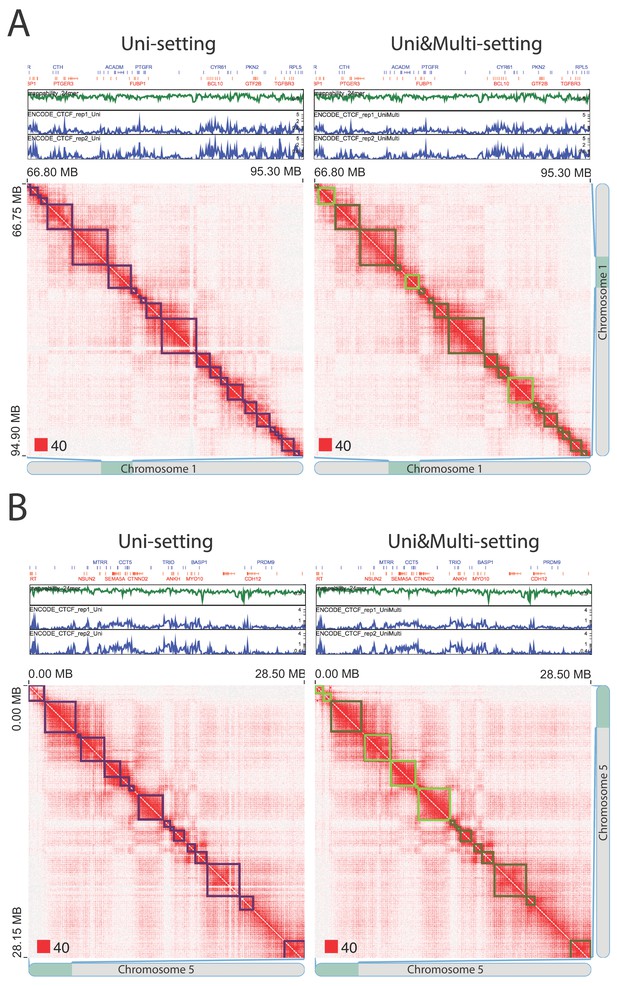
Existing topologically associating domains (TADs) with adjusted boundaries supported by CTCF peaks at the new TAD boundaries (IMR90).
(A) An example from chr1:66,800,000–95,300,000. (B) An example from chr5:0–28,500,000. Red squares at the left bottom of the matrices indicate the color scale.
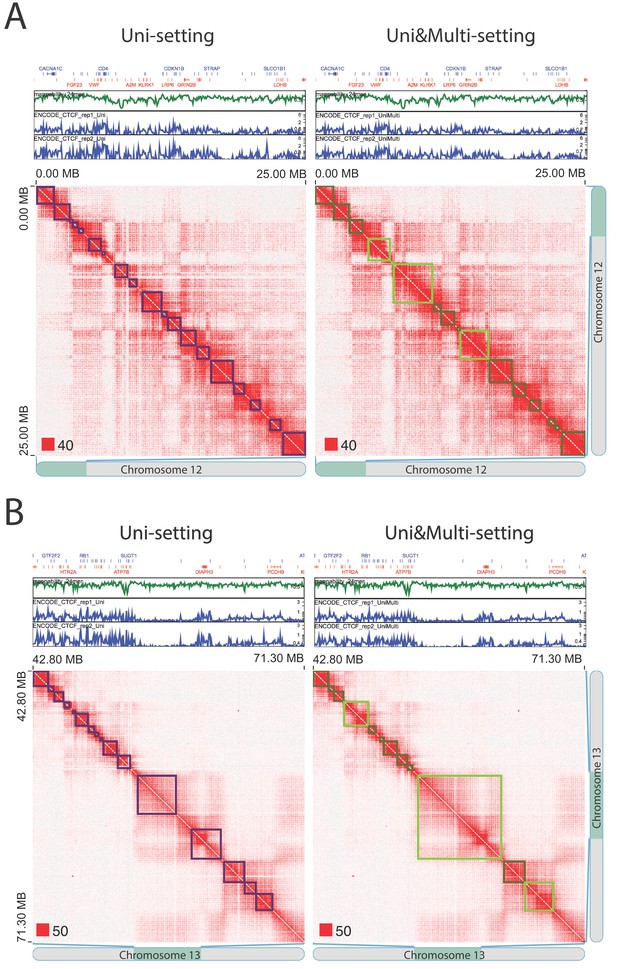
Existing topologically associating domains (TADs) with adjusted boundaries supported by CTCF peaks at the new TAD boundaries (IMR90).
(A) An example from chr12:0–25,000,000. (B) An example from chr13:42,800,000–71,300,000. Red squares at the left bottom of the matrices indicate the color scale.
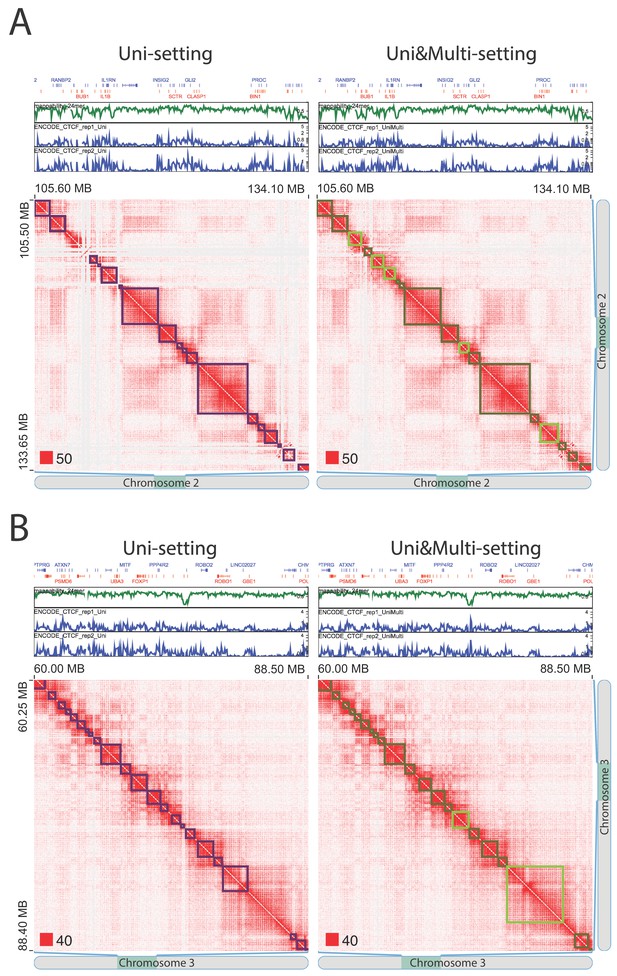
False positive topologically associating domains (TADs) detected by the Uni-setting due to the missing reads in low mappability regions (IMR90).
