CD95/Fas ligand mRNA is toxic to cells
Figures
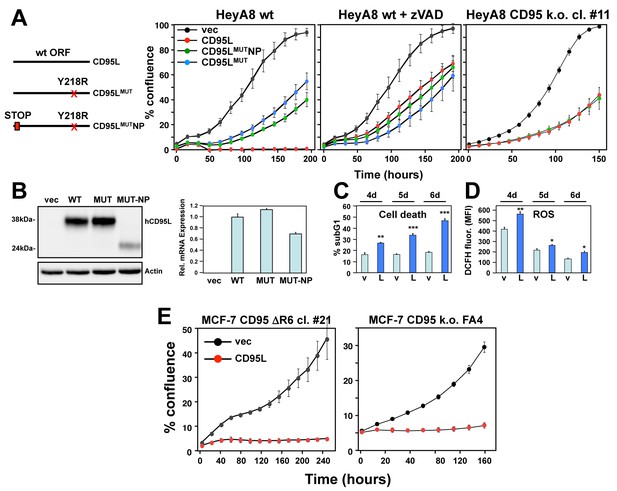
The CD95L mRNA is toxic to cells.
(A) Left: Schematic of the different CD95L mutants used. Right: Percent cell confluence over time of HeyA8 parental cells in the absence (left panel) or in the presence of 20 μM zVAD-fmk (center panel), or CD95 k.o. cells (right panel) after expression of CD95L constructs. Data are representative of one to three independent experiments. Values were calculated from samples done in triplicate or quadruplicate shown as mean ±SE. (B) Left: Western blot analysis of HeyA8 cells overexpressing different CD95L mutant RNAs. Cells expressing CD95LMUT or CD95L were pretreated with 20 μM zVAD-fmk. Note the small amount of truncated CD95L in cells infected with CD95L MUTNP does not have CD95 binding activity. Very similar data were obtained when the constructs were expressed in either CD95 k.o. HeyA8 cells (clone #11) or NB-7 cells, which lack expression of caspase-8, both without treatment with zVAD (data not shown). Right: RT-qPCR analysis for CD95L of the same samples. Data are representative of two independent experiments. Each bar represents mean ±S.D. of three replicates. (C, D) Quantification of cell death (C) and ROS production (D) in CD95 k.o. HeyA8 cells (clone #11) expressing either pLenti (v) or pLenti-CD95L (L) at different time points (days after infection). Data are representative of two independent experiments. Each bar represents mean ±SE of three replicates. *p<0.05, **p<0.001, ***p<0.0001, unpaired t-test. (E) Confluency over time of the MCF-7 complete CD95 k.o. FA4 clone (right) or a MCF-7 clone #21 in which we deleted the shR6 target site resulting in an out-of-frame shift after infection with either pLenti vector control (vec) or wt CD95L. Data are representative of two independent experiments. Each data point represents mean ±SE of three replicates.
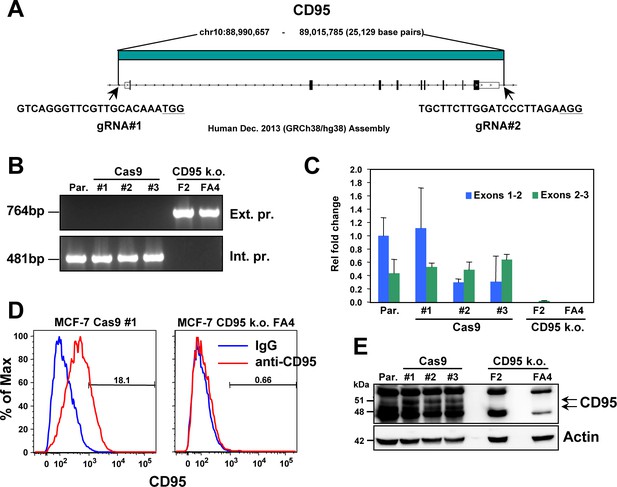
Generation of complete CD95 k.o. MCF-7 cells.
(A) Schematic of the genomic locations and sequences of the gRNAs used to excise the entire CD95 gene in MCF-7 cells. PAM sites are underlined. (B) PCR with flanking (top panels) and internal (bottom panels) primers used to confirm the absence of the CD95 gene in MCF-7 clones. Parental (Par.) cells and three clones infected with Cas9 only (Cas9) and two homozygous complete CD95 k.o. clones (F2 and FA4) are shown. (C) RT-qPCR analysis of the indicated clones using primers spanning either exon 1/2 or exon 2/3 of the CD95 gene. (D) Surface staining for CD95 of one wt and one k.o. clone. (E) Western blot analysis of all clones.
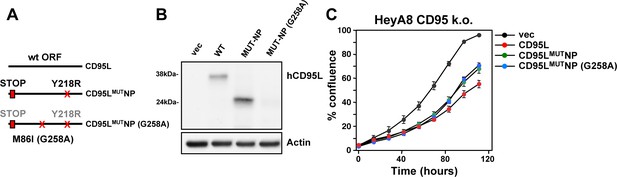
Mutation of the alternative start codon in CD95LMUTNP construct.
(A) Left: Schematic of the different CD95L mutants used. For the M86I mutant protein the point mutation on the DNA level is in parentheses. (B) Western blot analysis of HeyA8 CD95 k.o. cells overexpressing different CD95L mutants. Note, for unknown reasons in this experiment expression of the CD95LMUTNP protein was more efficient than in the experiment shown in Figure 1B. (C) Percent cell confluence over time of HeyA8 CD95 k.o. (cl. #11) cells after expression of the different CD95L constructs. Values were calculated from samples plated in triplicate shown as mean ±SE.
