Elucidating the mitochondrial proteome of Toxoplasma gondii reveals the presence of a divergent cytochrome c oxidase
Figures
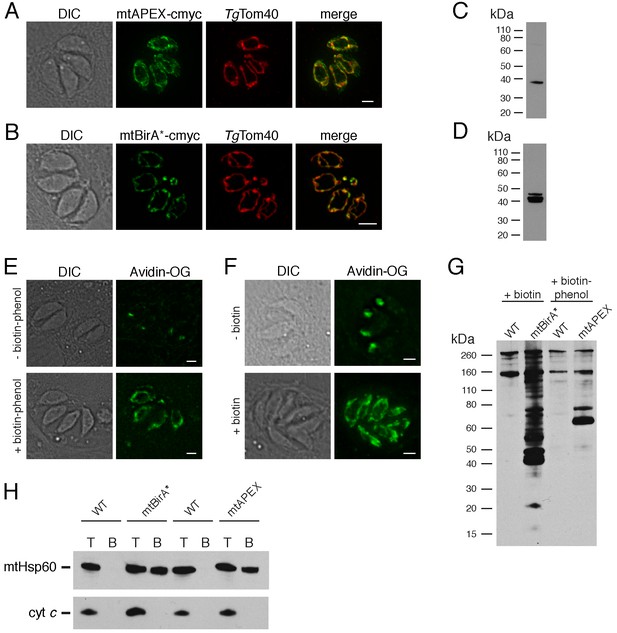
Biotinylation of mitochondrial matrix proteins in T.gondii parasites expressing mtAPEX and mtBirA*.T
(A–B) Immunofluorescence assays of parasites expressing c-myc-tagged, mitochondrially-targeted APEX (A) and BirA* (B), labelled with anti-c-myc (green) and the mitochondrial marker TgTom40 (red). Scale bars are 2 µm. (C–D) Western blots of parasites expressing c-myc-tagged, mitochondrially-targeted APEX (C) and BirA* (D), labelled with anti-c-myc. (E) Oregon Green-conjugated avidin (Avidin-OG) labelling of T. gondii parasites expressing mtAPEX, and cultured in the absence (top) or presence (bottom) of biotin-phenol and H2O2. Biotinylated proteins are labelled in green. (F) Avidin-OG labelling of T. gondii parasites expressing mtBirA*, and cultured in the absence (top) or presence (bottom) of biotin. Biotinylated proteins are labelled in green. Scale bars are 2 µm. (G) Neutravidin-HRP protein blot of WT, mtBirA* or mtAPEX parasites cultured in the presence of biotin or biotin-phenol. (H) Western blots of the mitochondrial matrix marker mtHsp60 and the mitochondrial intermembrane space marker cyt c in WT, mtBirA* or mtAPEX parasites cultured in the presence of biotin (lanes 1 – 4) or biotin-phenol (lanes 5 – 8). Parasites were either harvested following treatment to yield the total (T) protein fraction, or biotinylated proteins were purified on a streptavidin-agarose column to yield the bound (B) fraction.
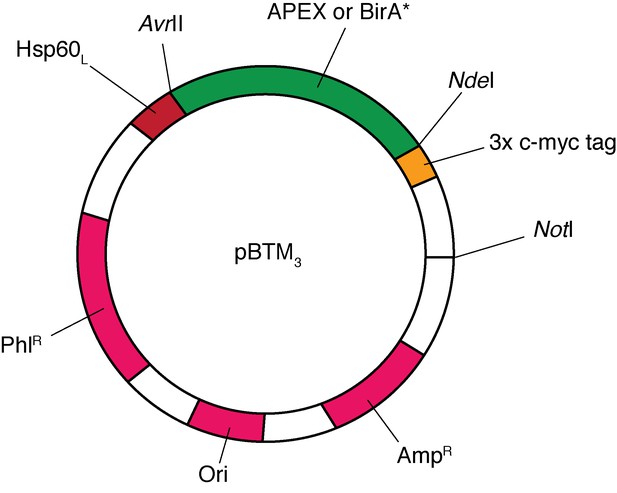
Map of the pBTM3 plasmid vector, showing the AvrII, NdeI and NotI cut sites between which the APEX and BirA* cassettes were ligated (open reading frame of enzyme shown in green), the position of mitochondrial targeting leader sequence of TgHsp60 (Hsp60L; red), the 3x c-myc tag (yellow), and the positions of the phleomycin resistance marker (PhlR) for T.gondii selection, the ampicillin resistance marker for E. coli selection (AmpR), and the origin of replication (Ori; all magenta).
Note: vector is not drawn to scale.
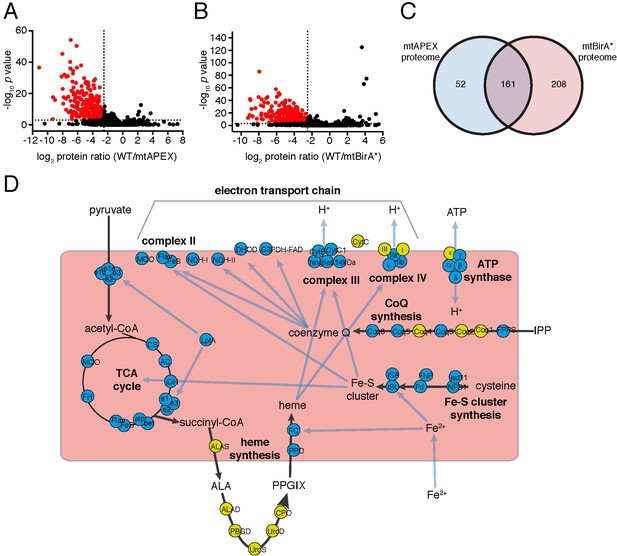
The mitochondrial proteome of T.gondii.
(A–B) Volcano plots showing the log2 protein ratios vs –log10 p values of biotinylated proteins in WT compared to mtAPEX (WT/APEX) samples (A) and in WT compared to mtBirA* samples (WT/BirA*) (B) following the quantitative pipeline analysis. Proteins were deemed to be enriched in the mitochondrion if the log2 fold change in protein expression was ≤−2.5 and the p value ≤ 0.001 (red). (C) Venn diagram of the mtAPEX and mtBirA* proteomes. 161 proteins were identified in both proteomes, while 52 were unique to the mtAPEX proteome and 208 unique to the mtBirA* proteome. (D) Metabolic map of expected mitochondrial proteins (circles), showing proteins present (blue) and absent (yellow) from the T. gondii mitochondrial proteome. Black arrows represent the flow of metabolites through metabolic pathways in the mitochondrion, and blue arrows depict the flow of ions, minerals or metabolic pathway products.