TADs that are split by white gaps are no longer detected once multi-reads are incorporated, indicating that they are highly likely false positives under the Uni-setting. (A) Example on chr2:105,600,000–134,100,000. (B) Example on chr3:60,000,000–88,500,000. Red squares at the left bottom of the matrices indicate the color scale.
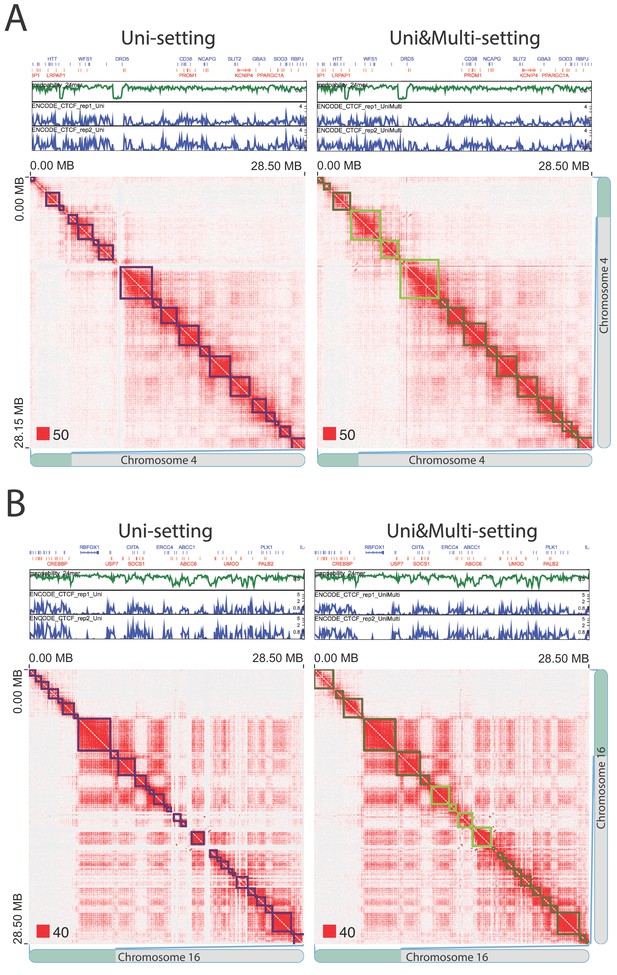
False positive topologically associating domains (TADs) detected by the Uni-setting due to the missing reads in low mappability regions (IMR90).
TADs that are split by white gaps are no longer detected once multi-reads are incorporated, indicating that they are highly likely false positives under the Uni-setting. (A) Example on chr4:0–28,500,000. (B) Example on chr16:0–28,500,000. Red squares at the left bottom of the matrices indicate the color scale.
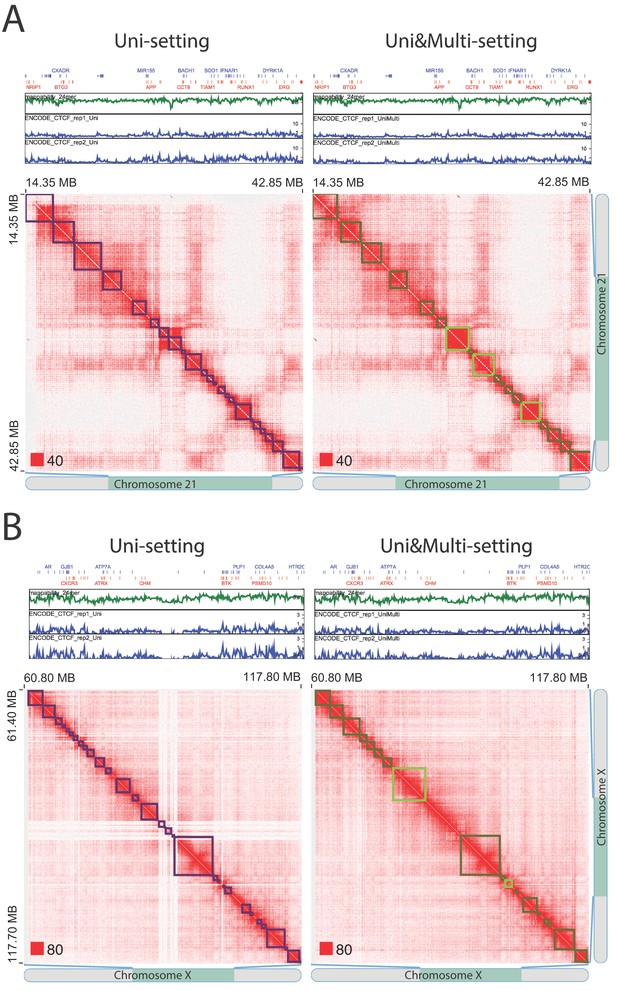
False positive topologically associating domains (TADs) detected by the Uni-setting due to the missing reads in low mappability regions (IMR90).
TADs that are split by white gaps are no longer detected once multi-reads are incorporated, indicating that they are highly likely false positives under the Uni-setting. (A) Example on chr21:14,350,000–42,850,000. (B) Example on chrX:60,800,000–117,800,000. Red squares at the left bottom of the matrices indicate the color scale.
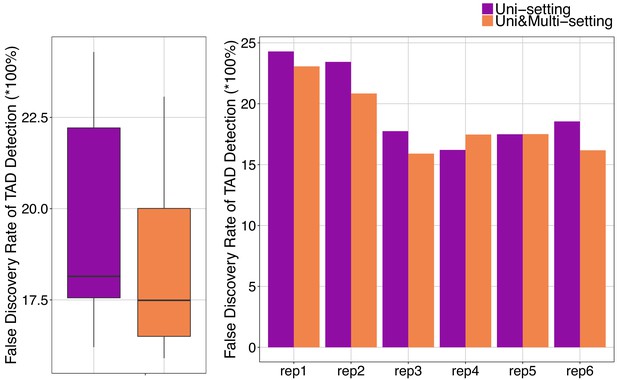
False discovery rate of TADs detected under Uni-setting and Uni&Multi-setting (IMR90).
TADs that are not reproducible are labeled as false positives without considering the CTCF peaks at the TAD boundaries.
