MDN brain descending neurons coordinately activate backward and inhibit forward locomotion
Figures
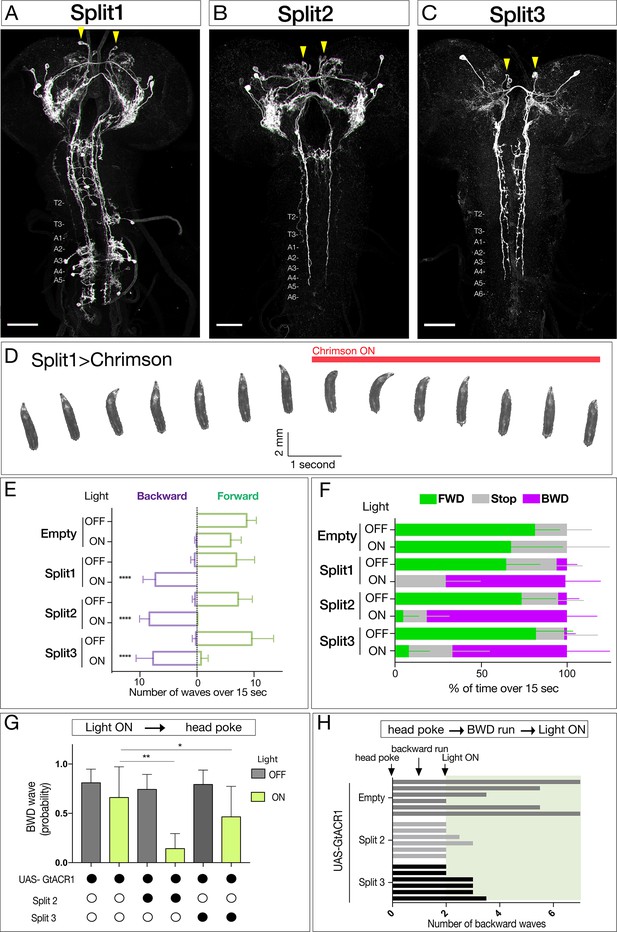
Neurons sufficient to induce backward larval locomotion.
(A–C) Split1-Split3 lines driving expression of membrane localized Venus in the third instar CNS. Corazonin (not shown) labels a single neuron in segments T2-A6, and was used to identify VNC segment identity. The only neurons potentially common to all three lines are a pair of bilateral ventral, anterior, medial neurons (arrowheads). Maximum intensity projection of entire CNS shown. Anterior, up; scale bars, 50 μm. Genotypes: R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus (Split1); R49F02-Gal4AD R53F07-Gal4DBD tsh-lexA lexAop-killer zipper UAS-Chrimson:mVenus (Split2); ss01613-Gal4 UAS-Chrimson:mVenus (Split3). (D) Split1 activation induces backward locomotion. Genotype: R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus. (E) Split1, Split2, or Split3 activation induces backward locomotion. Number of backward or forward waves in third instar larvae over 15 s with or without Chrimson activation. N = 10 for all genotypes. Genotypes: pBD-Gal4 UAS-Chrimson:mVenus (Empty) and see A-C above for Split1-3 genotypes. (F) Split1, Split2, or Split3 activation induces backward locomotion. Percentage of time performing forward locomotion (green), backward locomotion (magenta) or paused (grey) in third instar larvae over 15 s with or without Chrimson activation. N = 5 for all genotypes. Genotypes, see A-C. (G) Split2 or Split3 silencing reduces initiation of backward locomotion. Backward waves induced by a noxious head poke, with or without active GtACR1. Genotypes: pBD-Gal4 UAS-GtACR1:mVenus (first two bars, n = 20), R49F02-Gal4AD R53F07-Gal4DBD tsh-lexA lexAop-killer zipper UAS-GtACR1:mVenus (middle two bars, n = 8), ss01613-Gal4 UAS-GtACR1:mVenus (last two bars, n = 25). (H) Split2 or Split3 neuron silencing stops ongoing backward locomotion. After each larva initiated a backward run (two backward waves), light was used to activate GtACR1 or a no Gal4 control, and the number of backward waves was counted. n = 6 for both groups; each bar represents the average of two trials for the same larva. See G for genotypes.
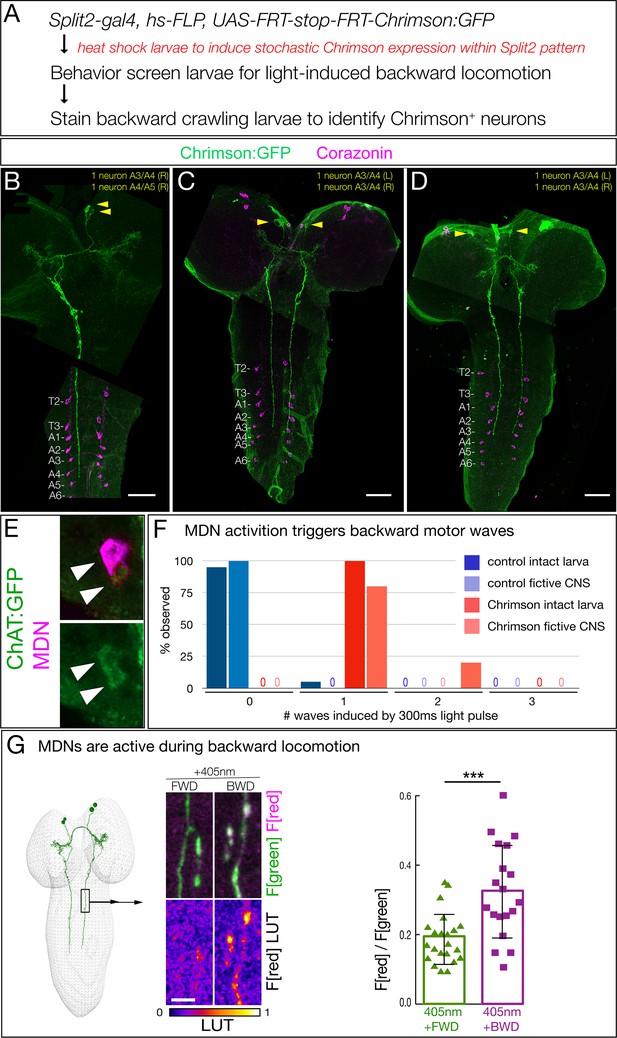
Two brain descending neurons are sufficient to induce backward larval locomotion.