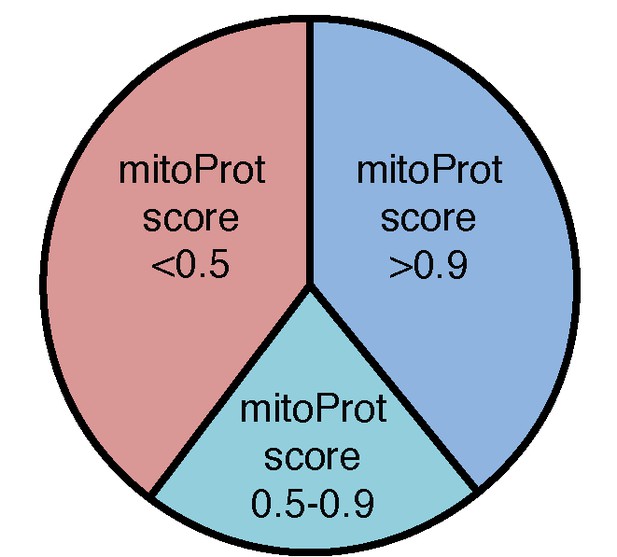
Analysis of putative mitochondrial targeting peptides in the T.gondii mitochondrial proteome.
Pie chart depicting mitochondrial targeting peptide predictions of the T. gondii mitochondrial proteome using MitoProt II. Proteins with high (>0.9; blue), medium (0.5 – 0.9; aqua) and low (<0.5; pink) prediction scores are shown.
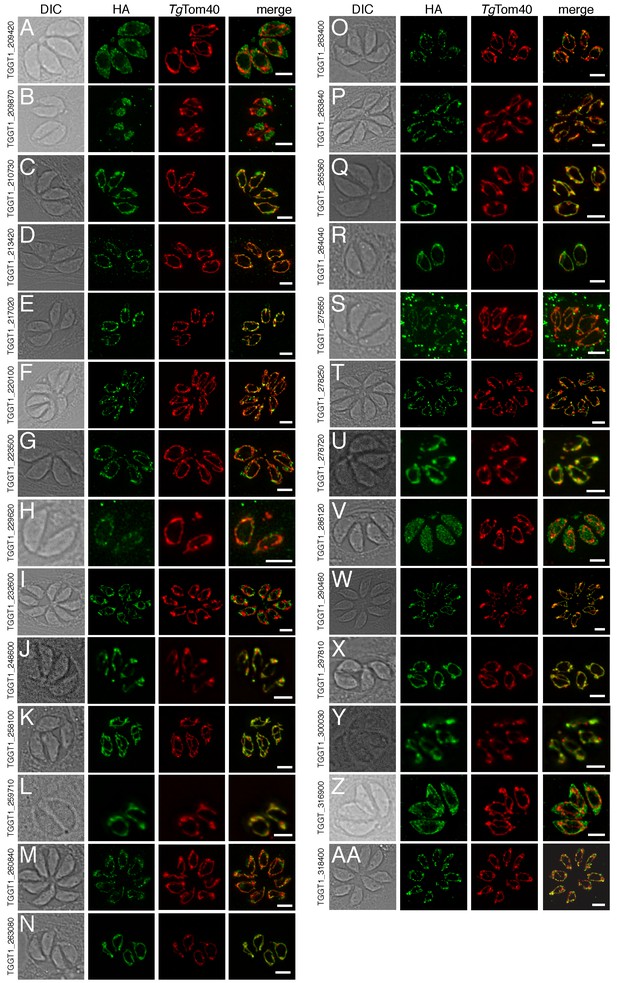
The localization of novel proteins from the T.gondii mitochondrial proteome.
(A–AA) Proteins with no previously determined localization in T. gondii were selected from the mitochondrial proteome, and the corresponding gene was tagged at the 3’-terminus of the open reading frame with a HA tag. Immunofluorescence assays depict HA-tagged proteins (green) co-labelled with the mitochondrial marker TgTom40 (red). The http://toxodb.org gene identification number is depicted for every gene that was tagged.
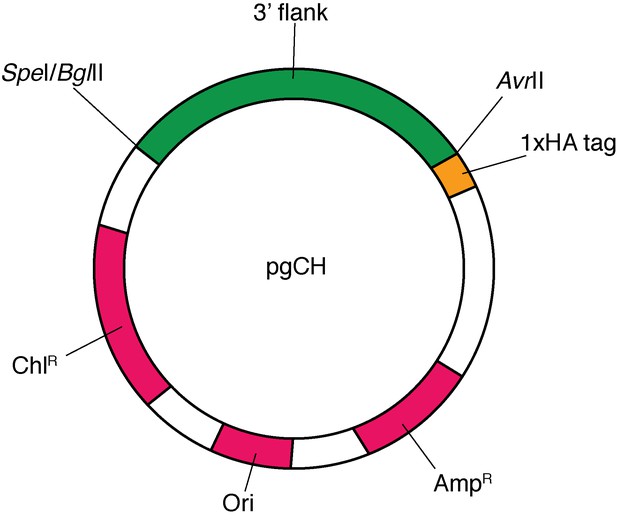
Map of the pgCH plasmid vector, showing the SpeI, BglII and AvrII cut sites between which the 3’ flanks of target genes were ligated (green), the position of the 1x HA tag (yellow), and the positions of the chloramphenicol resistance marker (ChlR) for T.gondii selection, the ampicillin resistance marker for E. coli selection (AmpR), and the origin of replication (Ori; all magenta).
Note: vector is not drawn to scale.
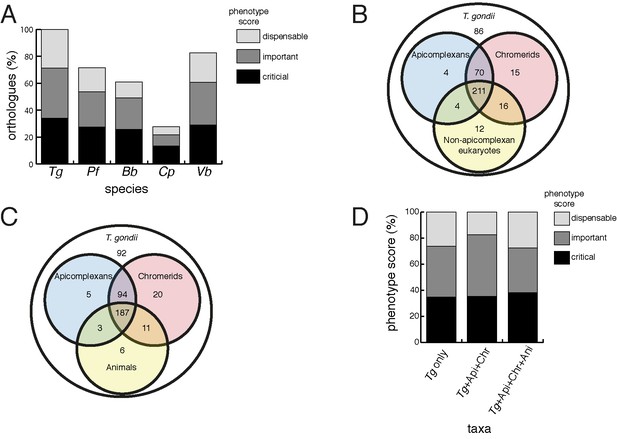
Orthology analyses of proteins from the T.gondii mitochondrial proteome reveal that many mitochondrial proteins are restricted to T. gondii and related organisms, and that most are important for parasite survival.