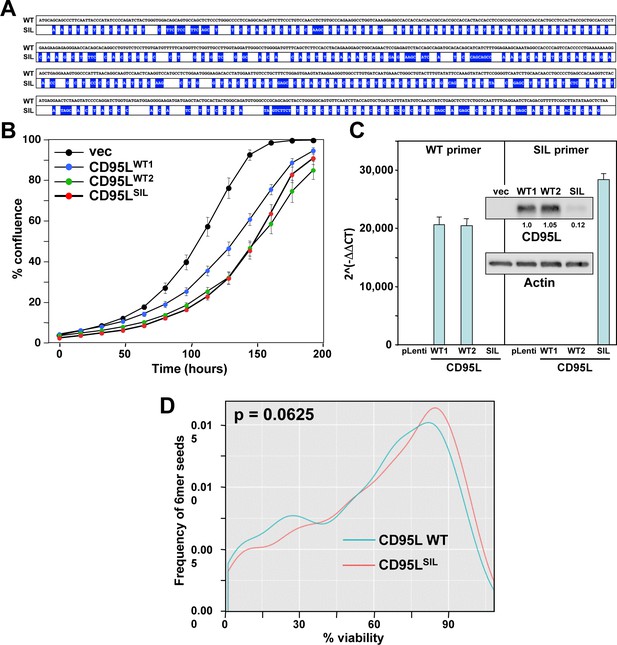
Toxicity of CD95L mRNA is independent of CD95L protein expression.
(A) Schematic showing the positions of the codon optimized silent mutations of the CD95LSIL mutant compared to wild type CD95L (mutations highlighted in blue). (B) Percent cell confluence over time of HeyA8 CD95 k.o. cells (cl. #11) over-expressing empty pLenti vector (vec), wild-type CD95L (from two separately cloned viruses), or the CD95LSIL. (C) RT-qPCR analysis and Western blot (inset) of wild type CD95L and CD95LSIL mutant mRNAs in the over-expressing cells shown in B. (D) Probability density plot comparing the toxicity of all possible 6mer seeds located in either the WT or SIL CD95L mRNA. p-value was calculated using a two-sample two-sided K-S test.
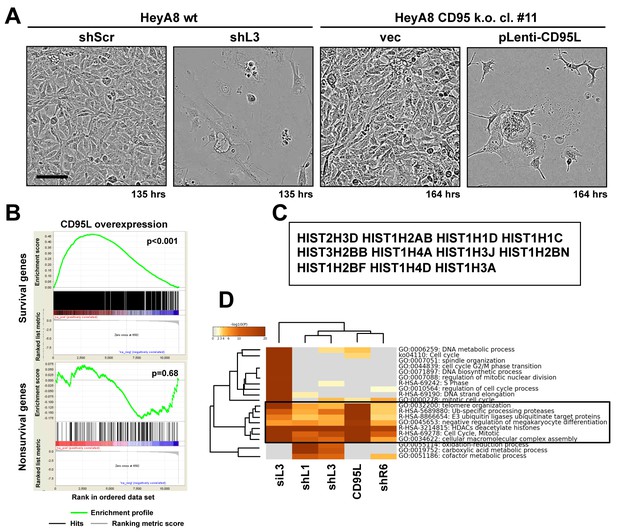
Toxicity induced by CD95L overexpression is reminiscent of DISE.
(A) Phase-contrast images of HeyA8 and HeyA8 CD95 k.o. cells (cl. #11) after infection with pLKO-shScr/shL3 or pLenti (vec)/pLenti-CD95L, respectively, at the indicated time point. (B) Gene set enrichment analysis for the 1846 survival genes (top panel) and the 416 nonsurvival genes (bottom panel) identified in the Sabatini study (Putzbach et al., 2017; Wang et al., 2015) of mRNAs downregulated in CD95L expressing HeyA8 CD95 k.o. cells compared to HeyA8 CD95 k.o. cells infected with pLenti virus. p-values indicate the significance of enrichment. (C) Common genes downregulated in all RNA-Seq experiments from (HeyA8) cells treated with either one of four si/shRNAs (Putzbach et al., 2017) derived from either CD95 or CD95L (see Figure 2D) and cells overexpressing CD95L ORF as described in (B). (D) Metascape analysis of 5 RNA Seq data sets analyzed. The boxed GO term clusters were highly enriched in all five data sets.
CD95 k.o. HeyA8 cells (cl. #11) infected with pLenti control virus.
https://doi.org/10.7554/eLife.38621.007CD95 k.o. HeyA8 cells (cl. #11) infected with pLenti-CD95L virus.
https://doi.org/10.7554/eLife.38621.008HeyA8 cells infected with pLKO-shScr.
https://doi.org/10.7554/eLife.38621.009HeyA8 cells infected with pLKO-shL3.
https://doi.org/10.7554/eLife.38621.010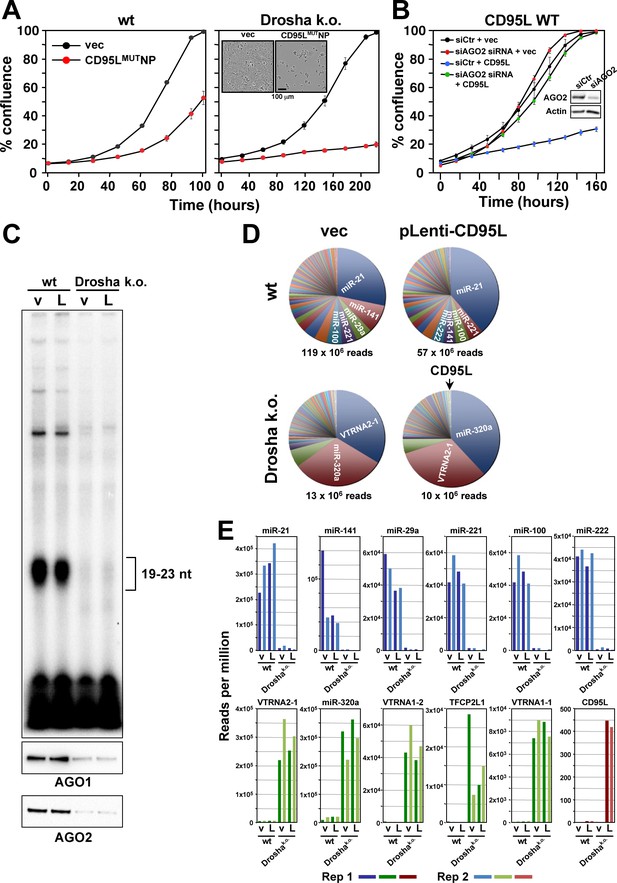
Small RNAs generated in cells expressing CD95L mRNA are loaded into the RISC.