(A) Experimental flow for generating sparse, stochastic patterns of Chrimson in subsets of the Split2 expression pattern. Genotype: hsFlpG5.PEST R49F02-Gal4AD R53F07-Gal4DBD tsh-lexA, lexAop-killer zipper UAS.dsFRT.Chrimson:mVenus. (B–D) The CNS from three larvae that crawled backward in response to Chrimson activation. All show expression in neurons with medial cell bodies, bilateral arbors, and a contralateral descending projection to A3-A5. (B) Note there is a tear in the CNS near segment T1. Chrimson:Venus, green; Corazonin (Crz; segmental marker), magenta. Scale bar, 50 μm. (E) MDNs are cholinergic. MDNs marked with mCherry (magenta) express ChAT:GFP (green). Genotype: R49F02-Gal4AD,UAS-Chrimson:mCherry; R53F07-Gal4DBD, mimic ChAT:GFP. (F) A brief pulse of MDN activity can trigger a backward wave. Intact larvae: individual L3 larvae were subjected to 300 ms of 561 nm light and the number of backward waves was quantified (n = 20). Genotype: ss01613-Gal4 (Split3); UAS-Chrimson::mVenus. Fictive CNS: isolated L3 CNS was subjected to 300 ms 561 nm light and the response of the downstream neuron A18b was monitored for a backward wave of GCaMP6f activity. All MDN activations led to at least one backward wave, but there are also backward waves that occur independently of MDN activation (see Discussion), and these may account for the observed second waves. Genotype: ss01613-Gal4 (Split3)/R94E10-lexA; lexAop-GCaMP6f,UAS-Chrimson:mCherry. (G) MDNs are preferentially active during backward (BWD) not forward (FWD) locomotion in the intact larva. CaMPARI in MDN descending projections within the SEZ of third instar larvae. Top, fluorescence emission following excitation by 488 nm (green) or 561 nm (magenta); bottom, emission from 561 nm imaging alone. Right, quantification of red fluorescence over green fluorescence, mean intensity. Each value represents data from an individual descending projection. See Materials and methods for details. n = 22 for FWD and 19 for BWD. Scale bar, 10 μm. Genotype: R49F02-Gal4AD R53F07-Gal4DBD UAS-CaMPARI.
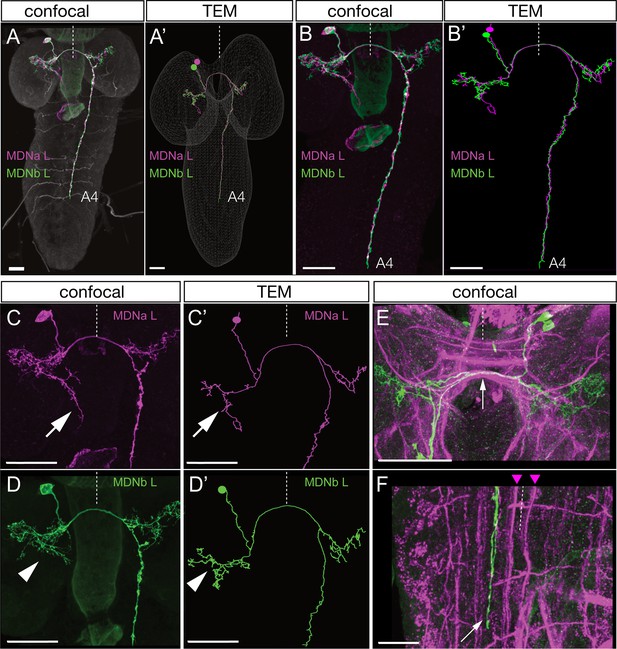
Identification of mooncrawler descending neurons by light and electron microscopy.
(A–F) Light microscopy. Multicolor FLP-out (MCFO) was used to visualize the morphology of individual neurons in the Split2 pattern in first instar larvae. Two neurons show morphology matching that seen in the Chrimson FLP-out experiments in Figure 2. Both neurons have anterior medial somata (A), ipsilateral and contralateral arbors (A–D), a contralateral projection in the posterior commissure (E, arrow), and descending neurons terminating in segments A3-A5 of the VNC (A–B). The neurons run lateral to the dorso-medial (DM) FasII tract in the VNC (F, DM tract marked with arrowheads). The two neurons can be distinguished by their ipsilateral arbor, which is either linear (C, arrow) or bushy (D, arrowhead). (A’–D’) Reconstructions from serial section transmission electron microscopy (TEM) of a first instar larva. Two neurons indistinguishable from the MDNs can be identified in the TEM reconstruction: MDNa (linear ipsilateral arbor) and MDNb (bushy ipsilateral arbor). We simply call them MDNs due to their similar morphology and connectivity. All panels show dorsal views with midline indicated (dashed line). Scale bars, 20 μm.