(A) Bar graph depicting the percentage of orthologs from the mitochondrial proteome of T. gondii (Tg) found in P. falciparum (Pf), B. bovis (Bb), C. parvum (Cp) and V. brassicaformis (Vb). Phenotype scores are indicated with shading, and reveal that most ortholog groups in each category are important or critical for tachyzoite growth. (B–C) Venn diagram depicting ortholog groupings from the mitochondrial proteome of T. gondii compared to (B) non-coccidian apicomplexans, chromerids and eukaryotes, or (C) non-coccidian apicomplexans, chromerids and animals. (D) Bar graph depicting distribution of phenotype scores in genes belonging to ortholog groups found only in T. gondii and other coccidians (Tg only), in T. gondii, non-coccidian apicomplexans and chromerids (Tg+ Api+ Chr), and in T. gondii, non-coccidian apicomplexans, chromerids and animals (Tg+ Api+ Chr+ Api). In (A) and (D), genes with phenotype scores of >-2 were considered dispensable, −2 to −4 were considered important, and <-4 were considered critical.
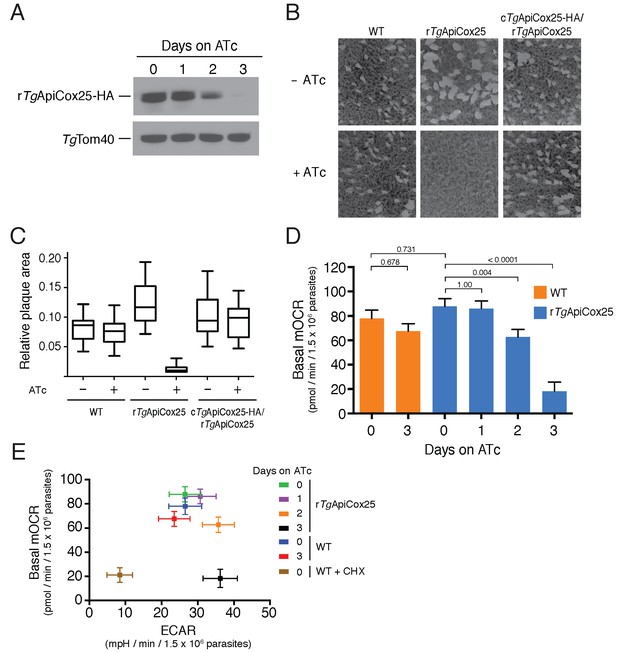
TgApiCox25 is important for parasite growth and mitochondrial O2 consumption.
(A) Western blot of proteins extracted from rTgApiCox25-HA parasites grown in the absence of ATc, or in ATc for 1–3 days, and detected using anti-HA antibodies (top) and anti-TgTom40 (as a loading control; bottom). (B) Plaque assays measuring growth of WT, rTgApiCox25 and complemented cTgApiCox25-HA/rTgApiCox25 parasites cultured in the absence (top) or presence (bottom) of ATc. Assays are from a single experiment and are representative of 3 independent experiments. (C) Quantification of plaque size from WT, rTgApiCox25 and complemented cTgApiCox25-HA/rTgApiCox25 parasites grown in the absence or presence of ATc for 9 days. Box and whisker plots depict the median plaque size (centre line), the 25th and 75th percentiles (box) and the 5th and 95th percentiles (lines). Data are from 30 plaques per flask from a single experiment, except in the case of the rTgApiCox25 strain, where only 18 plaques were discernible. (D) Basal mitochondrial oxygen consumption rates (mOCR) in WT parasites grown in the absence of ATc or in the presence of ATc for 3 days (orange), and rTgApiCox25 parasites grown in the absence of ATc, or in the presence of ATc for 1 – 3 days (blue). A linear mixed-effects model was fitted to the data, and the values depict the mean ± s.e.m. from three independent experiments. A one-way ANOVA followed by Tukey’s multiple pairwise comparison test was performed. Relevant p values are shown. (E) Basal mOCR plotted against basal extracellular acidification rate (ECAR) of WT cells grown in the absence of ATc, the presence of cycloheximide (CHX) for 1 day, or the presence of ATc for 3 days, and rTgApiCox25 parasites grown in the absence of ATc or presence of ATc for 1 – 3 days (mean ± s.e.m. of the linear mixed-effects model described above; n = 3).
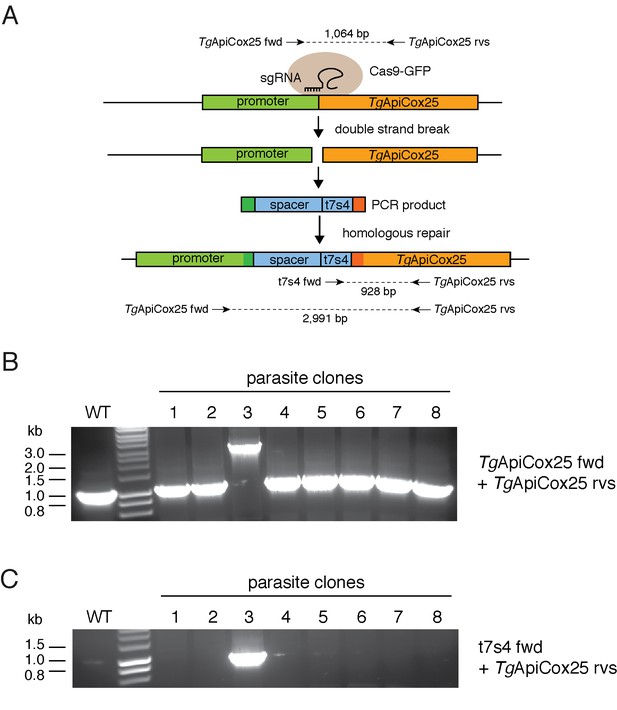
Generating an ATc-regulated promoter replacement strain of TgApiCox25.