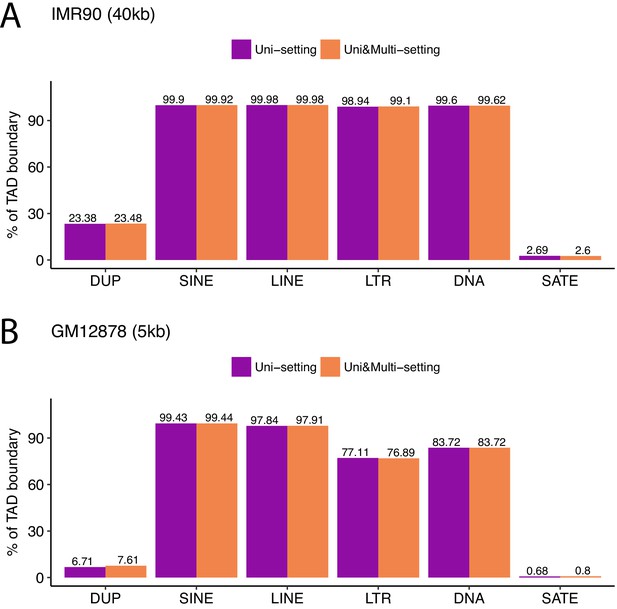
Percentage of TAD boundaries co-localized with different types of repetitive elements under Uni-setting and Uni&Multi-setting for IMR90 at 40 kb and GM12878 at 5 kb.
https://doi.org/10.7554/eLife.38070.062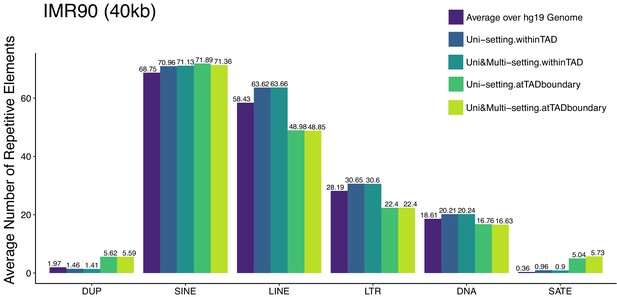
Average number of repetitive elements at the reproducible topologically associating domains detected under Uni-setting and Uni&Multi-setting for IMR90 at 40 kb.
Such enrichment is compared to those within TADs and genomewide intervals of the same size.
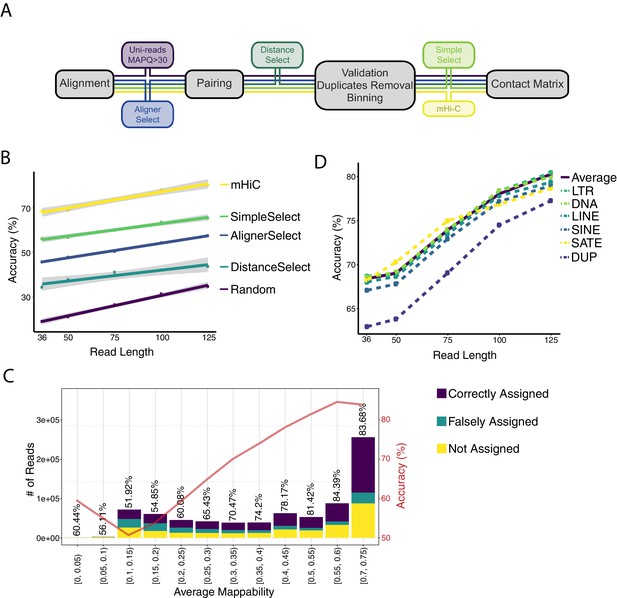
Assessing the accuracy of mHi-C allocation by trimming experiments with the A549 study set of 151 bp reads.
(A) Intuitive heuristic strategies (AlignerSelect, DistanceSelect, SimpleSelect) for model-free assignment of multi-reads at various stages the of Hi-C analysis pipeline. (B) Accuracy of mHi-C in allocating trimmed multi-reads with respect to trimmed read length, compared with model-free approaches as well as random selection as a baseline. (C) Allocation accuracy with respect to mappability for 75 bp reads. Red solid line depicts the overall accuracy trend. ‘Not assigned’ category refers to multi-reads with a maximum posterior probability of assignment 0.5. (D) mHi-C accuracy among different repetitive element classes.
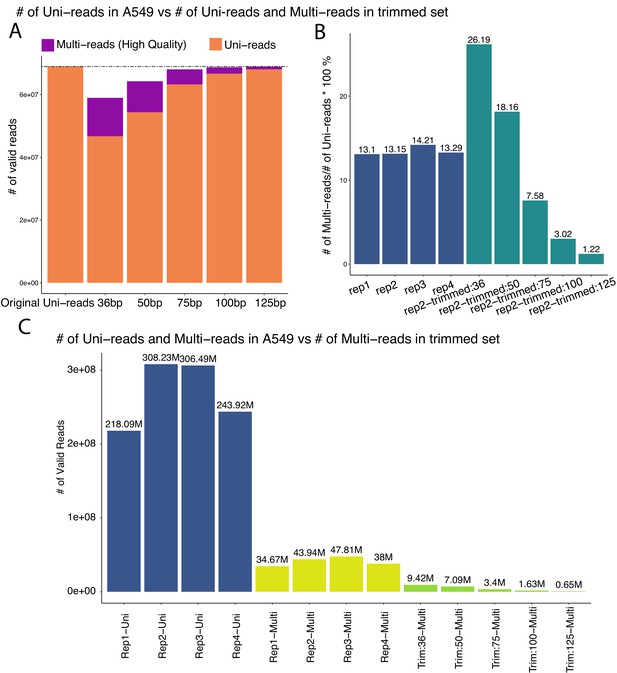
Summary of the sequencing depths of the full length and trimmed datasets of A549.
(A) Numbers of uni-reads and multi-reads across trimmed read lengths compared to the sequencing depth of uni-reads of full read length A549 dataset (replicate 2), not including uni-reads rescued from chimeric reads. (B) Percentage of multi-reads over uni-reads in the full-length A549 datasets (four replicates, excluding uni-reads from chimeric reads) and the trimmed datasets. Multi-to-uni percentages of the read sets in the trimmed datasets cover the range of the percentages observed in the study datasets. (C) Numbers of multi-reads from different trimming lengths compared to sequencing depths of the full read length A549 datasets. For the trimming setting (ii) described in Materials and methods, multi-reads (green bars) added to the uni-read datasets (blue bars) constitute a smaller percentage of the sequencing depth while enabling analysis at the higher resolution of 10 kb.