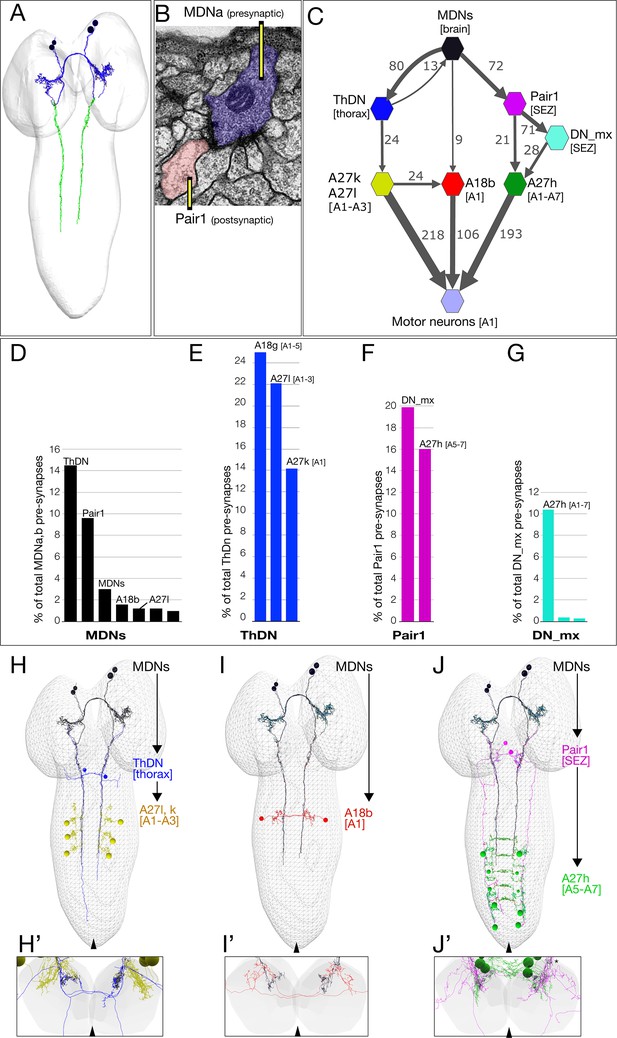
The MDN connectome: three pathways to distinct subsets of premotor neurons.
(A) TEM reconstruction of the bilateral MDNa,b neurons. Neuronal skeletons are colored to show post-synapses in the presumptive dendritic arbors of the brain (blue) and pre-synapses in the presumptive axonal descending process (green). Anterior, up. (B) Representative MDN output pre-synapse (blue) onto a post-synaptic Pair1 neuron (pink). (C) MDNs and their partners with the greatest number of synapses (synapse number shown next to connection arrows, and line width is proportional to synapse number). All connectivities are shown except unilateral synapses,<6 synapses, and the 15 synapses between MDNs. Each polygon represents pairs of the indicated neuron with the exception of these larger groups: A27k/A27l (six A27l neurons in A1-A3, four A27l neurons in A1-A2); A27h (14 neurons in A1-A7), and 30 pair of motor neurons in A1. This graph is provided as Supplementary file 1. json that can be opened in CATMAID. (D–G) Quantification of the percent of total pre-synapses that are targeted to the indicated neuron. All connectivities are shown except unilateral or <5 synapse connections. (H–J) The three MDN to premotor neuron pathways. (H) MDN-ThDN-A27l/k pathway. Only A27l is shown; A27k has a very similar morphology. (I) MDN-A18b pathway. (J) MDN-Pair1-A27h pathway. Dorsal view; anterior, up; midline, arrowhead. (H’–I’) Respective cross-sectional view of VNC neuropil (gray) and neurons in each pathway; note that synapses are primarily in the dorsal (motor) neuropil. Dorsal up, midline, arrowhead. Asterisk in J’ shows the approximate site of the synapse shown in panel B.
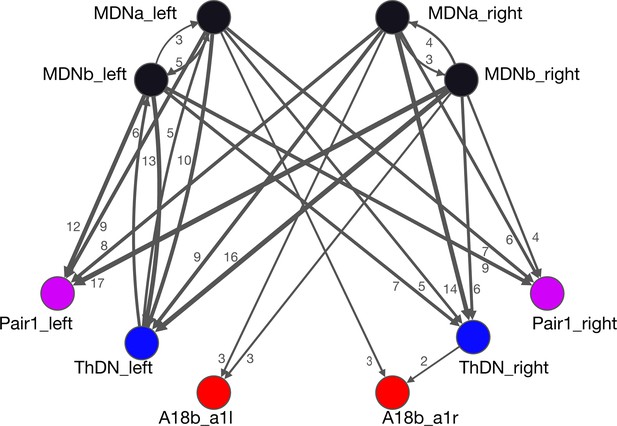
All MDNs have similar connectivity.
All synapses between the MDNs and output target neurons are shown, except those with single synapse connectivity. All MDNs have similar input connectivity as well (data not shown).
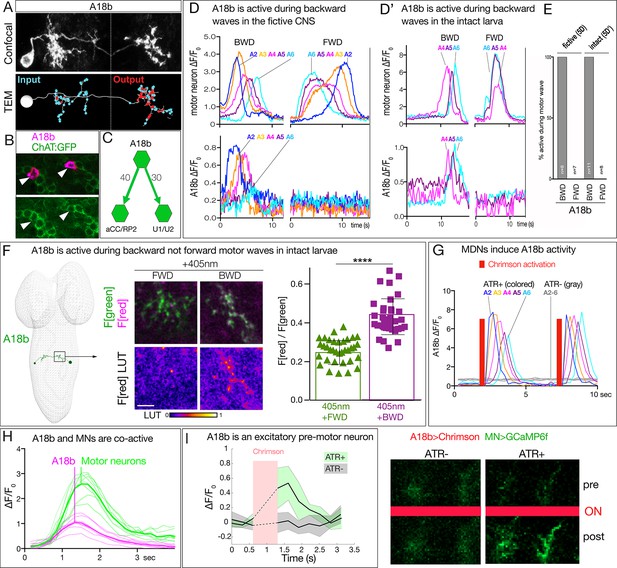
MDN activates the excitatory backward-active A18b premotor neuron.