(A) Diagram depicting the promoter replacement strategy to generated ATc-regulated TgApiCox25. A single guide RNA (sgRNA) was designed to target the T. gondii genome near the start codon of TgApiCox25, and mediate a double stranded break at the target site. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding the ATc regulated ‘t7s4’ promoter, which contains 7 copies of the Tet operon and a Sag4 minimal promoter, flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgApiCox25 start codon. The PCR product also contain a ‘spacer’ region that separates the regulatable promoter from the native promoter of TgApiCox25 gene to enable sufficient regulation. The parasite’s homologous repair pathway will mediate integration of the PCR product into the TgApiCox25 locus. The ‘TgApiCox25 fwd’, ‘TgApiCox25 rvs’ and ‘t7s4 fwd’ primers were used in screening parasite clones for successful integration of the regulatable promoter at the target site. (B–C) PCR screening analysis using genomic DNA extracted from parasite clones to identify clones that had successfully integrated the promoter. (B) Screening using the TgApiCox25 fwd and rvs primers. This will amplify a product of 1,064 bp if the locus is unmodified, and a product of 2,991 bp if the ATc-regulatable promoter has integrated successfully. (C) Screening using the TgApiCox25 rvs and t7s4 fwd primers. This will amplify a product of 928 bp if the ATc-regulatable promoter has integrated successfully. The analyses in B and C revealed that clone three had successfully integrated the ATc-regulatable promoter.
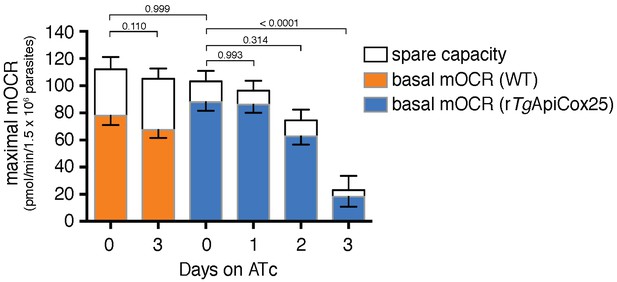
Knockdown of TgApiCox25 leads to defects in maximal mOCR.
Maximal mOCR, comprising of the sum of the basal mOCR (colored) and the spare capacity (white), of TATi/∆ku80 (WT) parasites grown in the absence of ATc or in the presence of ATc for 3 days (orange), and rTgApiCox25 cells grown in the absence of ATc, or in the presence of ATc for 1–3 days (blue). A linear mixed-effects model was fitted to the data, which are depicted as the mean ±s.e.m. from three independent experiments. A one-way ANOVA followed by Tukey’s multiple pairwise comparison test was performed on the maximal mOCR values. Relevant p values are shown.
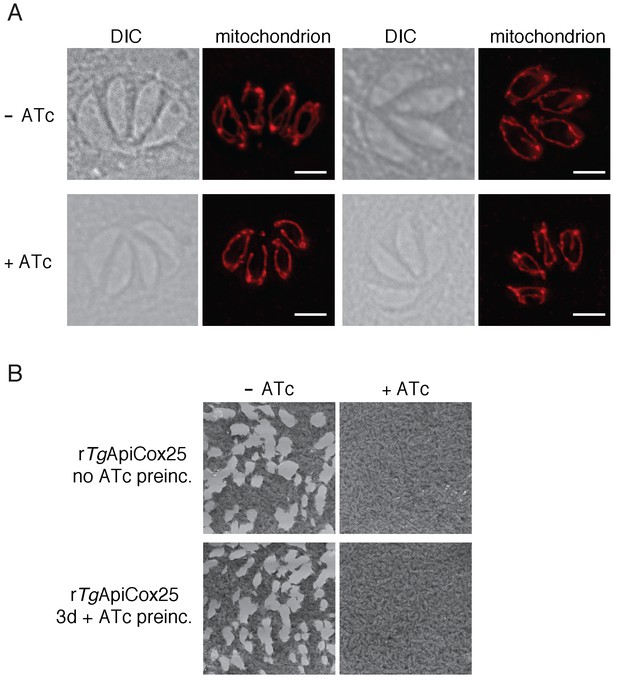
Defects in mOCR upon TgApiCox25 knockdown are not the result of general defects in mitochondrial morphology or parasite viability.
(A) Immunofluorescence assays assessing mitochondrial morphology in rTgApiCox25 parasites grown in the absence of ATc (top) or in the presence of ATc for 3 days (bottom). Mitochondria were labelled using antibodies against TgTom40 (red). Images are representative of 100 four-cell vacuoles examined in two independent experiments. The scale bar is 2 µm. (B) Plaque assays of rTgApiCox25 parasites grown for 9 days in the absence (left) or presence (right) of ATc. rTgApiCox25 parasites were not preincubated in ATc (no ATc preinc; top) or pre-incubated in ATc for 3 days (3d + ATc preinc; bottom) before commencing the experiment. Plaque assays are from a single experiment, representative of 3 independent experiments.
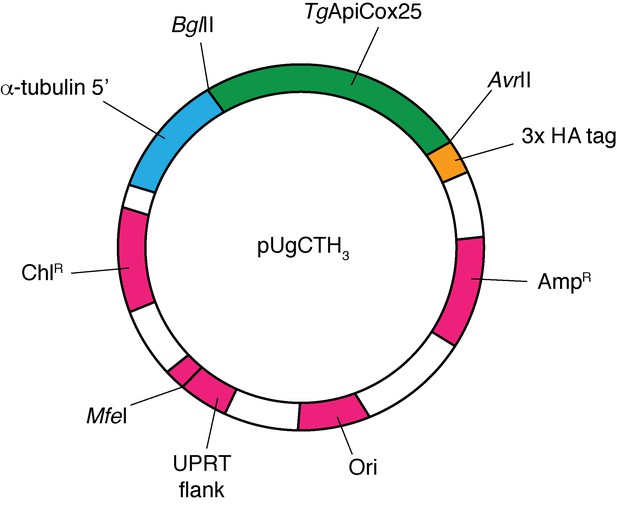
Map of the pUgCTH3 plasmid vector, showing the BglII and AvrII cut sites between which the TgApiCox25 open reading frame was ligated (green), the position of the 3x HA tag (yellow) and α-tubulin 5’ region (blue), and the positions of the chloramphenicol resistance marker (ChlR) for T.gondii selection, the UPRT flank (linearized at the indicated MfeI site before transfection), the ampicillin resistance marker for E. coli selection (AmpR), and the origin of replication (Ori; all magenta).
Note: vector is not drawn to scale.
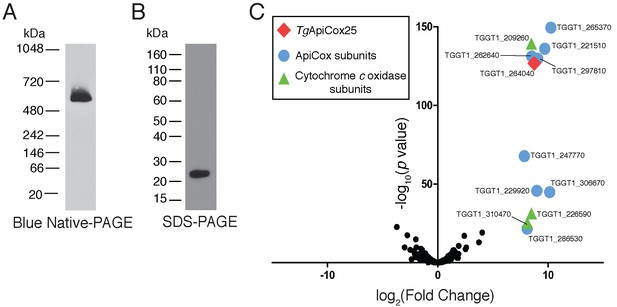
TgApiCox25 is part of a 600 kDa protein complex and co-purifies with canonical components of the cytochrome c oxidase complex.