(A) Percent cell confluence over time of HCT116 parental (left) or Drosha k.o. (right) cells after infection with CD95MUTNP. Data are representative of three independent experiments. Each data point represents the mean ±SE of three replicates. Inset: Phase contrast images of Drosha k.o. cells 9 days after infection with either empty vector or CD95LMUTNP. (B) Percent cell confluence of HeyA8 CD95 k.o. cells transfected with either non-targeting siRNA (siCtr) or a pool of 4 siRNAs targeting AGO2 following subsequent infection with either empty pLenti (vec) or pLenti CD95L. Inset: Western blot showing knock-down of human AGO2. (C) Top: autoradiograph on RNAs pulled down with the Ago binding peptide. Bottom: Western blot analysis of pulled down Ago proteins. v, pLenti; L, pLenti-CD95L expressing cells. (D) Pie charts showing the relative ratio of sRNAs pulled down with the Ago proteins in wt and Drosha k.o. cells. Depicted are all the amounts of all sRNAs that contributed at least 0.01% to the total RNA content. Only in the Drosha k.o. cells was a significant amount of CD95L derived Ago bound reads found. They represented the 75th most abundant sRNA species (arrow). The average number of total sequenced reads (of two duplicates) are shown for each condition. (E) Top: Number of reads (normalized per million) of the top six most abundant sRNAs in the RISC of either HCT116 wt-pLenti or -pLenti-CD95L cells. Bottom: Number of reads (per million) of the top five genes with sRNAs most abundant in the RISC or of CD95L in the RISC of either HCT116 Drosha k.o. pLenti (v), or -pLenti-CD95L (L) cells. Note: miR-21 is not included as it is already shown in the top row. Bottom right panel: Abundance of Ago bound CD95L derived sRNAs. Shown in all panels is the abundance of RNAs in the four samples. Rep 1 and Rep 2, replicate 1 and 2.
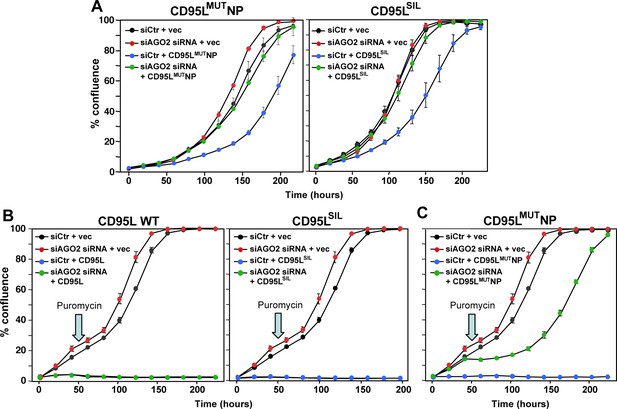
All CD95L mRNA mutants are toxic through RNAi.
(A) Percent cell confluence of HeyA8 CD95 k.o. cells (cl. #11) transfected with either non-targeting siRNAs (siCtr) or a pool of 4 siRNAs targeting AGO2 following subsequent infection with either empty pLenti (vec), pLenti CD95LMUTNP (left), or pLenti CD95LSIL (right). (B) Percent cell confluence of parental HeyA8 cells transfected with either a pool of 4 non-targeting siRNA (siCtr) or a pool of 4 siRNAs targeting AGO2 following subsequent infection with either empty pLenti (vec), pLenti CD95L WT (left), or pLenti CD95LSIL (right). (C) Percent cell confluence of parental HeyA8 cells transfected with either a pool of 4 non-targeting siRNA (siCtr) or a pool of 4 siRNAs targeting AGO2 following subsequent infection with either empty pLenti (vec), or pLenti CD95LMUTNP. In A cells were plated after puromycin selection. In (B) and (C) cells were plated directly after viral infection and puromycin was added 50 hr after infection (arrow).
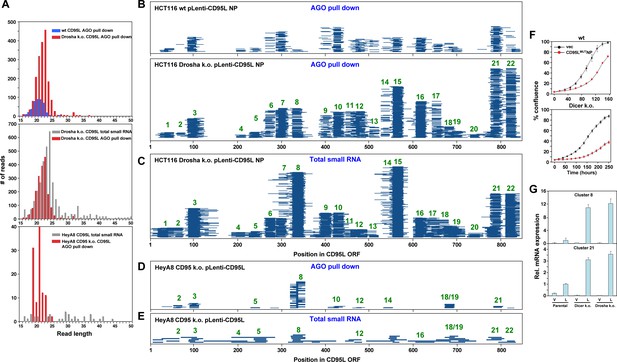
The entire CD95L mRNA gives rise to sRNAs that bind to the RISC.
(A) Length distribution of CD95L derived reads in various analyses. (B, C) Read alignment with CD95LMUTNP ORF of analyses of sRNAs pulled down with Ago proteins from HCT116 wt (B, top) and Drosha k.o. (B, bottom) cells and of total sRNAs from HCT116 Drosha k.o. cells (C) after infection with CD95LMUTNP. (D, E) Read alignment with wt CD95L ORF of analyses of sRNAs pulled down with Ago proteins (D) or total sRNAs (E) from HeyA8 CD95 k.o. cells after infection with wt CD95L. (F) Percent cell confluence over time of HCT116 parental (top) or Dicer k.o. (clone #43) (bottom) cells after infection with CD95MUTNP. (Dicer k.o. clone #45, gave a similar result, data not shown). Data are representative of two independent experiments. Each data point represents the mean ±SE of three replicates. (G) RT-qPCR analysis of clusters 8 and 21 in HCT116 parental, Dicer k.o. (clone #43), and Drosha k.o. cells after infection with CD95MUTNP. Each bar represents mean ± S.D. of three replicates. v, vector, L, CD95L expressing cells.