(A) A18b morphology by light (MCFO) and electron microscopy (TEM). Top: Dorsal view of an individual A18b neuron in a second instar larval CNS by light microscopy (R94E10 > MCFO). Bottom: Dorsal view of an individual A18b neuron in a first instar larva in the TEM reconstruction. Cyan dots, post-synaptic sites; red dots, pre-synaptic sites. Anterior, up. Midline, dashed line. Genotype: R94E10-gal4 UAS-MCFO2. (B) A18b is cholinergic. A18b cell body (mCherry; magenta) and ChAT:GFP (green). Genotype: R94E10-Gal4, UAS-Chrimson:mCherry; mimic ChAT:GFP. (C) Connectivity of A18b to neurons with the greatest number of A18b post-synapses: the dorsal-projecting motor neurons aCC/RP2 and U1/U2 in segment A1. Synapse number shown. (D) In fictive preparations, A18b neurons are active in backward but not forward locomotion. ΔF/F0 of GCaMP6m in U1-U5 motor neurons (top) or jRCaMP1b in A18b (bottom) of five segments executing a forward (FWD) and then a backward (BWD) wave. This experiment was performed on eight different isolated third instar CNSs with similar results; quantified in E. Genotype: CQ-lexA/+; lexAop-GCaMP6m/R94E10-Gal4 UAS-jRCaMP1b.. (D’) In intact larvae, A18b neurons are active in backward but not forward locomotion. ΔF/F0 of GCaMP6m in motor neurons (top) and jRCaMP1b in A18b (bottom) in three segments. Times of BWD and FWD motor waves indicated. This experiment was performed on 19 waves (11 BWD, 8 FWD) in seven third instar larvae, all with similar results; quantified in E. Genotype: CQ-lexA/+; lexAop-GCaMP6m/R94E10-Gal4 UAS-jRCaMP1b.. (E) Quantification of data in panels D and E. BWD, backward waves; FWD, forward waves. (F) In intact larvae, A18b is preferentially active during backward not forward locomotion. CaMPARI in A18b neurites in a third instar larval CNS. Top, fluorescence emission (F) following 488 nm (green) or 561 nm (magenta) illumination; bottom, emission from 561 nm alone. Left, photoconversion (405 nm) during FWD or BWD locomotion. Right, quantification of red fluorescence over green fluorescence mean intensity. Each value represents data from an individual neurite. n = 35 for FWD and 36 for BWD. LUT, 561 nm emission intensity look up table. Scale bar, 10 μm. Genotype: R94E10-Gal4 UAS-CaMPARI. (G) In fictive preparations, MDNs activate A18b neurons, and induce backward A18b activity waves. Chrimson is expressed in MDN, and GCaMP6f in A18b. Red bars, time of 561 nm Chrimson activation. Colored traces indicate the ΔF/F0 of A18b GCaMP6f signal in 5 segments of an ATR +brain; gray traces are from ATR- animal. This experiment was performed on five different animals with similar results. Genotype: R49F02-Gal4AD/R94E10-lexA; R53F07-Gal4DBD/lexAop-GCaMP6f UAS-Chrimson:mCherry. (H) Dual color calcium imaging of jRCaMP1b in A18b (magenta) and GCaMP6m in U1-U5 motor neurons (green). In fictive preparations, A18b and motor neurons are co-active during backward waves. Both show similar initiation of activity, but A18b peak activity precedes motor neuron peak activity (vertical lines). Data are acquired every 168 ms from eight A18b/motor neuron pairs from three animals; peak activity of the motor neurons followed that of A18b by 0 ms (two pair), 168 ms (four pair), or 336 ms (two pair). Dashed lines, individual neurons; solid lines, average. Genotype: CQ-lexA/+; lexAop-GCaMP6m/R94E10-Gal4 UAS-jRCaMP1b.’. (I) A18b is an excitatory pre-motor neuron. A18b expresses Chrimson and aCC/RP2 motor neurons express GCaMP6f. Left: ΔF/F0 traces of GCaMP6f before and after 561 nm Chrimson activation (red bar) of three aCC/RP2 axons/dendrites within an animal. Solid bars represent means and shaded regions represent standard deviation from the mean (SDM). ATR +is shaded in green and ATR- in grey. Five animals were used in each group. GCaMP6f signal was not acquired during the Chrimson activation (dashed lines); t-test analysis for the first ΔF/F0 value after Chrimson activation between +ATR and -ATR showed significance (p=0.0071). Right: images of motor neuron GCaMP6f fluorescence pre- and post-Chrimson activation in ATR +and ATR- larvae. Genotype: 94E10-lexA/+; lexAop-Chrimson:mCherry/RRa-Gal4 UAS-GCaMP6f..