(A) Western blot of proteins extracted from TgApiCox25-HA parasites, separated by blue native-PAGE, and detected with anti-HA antibodies. (B) Western blot of proteins extracted from TgApiCox25-HA parasites, separated by SDS-PAGE, and detected with anti-HA antibodies. (C) Volcano plot showing the log2 fold change vs –log10 p values of proteins purified from TgApiCox25-HA vs TgTom40-HA parasites using anti-HA immunoprecipitations and detected by mass spectrometry. Only proteins detected in each of the three independent experiments for both parasite lines are depicted. Proteins enriched in the TgApiCox25-HA samples (p<0.05; log2 fold change >5) have been coded according to whether they are orthologous to canonical cytochrome c oxidase subunits (green triangles), or restricted to the apicomplexan lineage (blue circles; ApiCox subunits). TgApiCox25 is also depicted (red diamond).
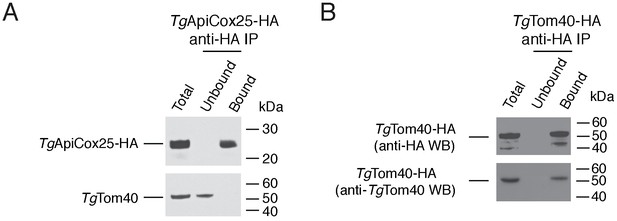
Immunopurification of the TgApiCox25 and TgTom40 protein complexes.
(A–B) Western blots of proteins extracted from parasites expressing TgApiCox25-HA (A) or TgTom40-HA (B). Extracts include samples before immunoprecipitation (Total), samples that did not bind to the anti-HA beads (Unbound), and samples that bound to the anti-HA beads (Bound). Samples were probed with anti-HA (top) and anti-TgTom40 (bottom) antibodies. Immunoprecipitations are representative of three independent experiments. Bound fractions from each experiment were subjected to mass spectrometry-based protein identification.
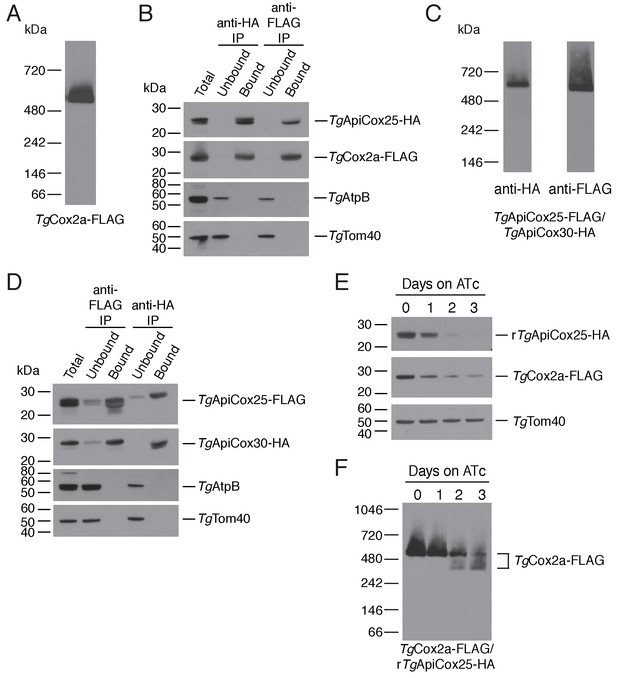
TgApiCox25 is a component of T.gondii cytochrome c oxidase and important for complex integrity.
(A) Anti-FLAG western blot of proteins from the TgCox2a-FLAG/TgApiCox25-HA strain separated by blue native-PAGE. (B) Western blots of proteins extracted from the TgCox2a-FLAG/TgApiCox25 HA strain and subjected to immunoprecipitation using anti-HA (anti-HA IP) or anti-FLAG (anti-FLAG IP) antibody-coupled beads. Extracts include samples before immunoprecipitation (Total), samples that did not bind to the anti-HA or anti-FLAG beads (Unbound), and samples that bound to the anti-HA or anti-FLAG beads (Bound). Samples were probed with anti-HA to detect TgApiCox25-HA, anti-FLAG to detect TgCox2a-FLAG, anti-AtpB to detect the β-subunit of T. gondii ATP synthase, and anti-TgTom40. (C) Anti-HA (left) and anti-FLAG (right) western blots of proteins from the TgApiCox25-FLAG/TgApiCox30-HA strain separated by blue native-PAGE. (D) Western blots of proteins extracted from the TgApiCox25-FLAG/TgApiCox30-HA strain and subjected to immunoprecipitation using anti-HA (anti-HA IP) or anti-FLAG (anti-FLAG IP) antibody-coupled beads. Extracts include samples before immunoprecipitation (Total), samples that did not bind to the anti-HA or anti-FLAG beads (Unbound), and samples that bound to the anti-HA or anti-FLAG beads (Bound). Samples were probed with anti-HA to detect TgApiCox30-HA, anti-FLAG to detect TgApiCox25-FLAG, anti-AtpB, and anti-TgTom40. (E) Western blot of proteins extracted from rTgApiCox25-HA/TgCox2a-FLAG parasites grown in the absence of ATc, or in ATc for 1 – 3 days, separated by SDS-PAGE and detected using anti-HA (top), anti-FLAG (middle) and anti-TgTom40 (as a loading control; bottom). (F) Western blot of proteins extracted from TgCox2a-FLAG/rTgApiCox25-HA parasites grown in the absence of ATc, or in ATc for 1 – 3 days, separated by blue native-PAGE, and detected using anti-FLAG antibodies.
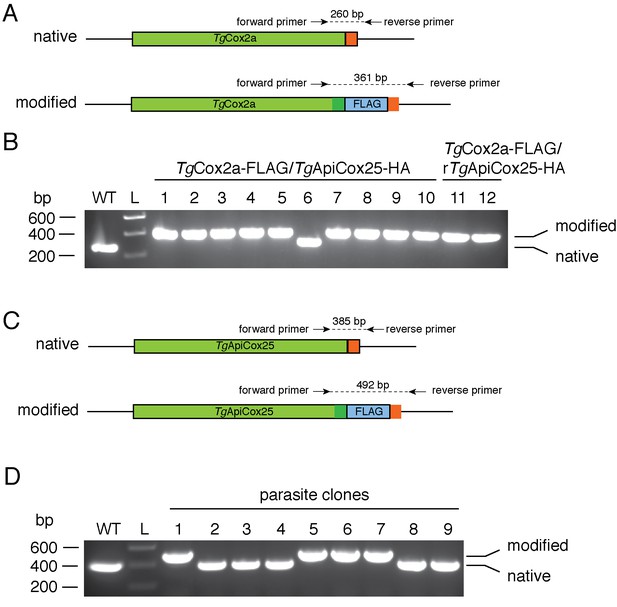
Generating FLAG tagged TgCox2a and TgApiCox25 strains.