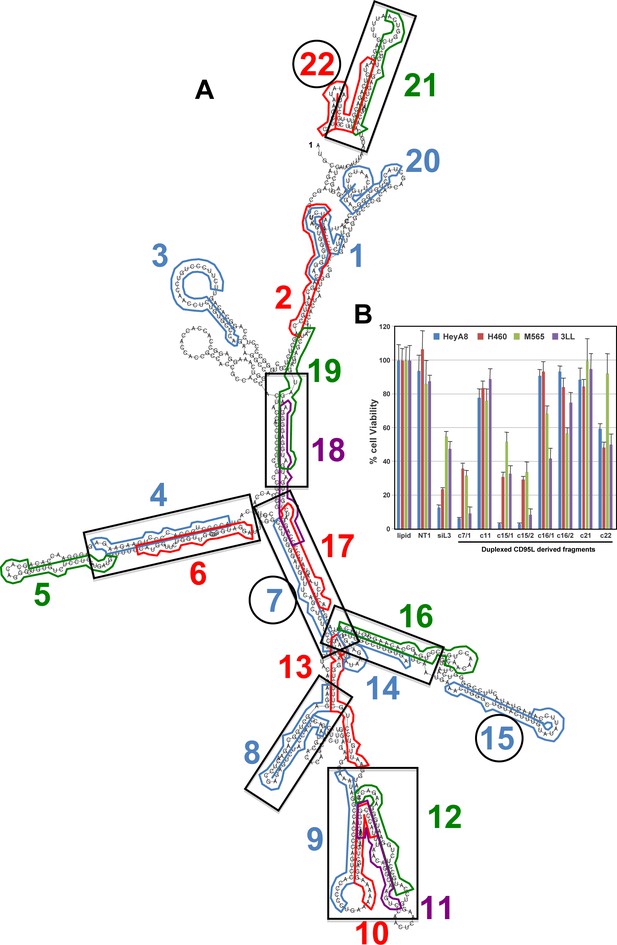
Predicted secondary structure of CD95L ORF and toxicity of CD95L-derived sRNAs after conversion to siRNAs.
(A) The CD95LMUTNP RNA was subjected to a RNA secondary structure analysis (http://rna.tbi.univie.ac.at) using default settings. The locations of 22 reads representative of the 22 read clusters are shown. Regions with potential duplex formation are boxed. The oligonucleotides that were found to be toxic when expressed as siRNAs are circled. (B) Toxicity of the eight siRNAs designed using the CD95L-derived small RNA fragments bound to Ago as the antisense strand sequences 96 hr post-transfection in the indicated cell lines. Each data point represents the mean ±SE of three replicates.
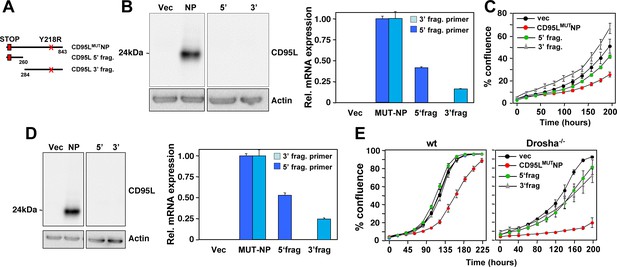
CD95L fragments are less toxic than full length CD95L mRNA.
(A) Schematic of the different CD95L fragments used. (B) Left: Western blot analysis of HeyA8 CD95 k.o. cells overexpressing different CD95L mutant RNAs. Right: RT-qPCR analysis for CD95L of the same samples using primers detecting either the 5' or the 3' half of the mRNA. Data are representative of two independent experiments. Each bar represents mean ±S.D. of three replicates. (C) Percent cell confluence over time of HeyA8 CD95 k.o. cells after expressing the CD95L mutant or fragments. Data are representative of two independent experiments. Values were calculated from samples done in triplicate shown as mean ±SE. (D) Left: Western blot analysis of HCT116 Drosha k.o. cells overexpressing different CD95L mutant RNAs. Right: RT-qPCR analysis for CD95L of the same samples using primers detecting either the 5' or the 3' half of the mRNA. Data are representative of two independent experiments. Each bar represents mean ±S.D. of three replicates. (E) Percent cell confluence over time of HCT116 Drosha k.o. cells after expressing the CD95L mutant or fragments. Data are representative of two independent experiments. Values were calculated from samples done in triplicate shown as mean ±SE.
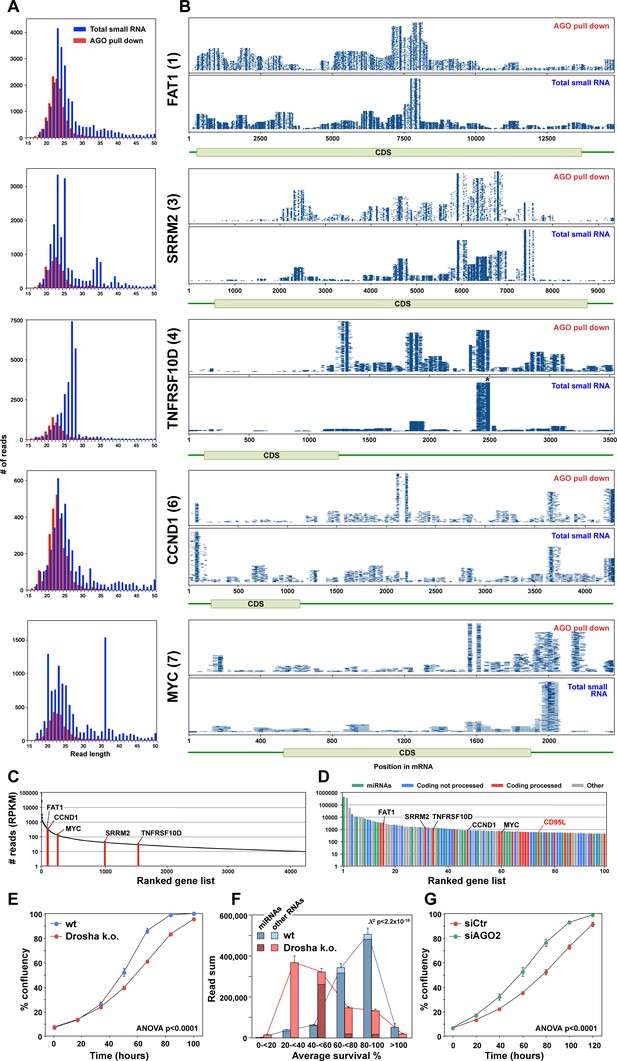
Endogenous mRNAs are processed and loaded into the RISC.