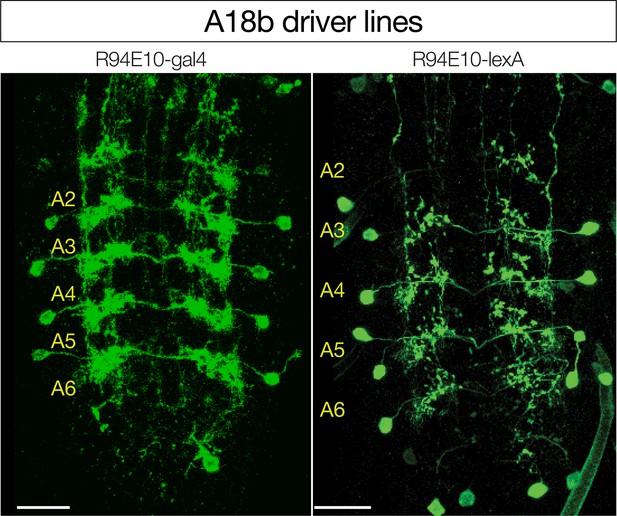
Driver lines for A18b.
R94E10-Gal4 and R94E10-LexA expression in A18b (shown); in addition, both lines have off-target expression in the brain (~10 neurons) and more medially in the VNC (~3 neurons) that are not shown. Note that both lines lack expression in A18b in A1 where MDNs form synaptic contacts, thus the increase in GCaMP6f fluorescence observed following MDN activation is either due to A18b activation in A1 triggering a posterior wave of A18b activity, or another indirect pathway. Dorsal view; anterior up; scale bars, 20 μm.
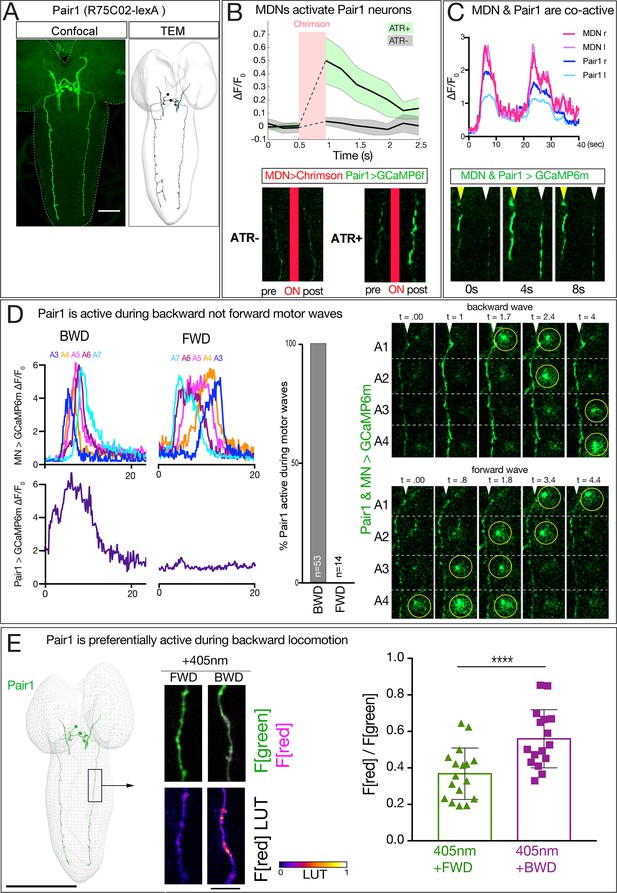
MDN activates Pair1 which is a backward-active descending neuron.
(A) Pair1 neurons by light (confocal) and electron microscopy (TEM). Confocal image is an L3 CNS, TEM reconstruction is from an L1 CNS. Anterior, up. Scale bar, 50 μm. Genotype: R75C02-lexA lexAop-myr:GFP.. (B) MDNs activate Pair1. MDN expresses Chrimson and Pair1 neurons express GCaMP6f. Top: ΔF/F0 traces of GCaMP6f before and after Chrimson activation (red bar) of Pair1 axons. Solid bars represent means and shaded regions represent standard deviation from the mean. ATR +is shaded in green and ATR- in grey. Six animals were used for ATR +and five for ATR-. GCaMP6f signal was not acquired during the Chrimson activation (dashed lines); t-test analysis for the first ΔF/F0 value after Chrimson activation between +ATR and -ATR showed significance (p=0.0004). Bottom: images of Pair1 GCaMP6f fluorescence pre- and post-Chrimson activation in ATR +and ATR- larvae. Genotype: R49F02-Gal4AD/R75 C02-lexA; R53F07-Gal4DBD/lexAop-GCaMP6f UAS-Chrimson:mCherry. (C) MDN and Pair one are co-active. Top: MDN and Pair1 expressing GCaMP6m in different regions of the neuropil, and show concurrent activity. Bottom: MDNs (white arrowhead) and Pair1 (yellow arrowhead) show similar timing of GCaMP6m fluorescence during a BWD wave. Anterior, up; midline, right side of panel. MDN and Pair1 co-activity was observed in 5 out of 10 brains examined; the other five brains showed Pair1 activity but no MDN activity (see Figure 6—figure supplement 1). Genotype: ss01613-Gal4/UAS-GCaMP6m; R75C02-Gal4. (D) Pair1 is active during backward (BWD) but not forward (FWD) waves in fictive preparations. Left: Pair1 GCaMP6m activity (bottom) and motor neuron activity (top) during fictive BWD and FWD waves in the same animal. Pair1 is not active during FWD waves. Center: quantification. N = 53 BWD waves from seven different animals, and 14 FWD waves from four different animals. Right: Pair1 GCaMP6m activity (arrowheads) precedes U1-U5 motor neuron activity (circled). Genotype: CQ-lexA/UAS-GCaMP6m; lexAop-GCaMP6m/R94E10-Gal4. (E) Pair1 is preferentially active during backward locomotion in the intact animal. CaMPARI was expressed in Pair1 and photoconversion was activated during FWD or BWD locomotion in intact third instar larvae. There is significantly more CaMPARI photoconversion during BWD locomotion. Graph, quantification of red fluorescence over green fluorescence mean intensity. Triangle or square, data from an individual axon. n = 36 for FWD and 34 for BWD. Scale bar, 10 μm. Genotype: R75C02-Gal4 UAS-CaMPARI..