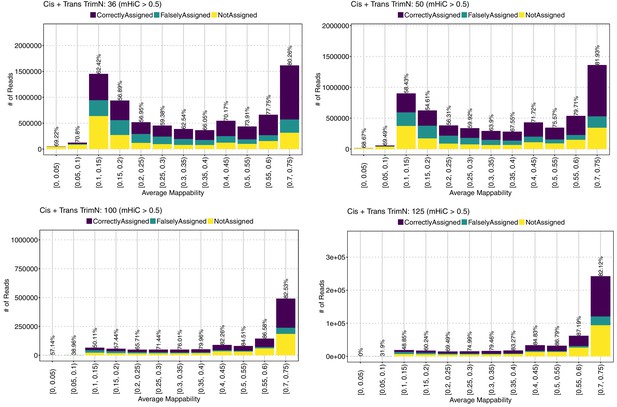
Allocation accuracy at the 40 kb resolution among different mappability regions for trimmed reads of varying lengths.
mHi-C > 0.5 refers to the fact that only the allocations with posterior probability of assignment greater than 0.5 are evaluated.
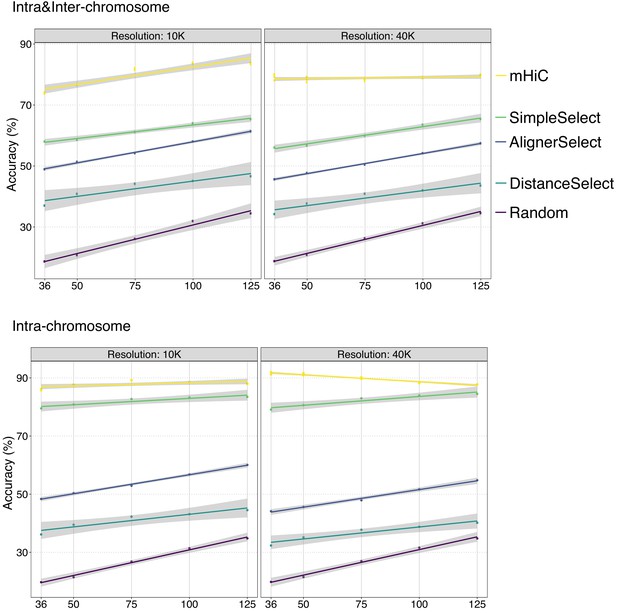
Intra-chromosomal and intra&inter-chromosomal allocation accuracy with respect to trimmed read length using uni-reads of replicate 1, 3, and 4 combined with multi-reads of replicate 2 (trimming setting (ii)).
https://doi.org/10.7554/eLife.38070.069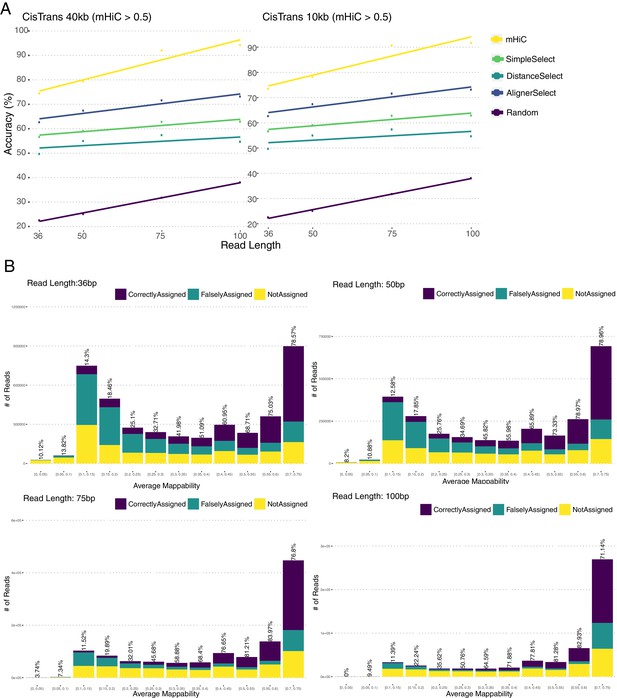
Evaluating accuracy of mHi-C allocation with simulations.
(A) Allocation accuracy of mHi-C at 40 kb and 10 kb resolutions. (B) Allocation accuracy for simulated reads of different lengths among regions of varying mappability.
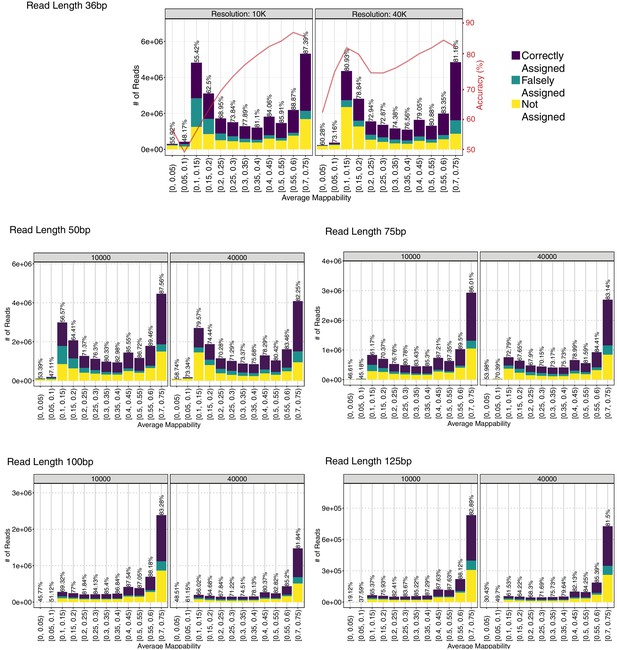
Allocation accuracy across different mappability regions for trimmed reads of 36 bp, 50 bp, 75 bp, 100 bp, and 125 bp, using uni-reads of replicate 1, 3, and 4, respectively.
https://doi.org/10.7554/eLife.38070.071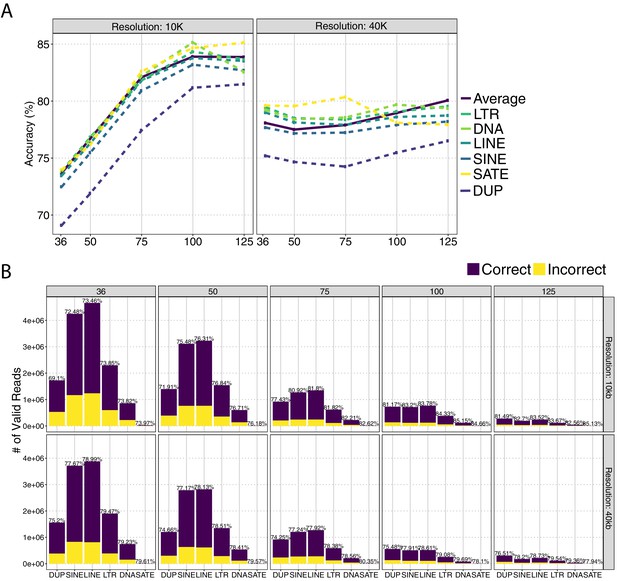
Allocation accuracy across different classes of repetitive elements at 10 kb and 40 kb resolutions using uni-reads of replicates one, three, and combined with multi-reads of replicate two (trimming setting (ii)).