(A) Length distribution of reads derived from five of the top ten most abundant genes loaded into the RISC of CD95L expressing HCT116 Drosha k.o. cells. The numbers in parentheses indicate the ranking in the top ten most abundant genes with Ago bound reads. (B) Alignment of the reads from the five genes shown in A with horizontal blue lines representing the mapped positions of the reads. Each blue line represents an individual read, with its length in the plot proportional to the read length. Small RNAs pulled down with Ago proteins (top) or total sRNAs (bottom) from HCT116 and Drosha k.o. cells after infection with wt CD95L. *This stack contains 14899 reads of which 3000 were randomly chosen and plotted. (C) All 4262 genes in HCT116 Drosha k.o. cells expressing CD95L ranked according to highest expression with more than 10 reads expressed as reads per kb per million (RPKM). The abundance of the six genes shown in A and B is labeled. (D) Genes ranked according to highest abundance in the RISC of Drosha k.o. cells. Reads derived from the five genes in A are labeled as well as the location of the reads derived from CD95L. (E) Percent cell confluence over time of parental HCT116 and Drosha k.o. cells. (F) Average seed toxicity of all Ago-bound miRNAs and non-miRNAs (Other) in parental HCT116 and Drosha k.o. cells. Reads are shown as reads per million (RPM). Chi squared test was used to calculate p-value. (G) Percent cell confluence over time of Drosha k.o. HCT116 cells 24 hr after transfection with 25 nM of either nontargeting SMARTpool (siCtr) or AGO2 SMARTpool siRNAs. Each data point represents mean ±SE of three replicates. The experiment is representative of three biological repeats.
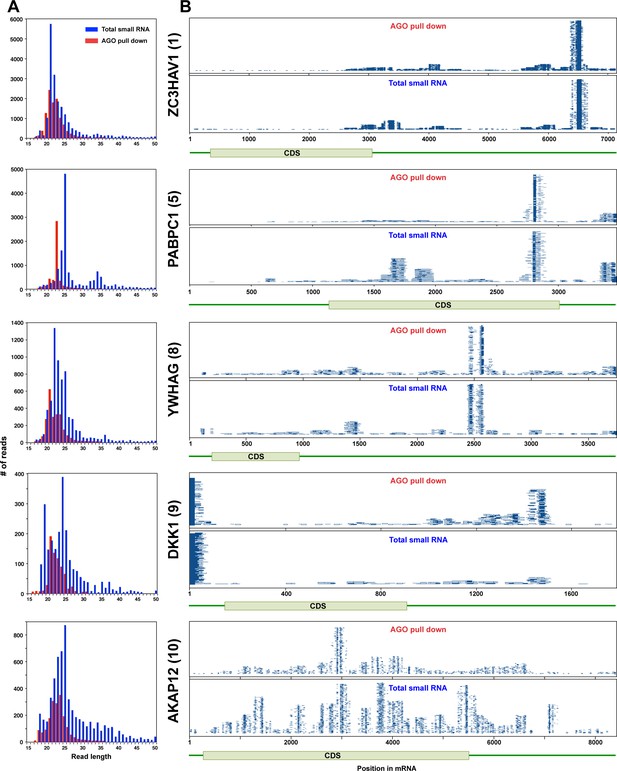
Endogenous mRNAs are processed and loaded into the RISC - additional genes.
(A) Length distribution of reads derived from 5 of the top ten most abundant genes loaded into RISC of CD95L expressing HCT116 Drosha k.o. cells. The numbers in brackets indicate the ranking in the top ten most abundant genes with Ago bound reads. (B) Alignment of the reads from the five genes shown in A with their respective mRNAs derived from small RNAs pulled down with Ago proteins (top) or total small RNAs (bottom) from HCT116 and Drosha k.o. cells after infection with WT CD95L.

Mapping of Ago bound reads from five processed genes to the human genome.
Alignment of all reads derived from the five genes shown in Figure 5A and B with the human genome. Shown are all 8 tracks of HCT116 wt and HCT116 Drosha k.o. cells infected with either pLenti control vector (vec) or pLenti-CD95L in duplicate. For each of the genes the Y axis was fixed to the same scale.