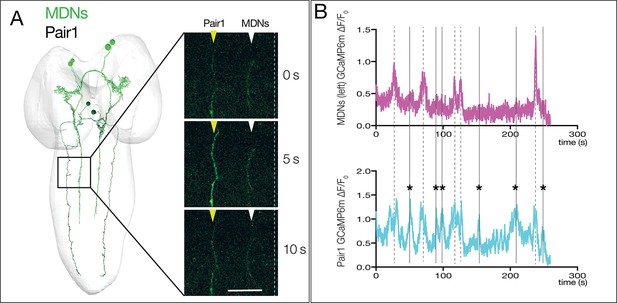
Pair1 can be activated independent of MDN activity.
(A) Left: dorsal view of TEM reconstruction of Pair1 and MDNs in a first instar larval CNS. Right: GCaMP6m activity in MDNs (white arrowheads) and Pair1 (yellow arrowheads) at three different time points in a third instar larval CNS fictive preparation. Pair1 is active while MDNs stay inactive. Anterior, up; midline, dashed line. Scale bar, 30 μm. (B) GCaMP6m activation in MDNs (top) and Pair1 (bottom) in a third instar larval CNS showing Pair1 activity concurrent with MDN activity (dashed lines) and Pair1 activity independent of MDN activity (solid lines, asterisk). MDNs were active in 46% of the Pair1 activity bouts (n = 48). Genotype: ss01613-Gal4/UAS-GCaMP6m; R75C02-Gal4.
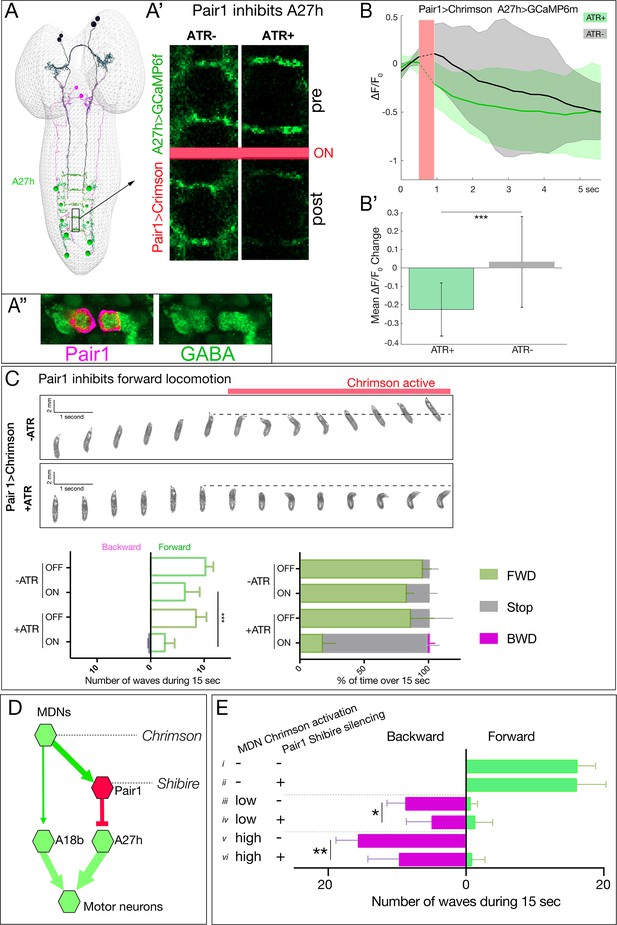
Pair1 inhibits the forward-active A27h premotor neuron, and arrests forward locomotion.
(A,B) Pair1 inhibits A27h. (A) Reconstruction of MDNs (black), Pair1 neurons (magenta) and A27h neurons (green) in the first instar CNS TEM volume. (A’) A27h GCaMP6m fluorescence is reduced following Pair1 Chrimson activation (red bar); two segments shown. (A’’) Pair1 is GABAergic. Pair1 cell body (mCherry; magenta, arrowheads) and GABA (green). Genotype: R75C02-Gal4, UAS-Chrimson:mCherry. (B) A27h GCaMP6m fluorescence is reduced following Pair1 Chrimson activation (red bar). (B’) ΔF/F0 was significantly inhibited in ATR +animals relative to ATR- controls. A total of 26 events from seven animals were averaged for ATR +and 16 events from four animals for ATR- group. See Materials and methods for further details. Genotype: R75C02-lexA/+; lexAop-Chrimson:mCherry/R36 G02-Gal4, UAS-GCaMP6m.. (C) Activation of Pair1 halts FWD locomotion for the duration of neuronal activation. Top, time-lapse images of ±ATR larvae expressing Chrimson in Pair1 neurons before and during light stimulation. Bottom left, backward and forward wave number over 15 s without Chrimson activation (Off) or during Chrimson activation (On) in third instar larvae. n = 12 for all groups. Bottom right, percent time performing forward locomotion (green), backward locomotion (magenta) or not moving (grey) over 15 s without Chrimson activation (Off) or during Chrimson activation (On) in third instar larvae. n = 5 for all groups. Genotype: R75C02-Gal4 UAS-Chrimson:mVenus.. (D) Schematic illustrating the experiment in (E). Arrows, excitatory connections; T-bar, inhibitory connection; line width proportional to synapse number. (E) Pair1 activity is necessary for efficient Chrimson-induced backward locomotion. Chrimson was expressed in MDNs, and shibirets was expressed in Pair1 neurons. Low (0.07 mW/mm2) or high (0.275 mW/mm2) light intensities were used to induce MDN activity; a temperature shift to 32°C was used to inactivate Shibirets and thus silence Pair1 neurons. Silencing of Pair1 alone had no detectable phenotype (i, ii). Silencing Pair1 decreased the efficacy of MDN-induced backward locomotion at low or high light levels (iii-vi). Genotypes: R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus pBD-lexA lexAop-Shibirets (i, iii and v) and R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus R75C02-lexA lexAop-Shits1 (ii, iv and vi).