(A) mHi-C accuracy among different types of repetitive element classes with respect to trimmed read length. (B) Allocation accuracy across different classes of repetitive elements at 10 kb and 40 kb resolutions using uni-reads of replicate 1, 3, and 4 combined with multi-reads of replicate 2 (trimming setting (ii)).
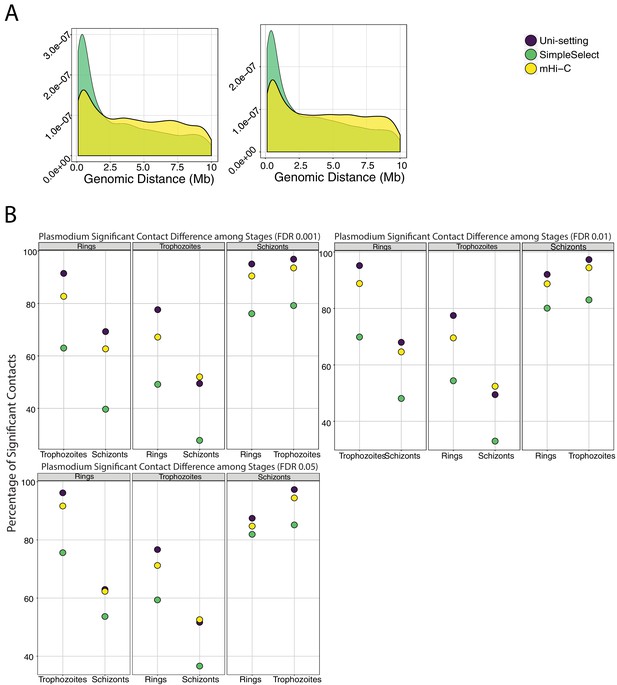
Comparison of significant interactions with respect to genomic distance and life stages between SimpleSelect and mHi-C.
(A) Comparison of genomic distance distributions of significant interactions between SimpleSelect and mHi-C for IMR90 rep5 (FDR 0.001). (B) Comparison of significant interactions among the three life stages of P. falciparum. Y-axis in each panel, namely rings, trophozoites, and schizonts, depicts the percentage of contacts that are significant only in the panel condition compared to the other two conditions. Under varying FDR thresholds, mHi-C and Uni-setting tend to have similar percentages of differential interactions among ring - trophozoites - schizonts plasmodium life stages. In contrast, SimpleSelect tends to underestimate differential interactions due to over-emphasizing contact distance prior.
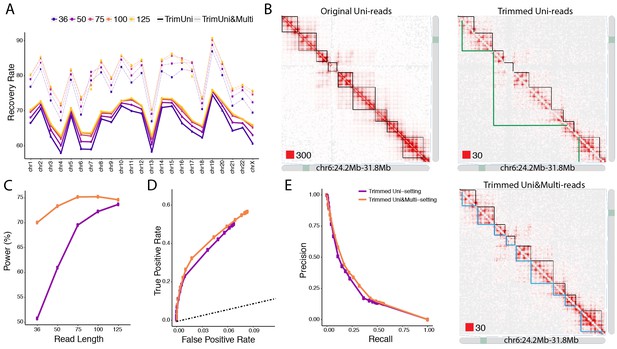
Trimmed uni- and multi-reads to recover the original contact matrix of the longer read dataset A549.
(A) mHi-C rescued multi-reads of the trimmed dataset along with trimmed uni-reads lead to contact matrices that are significantly more similar to original contact matrices compared to only using trimmed uni-reads. (B) TAD detection on chromosome six with the original longer uni-reads contact matrix with black TAD boundaries, trimmed uni-reads (36 bp) contact matrix with green TAD boundaries, and trimmed uni- and multi-reads (36 bp) contact matrix with blue TAD boundaries. (C) The power of recovering top 10,000 significant interactions of full read length dataset using trimmed reads under FDR10%. (D, E) Receiver Operating Characteristic (ROC) and Precision-Recall (PR) curves for trimmed Uni- and Uni&Multi-setting. The ground truth for these curves is based on the significant interactions identified by the full read length dataset at FDR of 10%. The dashed line is y = x.
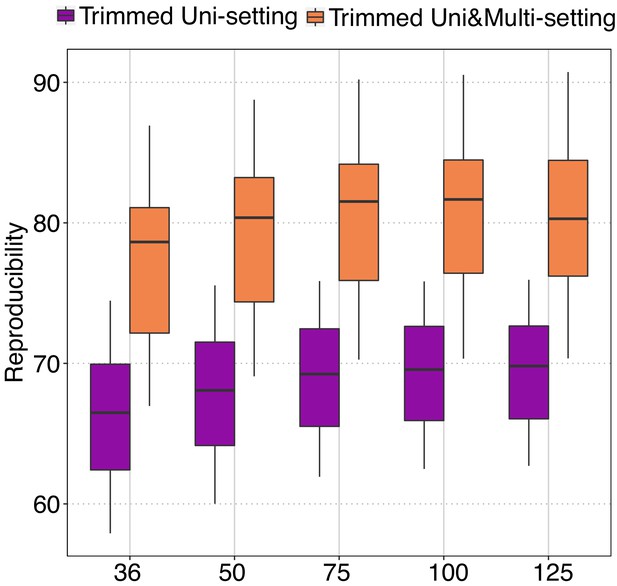
Reproducibility of trimmed Uni-setting and trimmed Uni&Multi-setting across different read lengths at 40 kb resolution.
https://doi.org/10.7554/eLife.38070.075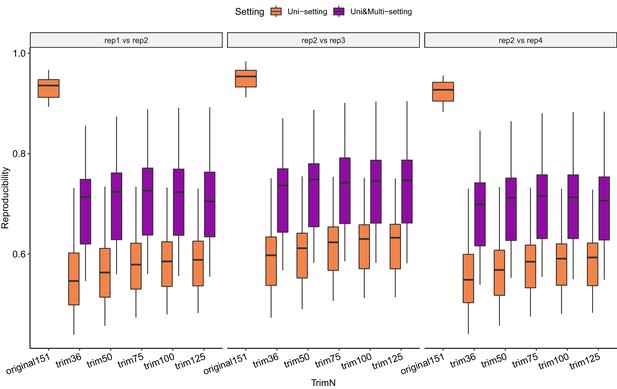
Reproducibility comparison between original Uni-setting and trimmed Uni-setting and Uni&Multi-setting across replicates at 40 kb resolution.