(A) Diagram depicting the 3’ replacement strategy to generate FLAG-tagged TgCox2a. An sgRNA was designed to target the T. gondii genome near the stop codon of TgCox2a, and mediate a double stranded break at the target site. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding a FLAG epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgCox2a stop codon. The parasite’s homologous repair pathway will mediate integration of the PCR product into the TgCox2a locus. Forward and reverse primers were used to screen parasite clones for successful integration of the FLAG tag at the target site, yielding a 260 bp product in the native locus and a 361 bp product in the modified locus. (B) PCR screening analysis using genomic DNA extracted from putative TgCox2a-FLAG/TgApiCox25-HA parasites (clones 1 – 10) and TgCox2a-FLAG/rTgApiCox25-HA parasites (clones 11 – 12). Clones 1 – 5 and 7 – 12 yielded PCR products that indicated that these clones had been successfully modified. (C) Diagram depicting the 3’ replacement strategy to generate FLAG-tagged TgApiCox25. An sgRNA was designed to target the T. gondii genome near the stop codon of TgApiCox25, and mediate a double stranded break at the target site. A plasmid containing the sgRNA and GFP-tagged Cas9 endonuclease was co-transfected into T. gondii parasites with a PCR product encoding a FLAG epitope tag flanked by 50 bp of sequence homologous to the regions immediately up- and down-stream of the TgApiCox25 stop codon. The parasite’s homologous repair pathway will mediate integration of the PCR product into the TgApiCox25 locus. Forward and reverse primers were used to screen parasite clones for successful integration of the FLAG tag at the target site, yielding a 385 bp product in the native locus and a 492 bp product in the modified locus. (B) PCR screening analysis using genomic DNA extracted from putative TgApiCox25-FLAG/TgApiCox30-HA parasites. Clones 1, 5 – 7 yielded PCR products that indicated that these clones had been successfully modified.
Tables
Summary of the features of proteins identified in proteomic analysis of the TgApiCox25 complex.
Similarity searches were performed using HMMER (https://www.ebi.ac.uk/Tools/hmmer/). The accession numbers listed were derived from http://EuPathDB.org (apicomplexan and chromerid species) or www.ncbi.nlm.nih.gov (all others). Abbreviations: Plasmodium falciparum (Pf), Cryptosporidium parvum (Cp), Vitrella brassicaformis (Vb), Saccharomyces cerevisiae (Sc), Homo sapiens (Hs), Arabidopsis thaliana (At).
| ToxoDB gene ID (http://toxodb.org) | Protein annotation | Predicted protein mass (kDa) | Mitochon-drial proteome (this study) | Phenotype score (Sidik et al., 2016) | Similarity search (E-value) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pf | Cp | Vb (Chromerid) | Sc (Fungi) | Hs (Animal) | At (Plant) | |||||
| TGGT1_264040 | Hypothetical protein (TgApiCox25) | 24.5 | ✓ | −2.54 | Conserved unknown protein PF3D7_1464000.1 (2.4e−53) | x | Hypothetical Protein Vbra_12326 .t1 (5.2e−29) | x | x | x |
| TGGT1_265370 | Hypothetical protein (TgApiCox16) | 16.0 | ✓ | 1.56 | x | x | x | x | x | x |
| TGGT1_209260 | Putative cytochrome c oxidase subunit (TgCox5b) | 34.8 | ✓ | −3.07 | Putative COX5B PF3D7_0927800.1 (4.3e−101) | x | COX5B-2 Vbra_9355 .t1 (1.6e−92) | Cox4p P04037 (0.32) | Cox5B NP_001853.2 (0.05) | COX5b At1g80230 (2.1e−08) |
| TGGT1_221510 | Hypothetical protein (TgApiCox18) | 17.9 | ✓ | −3.28 | Conserved unknown protein PF3D7_0523300.1 (1.5e−48) | x | Hypothetical Protein Vbra_21271 .t1 (5.2e−45) | x | x | x |
| TGGT1_262640 | Cg8 family protein (TgApiCox23) | 23.8 | ✓ | −3.49 | Cg8 protein PF3D7_0708700.1 (3.1e−64) | x | Hypothetical Protein Vbra_3012 .t1 (2.4e−53) | x | x | x |
| TGGT1_297810 | Hypothetical protein (TgApiCox30) | 29.6 | ✓ | −3.64 | Conserved unknown protein PF3D7_0915700.1 (1.2e−46) | x | Hypothetical Protein Vbra_17445 .t1 (6.7e−33) | x | x | x |
| TGGT1_247770 | Hypothetical protein (TgApiCox19) | 19.2 | ✓ | −2.61 | Conserved unknown protein PF3D7_1402200.1 (1.2e−34) | x | Hypothetical Protein Vbra_2065 .t1 (1.7e−27) | x | x | x |
| TGGT1_229920 | Hypothetical protein (TgApiCox35) | 35.0 | ✓ | −3.84 | Conserved unknown protein PF3D7_0306500.1 (1.5e−90) | x | Hypothetical Protein Vbra_6819 .t1 (1.6e−73) | x | x | x |
| TGGT1_306670 | Hypothetical protein (TgApiCox26) | 25.8 | ✓ | −3.68 | Conserved unknown protein PF3D7_1439600.1 (2.6e−43) | x | Hypothetical Protein Vbra_888 .t1 (1.2e−36) | x | x | x |
| TGGT1_226590 | Putative cytochrome c oxidase subunit (TgCox2a) | 34.5 | ✓ | −3.80 | Cytochrome c oxidase subunit 2 PF3D7_1361700.1 (4.9e−58) | x | Cytochrome c oxidase subunit 2 Vbra_8641 .t1 (3.6e−33) | Cox2 P00410 (2.6e−06) | Cox2 P00403 (0.0004) | Cox2 P93285 (3.3e−06) |
| TGGT1_310470 | Putative cytochrome c oxidase subunit (TgCox2b) | 21.2 | ✓ | −4.18 | Cytochrome c oxidase subunit 2 PF3D7_1430900.1 (7.6e−75) | x | Cytochrome c oxidase subunit 2 Vbra_14923 .t1 (4.2e−7) | Cox2 P00410 (4.3e−31) | Cox2 P00403 (9.2e−29) | Cox2 P93285 (3.8e−37) |
| TGGT1_286530 | Hypothetical protein (TgApiCox24) | 25.4 | ✓ | −2.82 | Conserved unknown protein PF3D7_1362000.1 (6.0e−45) | x | Hypothetical Protein Vbra_10089 .t1 (1.2e−11) | x | x | x |
| TGGT1_254030 | Zinc finger CDGSH-type domain-containing protein (TgApiCox13) | 13.2 | ✓ | −4.26 | CDGSH iron-sulfur domain-containing protein PF3D7_1022900.1 (7.8e−42) | x | CDGSH iron-sulfur domain-containing protein 3 Vbra_4701 .t1 (1.2e−44) | x | CDGSH iron-sulfur domain-containing protein 3 (2.7e−11) | x |
| TGGT1_242840 | Membrane protein (TgApiCox14) | 13.9 | ✓ | −3.58 | Conserved unknown protein PF3D7_1339400.1 (4.1e−16) | x | Hypothetical Protein Vbra_9996 .t1 (1.8e−9) | x | x | x |
| TGVEG_442760 | Cytochrome C family oxidase subunit III (TgCoxIII) | 16.8 | - | N/A | Cytochrome c oxidase subunit 3 mal_mito_1 (5.2e−5) | x | x | x | x | x |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Toxoplasma gondii) | RH∆hxgprt | PMID: 8662859 | Parental strain for mtAPEX and mtBirA* strains | |
| Strain, strain background (T. gondii) | mtAPEX-cmyc in RH∆hxgprt | This paper | mtAPEX-cmyc-expressing T. gondii | |
| Strain, strain background (T. gondii) | mtBirA*-cmyc in RH∆hxgprt | This paper | mtBirA*-cmyc-expressing T. gondii | |
| Strain, strain background (T. gondii) | TATi/∆ku80 | PMID: 22144892 | Parental for 3' HA tag integration strains and rTgApiCox25 strain and derivatives thereof | |