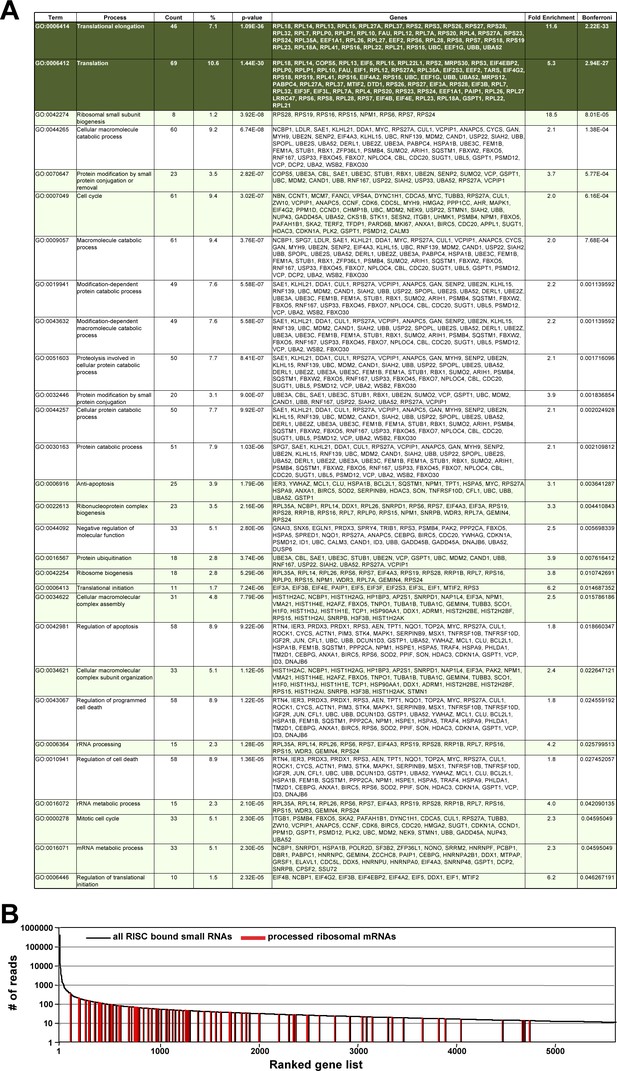
Genes with multiple reads bound to Ago proteins are involved in cell growth and protein translation.
(A) All 558 protein coding genes that were processed similar to CD95L and had reads bound to Ago proteins were subjected to a DAVID GO analysis. Shown are all significantly enriched gene clusters. The top two clusters (dark green) stood out with very low p-values of enrichment. Clusters highlighted in green are connected to cell proliferation (cell cycle, anti-apoptosis or protein translation). (B) All 5629 genes with Ago bound reads (10 or more counts) ranked according highest abundance. The positions of the mRNAs of ribosomal proteins are indicated in red in the ranked mRNAs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CD95L | NA | NM_000639 | |
| Gene (H. sapiens) | CD95 | NA | NM_000043 | |
| Cell line (H. sapiens) | MCF-7 | ATCC | ATCC: HTB-22 | Human adenocarcinoma of the mammary gland, breast; derived from metastatic site: pleural effusion |
| Cell line (H. sapiens) | MCF-7 CD95 ΔshR6 clone #21 | this paper | NA | MCF-7 CD95 ΔshR6 clone #21 with homozygous 227 nucleotide deletion of the shR6 target site in CD95 (chr10:89,008,920–89,009,146; Human Dec. 2013 GRCh38/hg38 assembly) produced using CRISPR/Cas9 technology; verified homozygous CD95 protein knockout |
| Cell line (H. sapiens) | MCF-7 CD95 deletion clone FA4 | this paper | NA | MCF-7 CD95 deletion clone FA4 with a homozygous deletion of the entire CD95 gene (chr10:88,990,657–89,015,785; Human Dec. 2013 GRCh38/hg38 assembly) produced using CRISPR/Cas9 technology; verified homozygous CD95 protein knockout |
| Cell line (H. sapiens) | HeyA8 | PMID: 4016745 | RRID: CVCL_8878 | Human high grade ovarian serous adenocarcinoma; derived from parent Hey cells (RRID: CVCL_0297) |
| Cell line (H. sapiens) | HeyA8 shR6 k.o. clone #11, HeyA8 CD95 k.o. | PMID: 29063830 | NA | HeyA8 CD95 k.o. clone with a homozygous 227 nucleotide deletion of the shR6 target site in CD95 (chr10:89,008,920–89,009,146; Human Dec. 2013 GRCh38/hg38 assembly) produced using CRISPR/Cas9 technology; verified homozygous CD95 protein knockout |
| Cell line (H. sapiens) | HCT116 | Korean Collection for Type Cultures (KCTC) | KCTC: cat#HC19023; ATCC: CCL_247 | Human colorectal carcinoma |
| Cell line (H. sapiens) | Drosha-/-; Drosha-/- clone #40 | Korean Collection for Type Cultures (KCTC); PMID: 26976605 | KCTC: cat#HC19020 | HCT116 clone #40 with homozygous knockout of Drosha protein; knockout achieved using CRISPR/Cas9 which resulted in a single nucleotide insertion in one allele and a 26 nucleotide deletion in the other |
| Cell line (H. sapiens) | Dicer-/-; Dicer-/- clone #43 | Korean Collection for Type Cultures (KCTC); PMID: 26976606 | KCTC: cat#HC19023 | HCT116 clone #43 with homozygous knockout of Dicer protein; knockout achieved using CRISPR/Cas9 which resulted in a three nucleotide insertion and 14 nucleotide deletion in one allele and a 35 nucleotide deletion in the other |
| Cell line (H. sapiens) | Dicer-/-; Dicer-/- clone #45 | Korean Collection for Type Cultures (KCTC); PMID: 26976607 | KCTC: cat#HC19024 | HCT116 clone #45 with homozygous knockout of Dicer protein; knockout achieved using CRISPR/Cas9 which resulted in a 53 nucleotide deletion in one allele and a 28 nucleotide deletion in the other |
| Cell line (H. sapiens) | 293T | ATCC | ATCC: CRL-3216 | Derived from HEK293 cells (ATCC: CRL-1573); express large T antigen; used for packaging viruses |
| Cell line (H. sapiens) | H460 | ATCC | ATCC: #HTB-177 | Human lung pleural effusion carcinoma |
| Cell line (Mus musculus) | 3LL | ATCC | ATCC #CRL-1642 | Mouse Lewis lung carcinoma |
| Cell line (Mus musculus) | M565 | PMID: 25366259 | NA | Mouse hepatocellular carcinoma isolated from naturally occurring tumor in a floxed CD95 background |
| Antibody | anti-human AGO1 (rabbit monoclonal) | Cell Signaling | Cell Signaling #5053 | 1:2000; for western blot; primary Ab |
| Antibody | anti-human AGO1 (rabbit polyclonal) | Abcam | Abcam #98056 | 1:2000; for western blot; primary Ab |
| Antibody | anti-human AGO2 (rabbit polyclonal) | Abcam | Abcam #32381 | 1:500; for western blot; primary Ab |
| Antibody | Goat anti-rabbit, IgG-HRP | Southern Biotech | Southern Biotech: cat#SB-4030–05 | 1:5000; for western blot; secondary Ab |
| Antibody | Anti-Argonaute-2 antibody (rabbit monoclonal) [EPR10411] | Abcam | Abcam #186733 | 1:1200; for western blot; primary Ab |
| Antibody | Anti-Human CD178 antibody (Mouse IgG1) Clone G247-4 | BD Pharmingen | BD Pharmingen #556387 | 1 μg/ml; for western blot; primary Ab |
| Antibody | Anti-CD95 (rabbit polyclonal, C-20) | Santa Cruz | Santa Cruz #sc-715 (since discontinued) | 1:1000; for western blot; primary Ab |