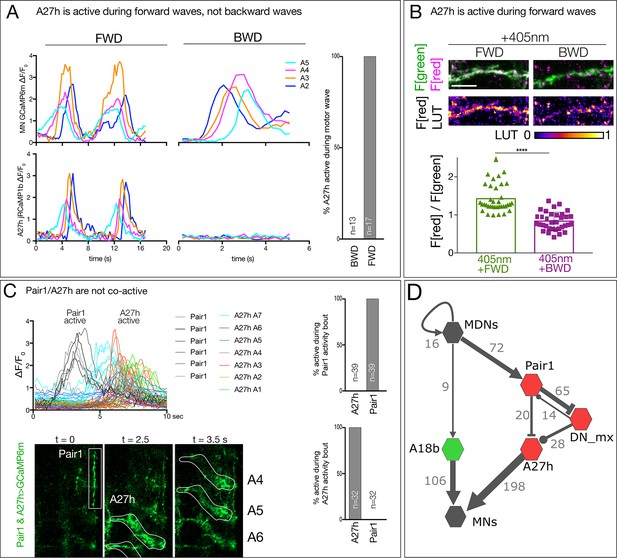
Timing of A27h neuronal activity.
(A) A27h is active during forward (FWD) but not backward (BWD) motor waves in fictive preparations. A27h expresses jRCaMP1b and U1-U5 motor neurons express GCaMP6m. Genotype: CQ-lexA/+ ; lexAop-GCaMP6m/R36G02-Gal4,UAS-jRCaMP1b. (B) A27h is preferentially active during forward locomotion in the intact animal. CaMPARI in A27h neurites of a third instar larval CNS. Top, fluorescence emission following 488 nm (green) or 561 nm (red) illumination. Bottom, graph represents quantification of red fluorescence over green fluorescence mean intensity. Each value represents data from an individual axon. n = 17 for FWD and 16 for BWD. Scale bar, 10 μm. Genotype: R36G02-Gal4 UAS-CaMPARI. (C) Pair1 and A27h activity is anti-correlated. GCaMP6m fluorescence was measured in distinct ROIs for Pair1 and A27h (bottom panels). Traces show one Pair1 neuron (gray lines) and seven A27h neurons (colored lines) before and during six A27h forward waves. Note that Pair1 activity precedes A27h activity. Quantification shown to the right; activity is defined as ΔF/F > 1.5. Genotype: R75C02-Gal4/R36G02-Gal4, UAS-GCaMP6m. (D) There is no direct anatomical connection between A27h and A18b pathways. Neurons with one or more synapse in the TEM reconstruction are shown (numbers next to arrows). Arrows, excitatory; T-bars, inhibitory; circles, unknown. Synapse numbers as of 5 July 2018.
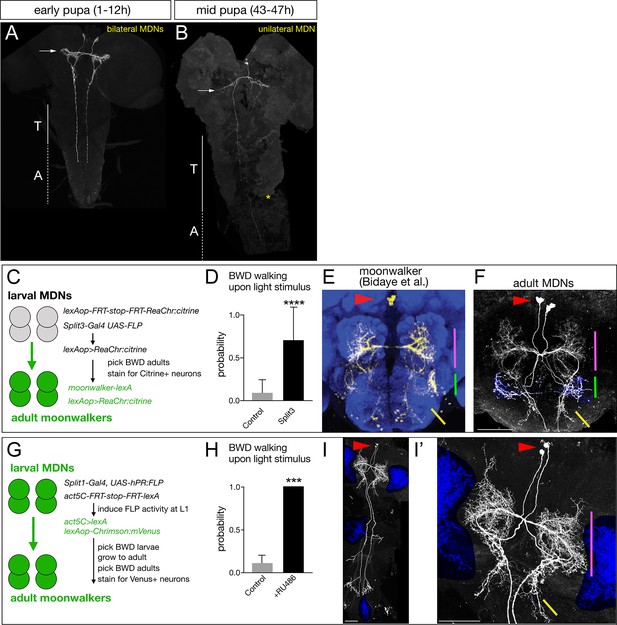
Larval MDNs persist into adulthood, match the moonwalker neuron morphology, and induce backward walking.