https://doi.org/10.7554/eLife.38070.076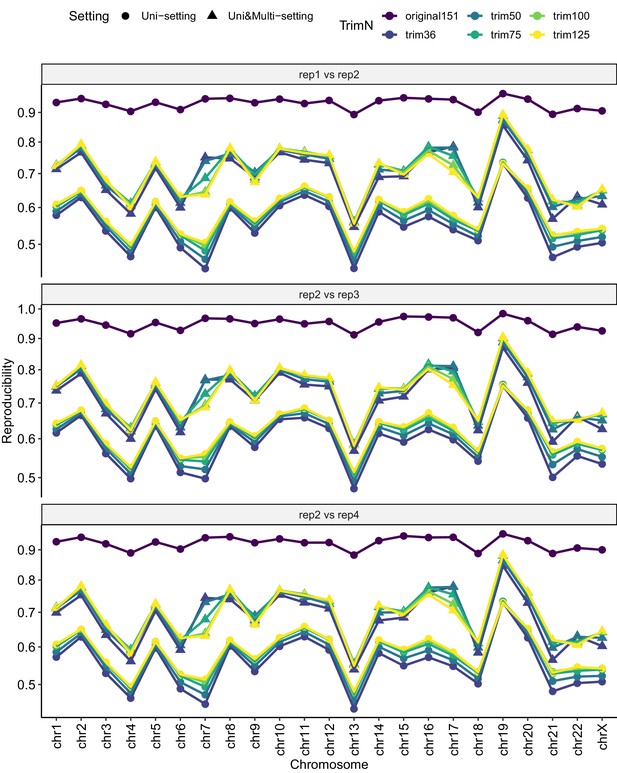
Reproducibility comparison between original Uni-setting and trimmed Uni- and Uni&Multi-settings across chromosomes at 40 kb resolution.
https://doi.org/10.7554/eLife.38070.077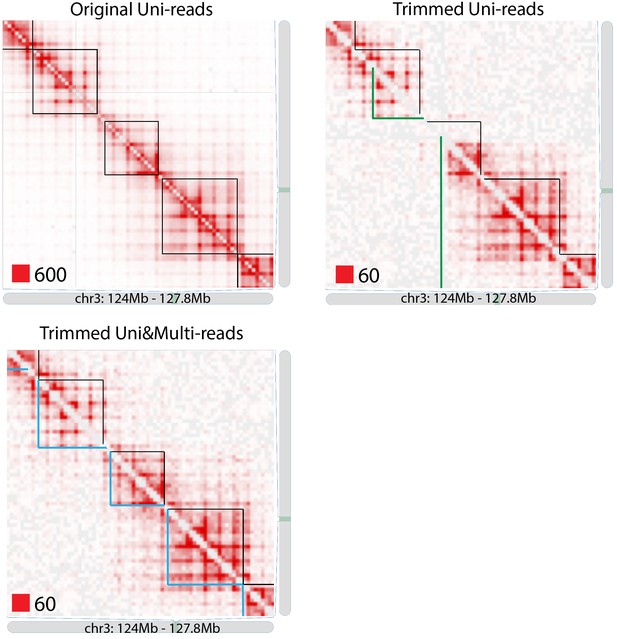
TAD detection on chromosome 3 of original longer uni-reads contact matrix with black TAD boundaries, trimmed uni-reads (36 bp) contact matrix with green TAD boundaries and trimmed uni- and multi-reads (36 bp) contact matrix with blue TAD boundaries.
https://doi.org/10.7554/eLife.38070.078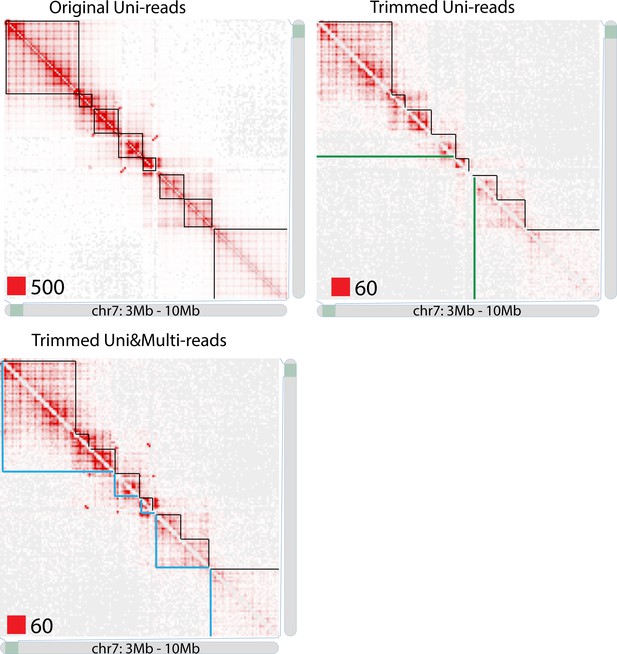
TAD detection on chromosome 7 of original longer uni-reads contact matrix, trimmed uni-reads (36 bp) contact matrix and trimmed uni- and multi-reads (36 bp) contact matrix.
https://doi.org/10.7554/eLife.38070.079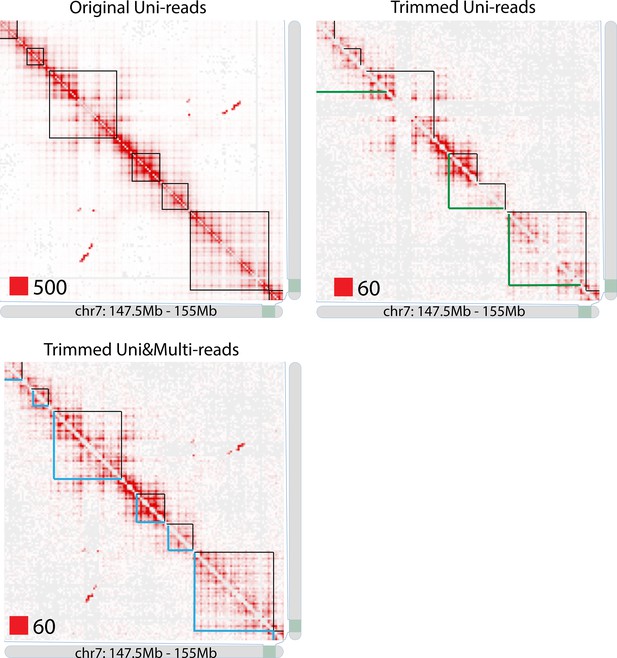
TAD detection on chromosome 7 of original longer uni-reads contact matrix, trimmed uni-reads (36 bp) contact matrix and trimmed uni- and multi-reads (36 bp) contact matrix.