| Antibody | Goat anti-rabbit, IgG-HRP | Cell Signaling | Cell Signaling: cat#7074 | 1:2000; for western blot; secondary Ab |
| Antibody | Goat anti-mouse; IgG1-HRP | Southern Biotech | Southern BioTech: cat#1070–05 | 1:5000; for western blot; secondary Ab |
| Recombinant protein reagent | LzCD95L | PMID: 14504390 | NA | Leucine zipper tagged CD95L; recombinant protein |
| Chemical compound | CellTiter-Glo | Promega | Promega #G7570 | Detects ATP release as a surrogate for cell death; read-out is fluorescence |
| Chemical compound | propidium iodide | Sigma-Aldrich | Sigma-Aldrich: cat#P4864 | Used for subG1 flow cytometry analysis |
| Chemical compound | puromycin | Sigma-Aldrich | Sigma-Aldrich: cat#P9620 | Used for selection of cells expressing puromycin resistance cassettes |
| Chemical compound | 2',7'-dichlorodihydrofluorescein diacetate | Thermofisher Scientific | Thermofisher Scientific #D399 | Dye used for detecting ROS production |
| Chemical compound | zVAD-fmk | Sigma-Aldrich | Sigma-Aldrich: cat#V116 | Used at 20 uM; pan caspase inhibitor |
| Recombinant DNA reagent | pLenti-GIII-CMV- RFP-2A-Puro vector; pLenti | ABM Inc | NA | pLenti control empty lentiviral vector; carries an RFP-2a-puromycin resistance cassette |
| Recombinant DNA reagent | pLenti-CD95L | this paper | NA | pLenti-GIII-CMV-RFP-2A-Puro vector that expresses human wild type CD95L cDNA (NM_000639.2); used to express wt human CD95L upon infection with lentiviral particles |
| Recombinant DNA reagent | pLenti-CD95LMUT | this paper | NA | pLenti-GIII-CMV-RFP-2A-Puro vector that expresses human CD95L cDNA (NM_000639.2) with two nucleotide substitutions in codon 218 (TAT - > CGT) resulting in replacement of tyrosine for arginine (Y218R mutation); unable to bind CD95 |
| Recombinant DNA reagent | pLenti-CD95LMUTNP | this paper | NA | pLenti-GIII-CMV-RFP-2A-Puro vector that expresses human CD95L cDNA (NM_000639.2) with both the Y218R mutation and a single nucleotide substitution at the second codon (CAG - > TAG), resulting in a premature stop codon right after the start codon |
| Recombinant DNA reagent | pLenti-CD95LSIL | this paper | NA | pLenti-GIII-CMV-RFP-2A-Puro vector that expresses human CD95L cDNA (NM_000639.2) with all codons containing synonymous mutations except for select codons in the proline-rich domain to meet IDT synthesis criteria |
| Transfected construct | gRNA scaffold | PMID: 23287722 | IDT: synthesized as gene block | 455 nucleotide CRISPR/Cas9 gRNA scaffold synthesized as a gene block; contains promoter, gRNA scaffold, target sequence, and termination sequence; scaffold transcribes gRNAs that target Cas9 endonuclease to cut at target sites; target sequences consist of 19 nucleotides that are complementary to the target site of choice; co-transfected with Cas9 to catalyze cleavage. |
| Recombinant DNA reagent | pLenti-CD95LMUTNP (G258A) | This paper | NA | pLenti-GIII-CMV-RFP-2A-Puro vector that expresses human CD95L cDNA (NM_000639.2) with the Y218R mutation, and two additional single nucleotide substitutions; one at the second codon (CAG - > TAG), resulting in a premature stop codon right after the start codon, and another, G258A, resulting in the replacement of a methionine with an isoleucine, thus removing the alternative translational start site. |
| Transfected construct | pMJ920 Cas9 plasmid | Addgene; PMID: 23386978 | Addgene: cat#42234 | Plasmid that expresses a human codon-optimized Cas9 tagged with GFP and HA; used to express Cas9 for CRISPR-mediated deletions. |
| Chemical compound | Lipofectamine 2000 | ThermoFisher Scientific | ThermoFisher Scientific: cat#11668019 | Transfection reagent |
| Chemical compound | Lipofectamine RNAiMAX | ThermoFisher Scientific | ThermoFisher Scientific: cat#13778150 | Transfection reagent; used for transfection of small RNAs such as siRNAs |
| Commercial assay or kit | StrataClone Blunt PCR Cloning Kit | Agilent Technologies | Agilent Technologies: cat#240207 | Used to blunt-end clone the gRNA scaffolds into the pSC-B plasmid |
| Genetic reagent | Taqman Gene expression master mix | ThermoFisher Scientific | #4369016 | |
| Sequence-based reagent | shR6 flanking Fr primer | IDT | IDT: custom DNA oligo | Fr primer that flanks shR6 site; used to detect 227 nt shR6 deletion; 5’-GGTGTCATGCTGTGACTGTTG-3’ |
| Sequence-based reagent | shR6 flanking Rev primer | IDT | IDT: custom DNA oligo | Rev primer that flanks shR6 site; used to detect 227 nt shR6 deletion; 5’-TTTAGCTTAAGTGGCCAGCAA-3’ |
| Sequence-based reagent | shR6 internal Rev primer | IDT | IDT: custom DNA oligo | Rev primer that overlaps with the shR6 site; used to detect 227 nt shR6 deletion; 5’-AAGTTGGTTTACATCTGCAC-3’ |