| Strain, strain background (T. gondii) | TGGT1_209420 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_209870 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_210730 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_213420 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_217020 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_220100 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_223500 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_229620 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_232600 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_248600 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_258100 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_259710 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_260840 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_263080 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_263400 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_263840 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_265360 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_264040 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag; termedTgApiCox25-HA strain | |
| Strain, strain background (T. gondii) | TGGT1_275650 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_278250 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_278720 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_286120 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_290460 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_297810 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag; termed TgApiCox30-HA strain | |
| Strain, strain background (T. gondii) | TGGT1_300030 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_316900 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | TGGT1_318400 HA in TATi/∆ku80 | This paper | Gene contains integrated 3' HA tag | |
| Strain, strain background (T. gondii) | rTgApiCox25 in TATi/∆ku80 | This paper | ATc-regulated promoter driving TgApiCox25 expression | |
| Strain, strain background (T. gondii) | rTgApiCox25-HA | This paper | Regulatable TgApiCox25 gene with integrated 3' HA tag | |
| Strain, strain background (T. gondii) | cTgApiCox25-HA/rTgApiCox25 | This paper | rTgApiCox25 strain complemented with constitutive TgApiCox25-HA | |
| Strain, strain background (T. gondii) | TgCox2A-FLAG inTgApiCox25-HA | This paper | Integrated 3' FLAG tag in TgCox2a locus of TgApiCox25-HA strain | |
| Strain, strain background (T. gondii) | TgCox2A-FLAG in rTgApiCox25-HA | This paper | Integrated 3' FLAG tag in TgCox2a locus of rTgApiCox25-HA strain | |
| Strain, strain background (T. gondii) | TgApiCox25-FLAG in TgApiCox30-HA | This paper | Integrated 3' FLAG tag in TgApiCox25 locus of TgApiCox30-HA strain | |
| Cell line (Homo sapians) | Human Foreskin Fibroblasts | Gift from Holger Schülter, Peter MacCallum Cancer Centre | ||
| Antibody | mouse anti-cmyc | Santa Cruz | Clone 9E10 | (1:200 to 1:500) |
| Antibody | rat anti-HA | Sigma | Clone 3F10 | (1:200 to 1:1,000) |
| Antibody | mouse anti-FLAG | Sigma | Clone M2 | (1:500 to 1:2,000) |
| Antibody | rabbit anti-AtpB | Agrisera | cat #: AS05 085 | (1:500) |
| Antibody | rabbit anti-TgTom40 | PMID: 27458014 | (1:2,000) | |
| Antibody | rabbit anti-TgCytC | This paper | Peptide antibody made against residues 1–14 (MSRAEPDVQVPSGD) ofT. gondii cytochrome c (TGGT1_219750) (1:500) | |
| Antibody | rabbit anti-Hsp60 | PMID: 15279947 | (1:1,000) | |
| Antibody | goat anti-mouse Alexa Fluor 488 | Life Technologies | cat #: A11029 | (1:500) |
| Antibody | goat anti-rat Alexa Fluor 488 | Life Technologies | cat #: A11006 | (1:100 to 1:500) |
| Antibody | goat anti-rat CF 488A | Sigma | cat #: SAB4600046 | (1:100 to 1:500) |
| Antibody | goat anti-rabbit Alexa Fluor 546 | Life Technologies | cat #: A11035 | (1:500) |
| Antibody | goat anti-mouse HRP-conjugated | Santa Cruz | cat #: sc-2005 | (1:5,000) |
| Antibody | goat anti-rat HRP-conjugated | Santa Cruz | cat #: sc-2006 | (1:5,000) |
| Antibody | goat anti-rabbit HRP-conjugated | Santa Cruz | cat #: sc-2004 | (1:5,000) |
| Antibody | anti-mouse HRP-conjugated TrueBlot Ultra | eBioscience | cat #: 18-8817-31 | (1:5,000) |
| Antibody | anti-FLAG M2 affinity gel | Sigma | cat #: A2220 | |
| Antibody | anti-HA affinity matrix | Sigma | cat #: 11815016001 | |
| Peptide, recombinant protein | Avidin, Oregon Green-conjugated | Life Technologies | cat #: A6374 | (1:1,000) |
| Peptide, recombinant protein | NeutrAvidin, HRP-conjugated | Life Technologies | cat #: A2664 | (1:10,000) |
| Peptide, recombinant protein | Streptavidin magnetic beads | Thermo Scientific | cat #: PIE88817 | |
| Recombinant DNA reagent | pcDNA3-mito-APEX | PMID: 23086203 | Addgene cat # 42607 | |
| Recombinant DNA reagent | pBirA*−3XhA-LIC-DHFR | PMID: 25691595 | ||
| Recombinant DNA reagent | pSAG1::Cas9-U6::sgUPRT | PMID: 24825012 | Addgene cat # 54467 | |
| Recombinant DNA reagent | pUgCTH3 | PMID: 28205520 | ||
| Recombinant DNA reagent | mtAPEX-cmyc in pBTM3 | This paper | T. gondii expression vector encoding mitochondrially targeted APEX | |
| Recombinant DNA reagent | mtBirA*-cmyc in pBTM3 | This paper | T. gondii expression vector encoding mitochondrially targeted APEX | |
| Recombinant DNA reagent | TGGT1_209420 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_209870 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_210730 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_213420 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_217020 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_220100 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_223500 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_229620 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_232600 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_248600 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_258100 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_259710 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_260840 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_263080 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_263400 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_263840 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_265360 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_264040 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene (TgApiCox25) | |