https://doi.org/10.7554/eLife.38070.080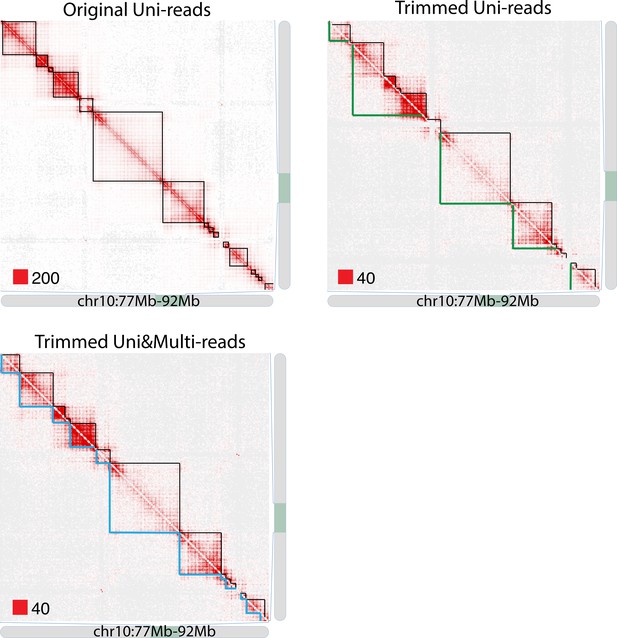
TAD detection on chromosome 10 of original longer uni-reads contact matrix with black TAD boundaries, trimmed uni-reads (36 bp) contact matrix with green TAD boundaries and trimmed uni- and multi-reads (36 bp) contact matrix with blue TAD bounaries.
https://doi.org/10.7554/eLife.38070.081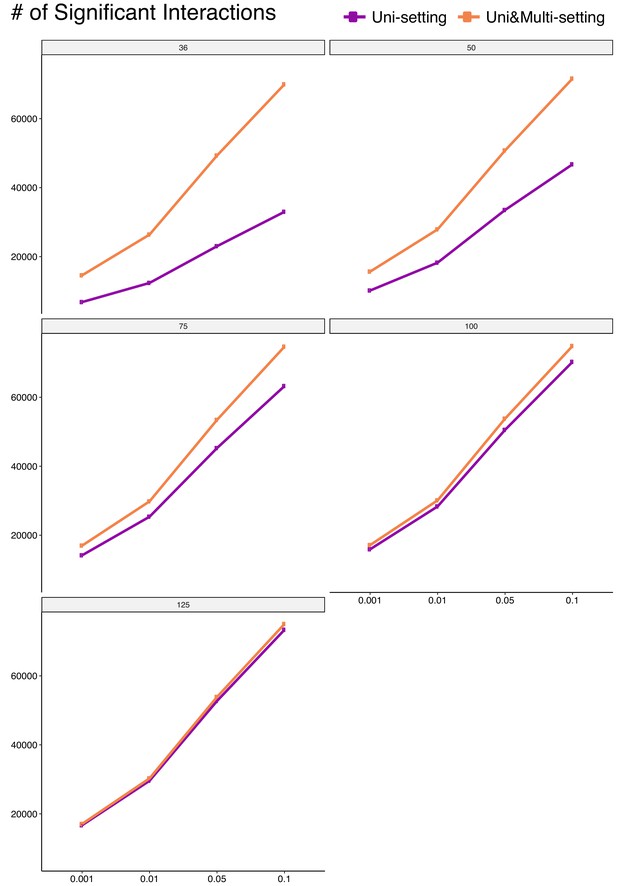
Numbers of significant interactions identified with trimmed reads under Uni- and Uni&Multi-settings at FDR 0.1%, 1%, 5%, 10%.
https://doi.org/10.7554/eLife.38070.082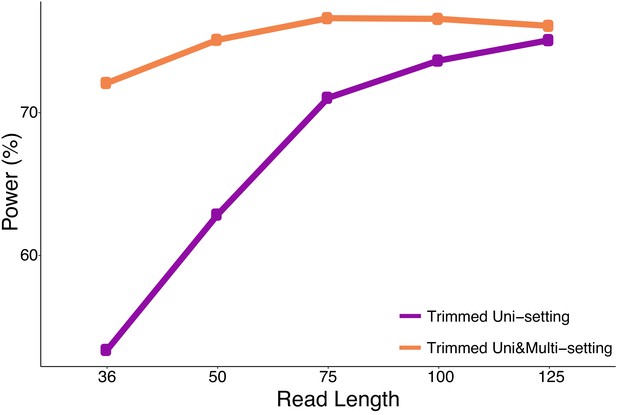
Power is computed as the percentage of top 10,000 significant interactions of the full read length dataset detected by the analysis of trimmed read datasets under FDR of 10%.
https://doi.org/10.7554/eLife.38070.083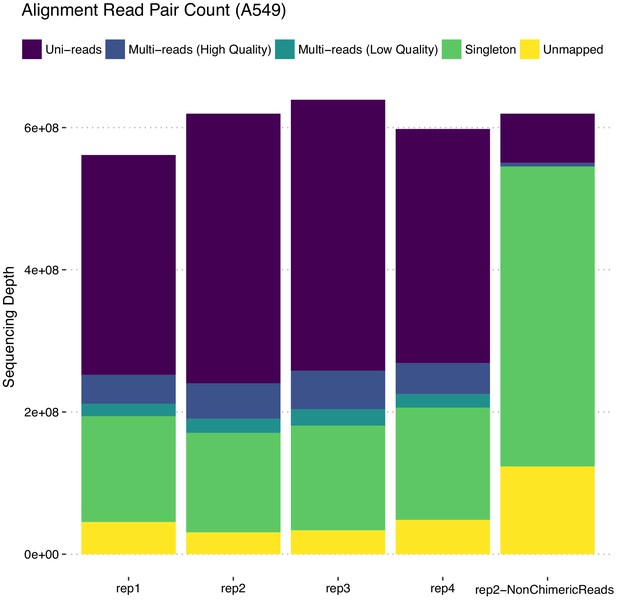
Comparison of read decompositions of ‘rep2’ with ‘rep2-NonChimericReads’ sequencing depths indicates that in the full read length dataset, a large proportion of the uni-reads are due to rescued chimeric reads (i.e., these are Uni- or Multi-reads in rep2 and Singletons in rep2-NonChimericReads).
Downstream analysis of trimming experiments beyond evaluating the accuracy of multi-read assignments utilized Uni-reads of rep2 displayed here for deriving gold standard interaction sets.