(A–B) MDN neurons labeled by Split1 MCFO are similar in morphology to larval neurons during early pupal stages (A), but prune their brain and SEZ arbors by mid-pupal stages (B, arrow). T, thoracic segments; A, abdominal segments. Asterisk, tissue damage from dissection. (C–F) Larval MDNs persist to adulthood, match adult moonwalker morphology, and can induce backward walking in adults. (C) Genetic scheme for the experiment. Note that Split3 has no adult central brain expression (data not shown), and thus only the Split3 larval neurons will have the ‘flipped out’ lexAOP-ReaChr:citrine transgene. (D) Probability of adult backward walking upon light activation of Split3 immortalized neurons (split3) or controls lacking the DBD half of Split3 genotype (control). (E) Adult moonwalker neurons from Bidaye et al., 2014. Red arrowhead, cell bodies; colored lines, distinctive arbors. (F) One example of ‘immortalized’ larval MDNs showing the same cell body location (red arrowhead) and same distinctive arbors (colored lines); the arbor marked by the green line is an off-target projection not connected to the MDN neurons. Genotypes: Control: UAS-FLP.PEST ss01613-(AD)-Gal4/TM3 VT044845-lexA lexAop-FRT-stop-FRT-ReaChr:citrine. Split3: UAS-FLP.PEST ss01613-(AD + DBD)-Gal4 VT044845-lexA lexAop-FRT-stop-FRT-ReaChr:citrine.. (G–I) Larval MDNs persist to adulthood and induce backward walking. (G) Intersectional genetics used in this experiment. (H) Probability of adult backward walking upon light activation of the neurons immortalized by RU486-induced Flp activity (RU486+) or controls not given RU486 and thus lacking Chrimson expression in adult MDNs (control). (I) One example of an adult CNS plus VNC showing two MDNs (arrowhead) and four off target neurons (blue shading). (I’) Enlargement of brain showing MDNs and parts of two off target neurons (blue shading). Red arrowhead, cell bodies; colored lines, distinctive arbors in the protocerebrum (magenta line) and SEZ (green line). Scale bars, 50 μm.
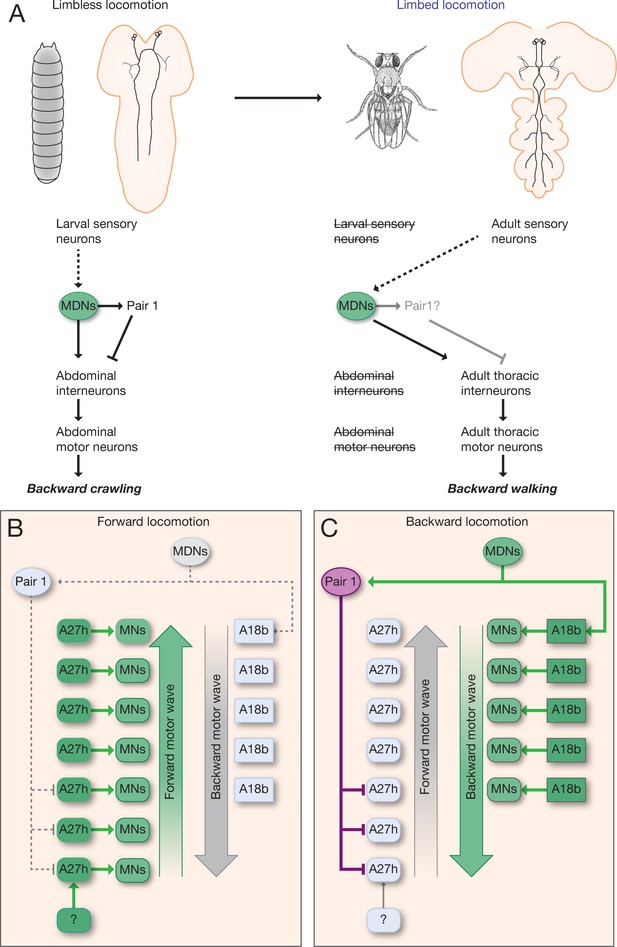
Model describing the MDN-mediated backward crawling.
(A) MDN neurons are present in larval stages where they promote backward peristaltic crawling via abdominal premotor and motor neurons; MDNs subsequently persist into the adult fly where they promote backward walking of the six-limbed adult fly, using a different pool of thoracic motor neurons. Solid arrows represent direct excitatory connectivity; solid T-bars represent direct inhibitory connectivity; dashed arrows represent known (larva) or predicted (adult) polysynaptic connectivity. (B,C) Model. (B) During forward locomotion the MDNs, Pair1 and A18b are silent; an unknown neuron (?) may initiate forward locomotion. (C) To initiate backward locomotion, MDNs activate Pair1 descending neuron which inhibits the forward-active A27h premotor neuron in segments A5-A7 to halt forward locomotion. MDNs also activate A18b in A1 and/or in more anterior segments which triggers a backward motor wave.
Videos
MDN activation induces backward larval locomotion.
Crawling behavior of third instar larvae expressing Chrimson in MDNs (Split1 > Chrimson:mVenus) with ATR. During the first 15 s, the animals are not under optogenetic light followed by 15 s under 0.5 mW/mm2 of green light.
MDN activation induces backward larval locomotion.
Crawling behavior of third instar larvae expressing Chrimson in MDNs (Split1 >Chrimson:mVenus) without ATR. During the first 15 s, the animals are not under optogenetic light followed by 15 s under 0.5 mW/mm2 of green light.
Pair1 activation blocks forward locomotion.
Crawling behavior of third instar larvae expressing Chrimson in Pair1 (75C02 > Chrimson:mVenus) with ATR. During the first 10 s, the animals are not under optogenetic light followed by 10 s under 0.28 mW/mm2 of green light.
Pair1 activation blocks forward locomotion.
Crawling behavior of third instar larvae expressing Chrimson in Pair1 (75C02 > Chrimson:mVenus) without ATR. During the first 10 s, the animals are not under optogenetic light followed by 10 s under 0.28 mW/mm2 of green light.
Larval MDNs persist into adulthood and induce backward walking.
Walking behavior of adult flies carrying all the components showed in 8F (split3, right) or all the genetic components except the DBD half of Split3 (control, left). During the first 10 s, the animals are not under optogenetic light followed by 10 s under 0.28 mW/mm2 of red light.
Additional files
-
Supplementary file 1
Graph view of the MDN and downstream neurons.
File can be opened in CATMAID using the graph widget.
- https://doi.org/10.7554/eLife.38554.021
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38554.022