| Recombinant DNA reagent | TGGT1_275650 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_278250 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_278720 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_286120 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_290460 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_297810 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene (TgApiCox30) | |
| Recombinant DNA reagent | TGGT1_300030 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_316900 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TGGT1_318400 HA in pgCH | This paper | T. gondii 3' replacment vector that introduces 1xHA tag into target gene | |
| Recombinant DNA reagent | TgApiCox25 5' sgRNA in pSAG1::Cas9-U6 | This paper | pSAG1::Cas9-U6 vector expressing sgRNA that targets 5' region of TgApiCox25 | |
| Recombinant DNA reagent | TgApiCox25 3' sgRNA in pSAG1::Cas9-U6 | This paper | pSAG1::Cas9-U6 vector expressing sgRNA that targets 3' region of TgApiCox25 | |
| Recombinant DNA reagent | TgCox2a 3' sgRNA in pSAG1::Cas9-U6 | This paper | pSAG1::Cas9-U6 vector expressing sgRNA that targets 3' region of TgCox2a | |
| Recombinant DNA reagent | TgApiCox25 in pUgCTH3 | This paper | Vector that expresses TgApiCox25-HA from the constitutive α-tubulin promoter | |
| Software, algorithm | Mitochondrial Matrix Quantitative Proteome search tool | This paper | https://bit.ly/2FySSmU | Link to region of ToxoDB website containing the T. gondii mitochondrial proteome search tool |
Additional files
-
Supplementary file 1
List of genes encoding putative T. gondii mitochondrial proteins.
Table 1. List of peptides identified in the APEX and BirA* proteomic analyses. Included are the ToxoDB accession numbers, the identified peptide, the experiment in which the peptide was identified, and the charge, m/z ratio, mass error, posterior error probability, score, delta score and intensity of each peptide from each experiment. Table 2. List of proteins identified in the RH control and mtAPEX proteomes, including the ToxoDB accession numbers, the log2 protein ratios, p value, and unique sequence counts. Table 3. List of proteins identified in the RH control and mtBirA* proteomes, including the ToxoDB accession numbers, the log2 protein ratios, p value, and unique sequence counts. Table 4. Summary of putative mitochondrial proteins identified in this study. The summary includes the ToxoDB accession numbers and annotation of proteins identified from the combined list (colums A and B), proteins identified in both lists (columns D and E), and proteins identified in the mtAPEX proteome (columns G and H) or mtBirA* (columns J and K) proteomes only. Proteins highlighted in green were demonstrated by this study to localise to the mitochondrion, while those highlighted in red did not localise to the mitochondrion. Table 5. Annotated protein list of the T. gondii mitochondrial proteome, noting the ToxoDB accession number of the corresponding gene, the protein annotation, mean phenotype score, molecular mass, number of transmembrane domains, amino acid sequence, MitoFates and MitoProt II prediction scores, the ortholog grouping, and the accession number of orthologous genes in P. falciparum, C. parvum, B. bovis, V. brassicaformis, and S. cerevisiae based on reciprocal BLAST searches. Homologs identified in S. cerevisiae were queried against the ‘high confidence’ mitochondrial proteome (Sc mito proteome) described in (Morgenstern et al., 2017). Table 6. Summary of the OrthoMCL analysis of the T. gondii mitochondrial proteome, depicting the gene annotation, mean phenotype score, and the relevant orthology grouping.
- https://doi.org/10.7554/eLife.38131.019
-
Supplementary file 2
Summary of metabolic pathway enrichment in the T. gondii mitochondrial proteome.
- https://doi.org/10.7554/eLife.38131.020
-
Supplementary file 3
Expected mitochondrial proteins and false negatives identified from the T. gondii mitochondrial proteome.
List of proteins identified in the T. gondii mitochondrial proteome that previous studies have demonstrated or predicted to localize to the mitochondrion, and proteins that previous studies have demonstrated do not localize to the mitochondrion. Included are the protein annotation, the process in which it functions, the proteome in which it was detected, and the ToxoDB gene ID. Note that natively biotinylated proteins (including the mitochondrially-localized pyruvate carboxylase; Nitzsche et al., 2017) were excluded from these analyses because of their biased (i.e. APEX and BirA*-independent) enrichment in control and experimental conditions. Color coding: green, predicted mitochondrial protein present in proteome; pink, predicted mitochondrial protein absent from proteome.
- https://doi.org/10.7554/eLife.38131.021
-
Supplementary file 4
List of primers and templates used in this study.
Table 1. Primers and templates used in general cloning. Table 2. Primers used in 3’ replacement localization studies. 3’ fragments of target genes (ToxoDB gene ID) were amplified using the listed forward and reverse primers. The resulting PCR product was digested and ligated into the vector pgCH as outlined in the cloning strategy. The final vector was linearized with the indicated restriction enzyme before transfection.
- https://doi.org/10.7554/eLife.38131.022
-
Supplementary file 5
Table 1: List of proteins identified in the TgApiCox25 and TgTom40 immunoprecipitations.
Included is a description of each identified protein, the UniProt accession number, the predicted molecular mass, the fold change, the normalized total precursor intensity for each biological replicate, and the Cox or ApiCox designation of the identified protein. Table 2: A list of the log fold change (logFC) and p values calculated for each protein identified in all replicates of the TgApiCox25 and TgTom40 immunoprecipitations following EdgeR analysis.
- https://doi.org/10.7554/eLife.38131.023
-
Source code 1
R script used in the analysis of the Seahorse XFe96 data.
- https://doi.org/10.7554/eLife.38131.024
-
Source code 2
R script used in the analysis of proteomic data from the TgApiCox25 and TgTom40 immunoprecipitations.
- https://doi.org/10.7554/eLife.38131.025
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38131.026