| Sequence-based reagent | CD95 flanking Fr primer | IDT | IDT: custom DNA oligo | Fr primer that flanks the CD95 gene; used to detect CD95 gene deletion; 5’-TGTTTAATATAGCTGGGGCTATGC-3' |
| Sequence-based reagent | CD95 flanking Rev primer | IDT | IDT: custom DNA oligo | Rev primer that flanks the CD95 gene; used to detect CD95 gene deletion; 5’-TGGGACTCATGGGTTAAATAGAAT-3’ |
| Sequence-based reagent | CD95 internal Rev primer | IDT | IDT: custom DNA oligo | Rev internal primer that targets within the CD95 gene; used to detect CD95 gene deletion; 5’-GACCAGTCTTCTCATTTCAGAGGT-3’ |
| Sequence-based reagent | siScr/siNT1 | IDT; | IDT: custom DNA oligo | control non-targeting siRNA; sense: UGGUUUACAUGUCGACUAA-3' |
| Sequence-based reagent | c7/1 | IDT | custom siRNA; antisense strand corresponds to cluster 7 CD95L sequence | antisense: 5’-AUUGGGCCUG GGGAUGUUU-3'; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c7/2 | IDT | custom siRNA; antisense strand corresponds to cluster 7 CD95L sequence | antisense: 5’-CCUGGGGAU GUUUCAGCUC-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c11 | IDT | custom siRNA; antisense strand corresponds to cluster 11 CD95L sequence | antisense: 5’-CCAACUCAAGG UCCAUGCC-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c15/1 | IDT | custom siRNA; antisense strand corresponds to cluster 15 CD95L sequence | antisense: 5’-AAACUGGGCUGU ACUUUGU-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c15/2 | IDT | custom siRNA; antisense strand corresponds to cluster 15 CD95L sequence | antisense: 5’- AACUGGGCUGU ACUUUGUA-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c16/1 | IDT | custom siRNA; antisense strand corresponds to cluster 16 CD95L sequence | antisense: 5’- CAACAACCUGCC CCUGAGC-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c16/2 | IDT | custom siRNA; antisense strand corresponds to cluster 16 CD95L sequence | antisense: 5’- AACUCUAAGCG UCCCCAGG-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c21 | IDT | custom siRNA; antisense strand corresponds to cluster 21 CD95L sequence | antisense: 5’- UCAACGUAUC UGAGCUCUC-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | c22 | IDT | custom siRNA; antisense strand corresponds to cluster 22 CD95L sequence | antisense: 5’- AAUCUCAGACG UUUUUCGG-3’; antisense strand designed with 3' deoxy AA; complementary sense strand has 3' deoxy TT and 2'-O-methylation at the first two positions |
| Sequence-based reagent | siCtr pool | Dharmacon | D-001810–10 | control non-targeting siRNA pool |
| Sequence-based reagent | SMARTpool siRNA targeting AGO2 | Dharmacon | L-004639-00-0005 | siRNA pool designed to target AGO2 |
| Sequence based reagent (human) | GAPDH primer | Thermofisher Scientific | Hs00266705_g1 | RT-qPCR; control probe |
| Sequence based reagent (human) | CD95L primers | Thermofisher Scientific | Hs00181226_g1; Hs00181225_m1 | RT-qPCR |
| Sequence based reagent (human) | CD95 primers | Thermofisher Scientific | Hs00531110_m1; Hs00236330_m1 | RT-qPCR |
| Sequence based reagent (human) | CD95LSIL primer | Thermofisher Scientific | assay ID: APNKTUD | Custom RT-qPCR primer designed using the Thermofisher Scientific design tool to detect CD95LSIL mRNA |
| Sequence based reagent (human) | Cluster 8 CD95L small RNA primer | Thermofisher Scientific | custom probe Assay ID: CT7DPEM | Custom RT-qPCR primer designed using the Thermofisher Scientific design tool at https://www.thermofisher.com/order/custom- genomic-products/tools/small-rna to specifically detect small RNAs from cluster 8 of CD95L (5’- AAGGAGCTGGCAGAACTCCGAGA-3’) |
| Sequence based reagent (human) | Cluster 21 CD95L small RNA primer | Thermofisher Scientific | custom probe Assay ID: CTAAADA | Custom RT-qPCR primer designed using the Thermofisher Scientific design tool at https://www.thermofisher.com/order/custom- genomic-products/tools/small-rna to specifically detect small RNAs from cluster 21 of CD95L (5’- TCAACGTATCTGAGCTCTCTC-3’) |
| Sequence based reagent (human) | z30 primer | Thermofisher Scientific | ThermoFisher Scientific #4427975 | RT-qPCR for small RNA; control probe |
| Peptide, recombinant protein | Flag-GST-T6B peptide | PMID: 26351695 | NA | Peptide derived from GW182/TNRC6B used to pull down AGO1 to 4 |
| Commercial assay or kit | anti-Flag M2 magnetic beads | Sigma-Aldrich | Sigma-Aldrich #M8823 |
Additional files
-
Supplementary file 1
Reads from coding and noncoding genes pulled-down with Ago proteins in HCT116 Drosha k.o. pLenti-CD95L cells.
Tab 1: Reads from all genes; Tab 2: Reads from processed protein coding genes (>10 reads), Tab 3: Reads from unprocessed protein coding genes (>10 reads).
- https://doi.org/10.7554/eLife.38621.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38621.021