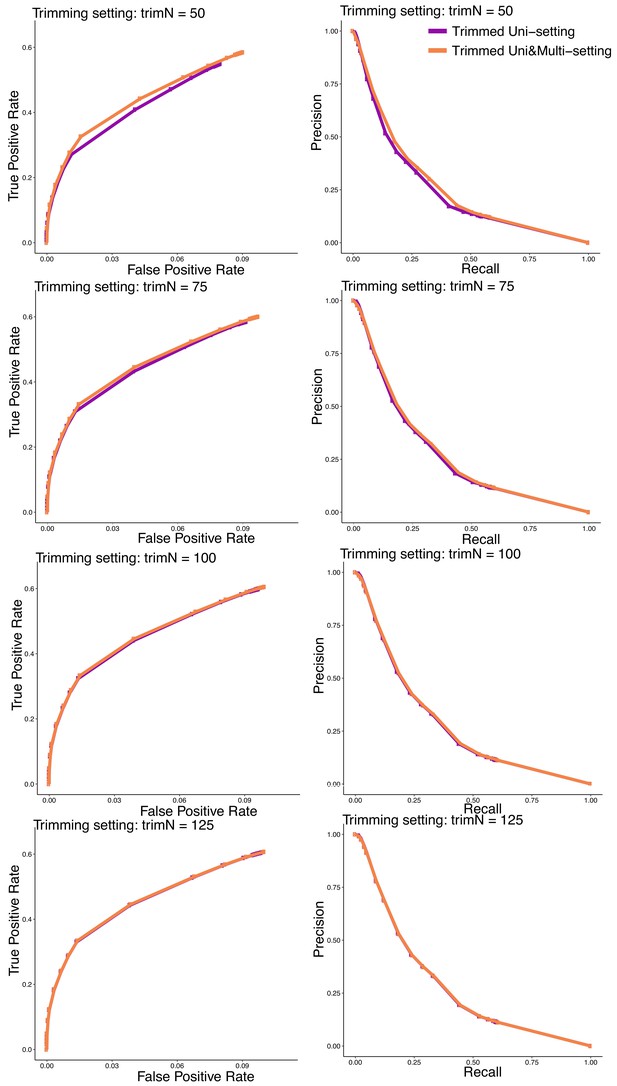
ROC and PR curves for detection of full read length dataset significant interactions by the analysis of trimmed read datasets under the trimmed Uni- and Uni&Multi-settings at read lengths of 50 bp, 75 bp, 100 bp, and 125 bp.
In the ROC curves, the false positive rates are displayed up until 10% as the values of both the x- and y-axis rates shoot up to one after this value. Ground truth for these curves is based on the significant interactions identified by the full read length dataset at FDR of 10%.
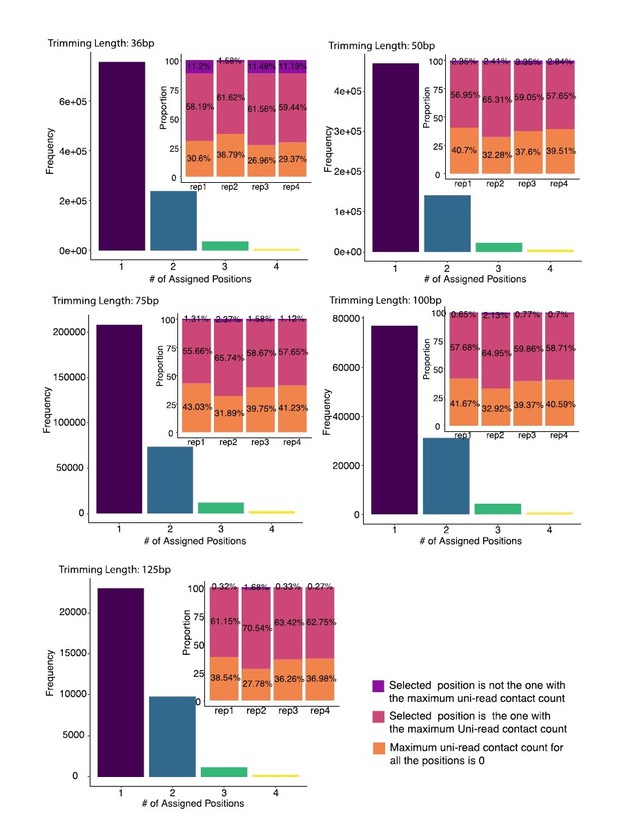
Distribution of numbers of distinct assignment positions for incorrectly assigned multi-reads across 4 replicates of A549 at 40 kb resolution.
mHi-C utilizes the trimmed uni-reads individually from four replicates to assign the same trimmed multi-reads from replicate 2 in the analysis of each trimmed replicate experiment. Each multi-read is either assigned to its true origin or can be incorrectly assigned to up to four different positions. Multi-reads that were incorrectly assigned to the same position are further stratified into three categories as: (i) assigned to the candidate position with the highest uni-reads signal (pink) (7.77% of all the multi-reads for trimming at 36bp); (ii) not assigned to the highest uni-reads enriched position (purple) (1.45% of all the multi-reads for trimming at 36bp); (iii) none of the candidate positions have any uni-reads (orange) (4.64% of all the multi-reads for trimming at 36bp).
Tables
Hi-C Data Summary.
https://doi.org/10.7554/eLife.38070.010| Cell line | Replicate | Read length (bp) | Restriction enzyme | HiC protocol | Source | Resolution (kb) |
|---|---|---|---|---|---|---|
| IMR90 | rep1-6 | 36 | HindIII | dilution | (Jin et al., 2013) | 40 |
| GM12878 | rep2-9 | 101 | MboI | in situ | (Rao et al., 2014) | 5, 10*, 40* |
| GM12878 | rep32, rep33 | 101 | DpnII | in situ | (Rao et al., 2014) | 5 |
| A549 | rep1-4 | 151 | MboI | in situ | (Dixon et al., 2018) | 10, 40 |
| ESC(2012) | rep1, rep2 | 36 | HindIII | dilution | (Dixon et al., 2012) | 40 |
| ESC(2017) | rep1-4 | 50 | DpnII | in situ | (Bonev et al., 2017) | 10, 40 |
| Cortex | rep1-4 | 50 | DpnII | in situ | (Bonev et al., 2017) | 10, 40 |
| P. falciparum | three stages | 40 | MboI | dilution | (Ay et al., 2014b) | 10, 40 |
-
*Replicates 2, 3, 4, and 6 of the GM12878 cell line datasets were process at 10 kb and 40 kb resolutions.
Additional files
-
Supplementary file 1
Hi-C and mHi-C terminology.
- https://doi.org/10.7554/eLife.38070.086
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38070.087





