An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics
Figures
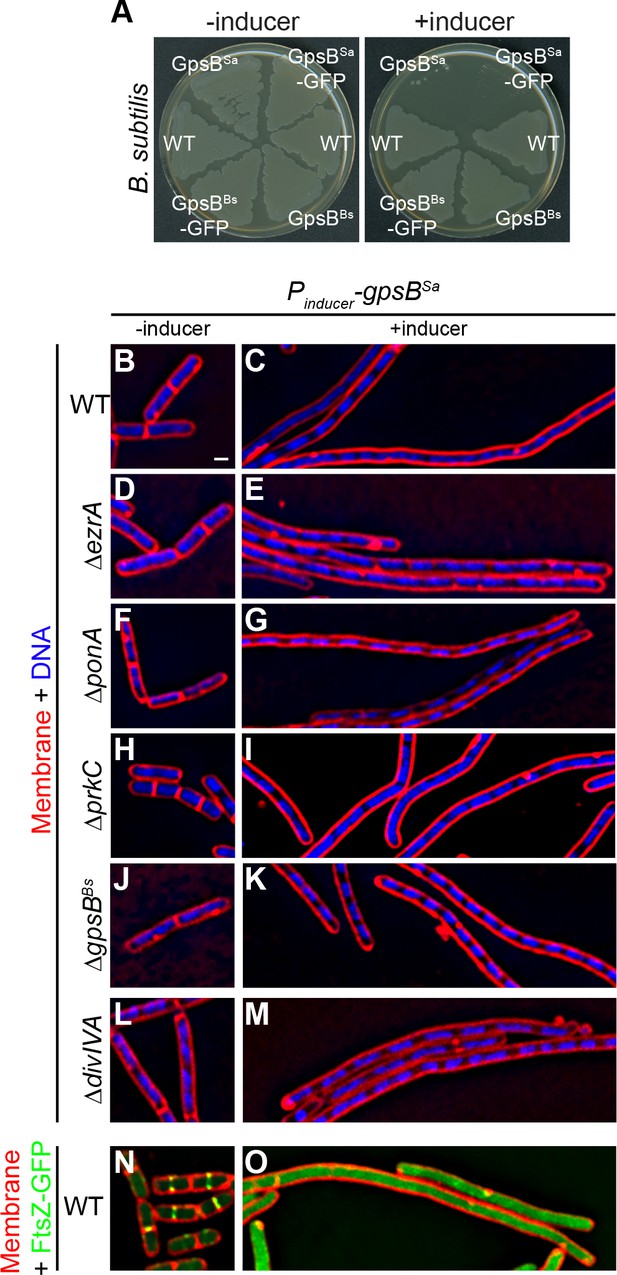
Overexpression of S.aureus gpsB inhibits cell division in B. subtilis.
(A) Luria-Bertani agar plates streaked with wild type B. subtilis (WT, strain PY79), or otherwise wild type B. subtilis harboring an inducible copy of gpsBBs (GG18), gpsBBs-gfp (GG19), gpsBSa (GG7), or gpsBSa-gfp (GG8) integrated into the chromosome, in the absence (left) or presence (right) of inducer. (B–M) Morphology of cells of different deletion mutants of B. subtilis (ΔezrA, GG35; ΔponA, CS26; ΔprkC, CS24; ΔgpsB, CS40; ΔdivIVA, CS94) harboring an inducible copy of gpsBSa grown in the absence (B, D, F; H, J, L) or presence (C, E, G, I, K, M) of inducer. (N–O) Localization of FtsZ-GFP in a strain (GG9) harboring an inducible copy of gpsBSa grown in the absence (N) or presence (O) of inducer. Membranes (red; B–O) visualized using the fluorescent dye FM4-64; chromosomes (blue; B–M) visualized using DAPI; FtsZ-GFP localization (green; N–O). Scale bar: 1 μm. Genotypes are listed in Key Resources Table.
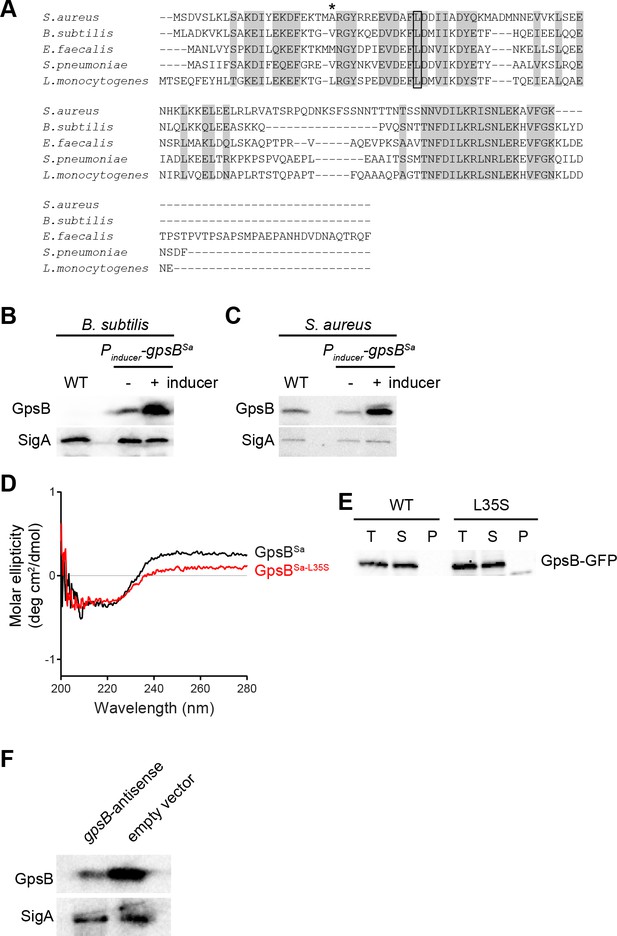
GpsB sequence and subcellular distribution in S.aureus.
(A) Alignment of amino acid sequences of GpsB from various strains. Identical residues are highlighted in gray; Leu35, (site of loss-of-function suppressor mutation in gpsBSa) is boxed; Ala23 (which corresponds to Leu24 in the L. monocytogenes ortholog and mediates membrane association) is denoted with an asterisk. (B) Immunoblot of cell extracts from B. subtilis cells (WT; PY79) or cells harboring an inducible copy of gpsBSa (GG7) in the absence (-) or presence (+) of inducer using rabbit antisera raised against purified GpsBSa or SigA (loading control). Note: anti-GpsBSa antiserum does not detect B. subtilis GpsB. (C) Immunoblot of cell extracts from S. aureus cells (WT; SH1000) or cells harboring a plasmid-encoded inducible copy of gpsBSa (SH1000 pPE45) in the absence (-) or presence (+) of inducer using rabbit antisera raised against purified GpsBSa or B. subtilis SigA (which also recognizes S. aureus SigA). (D) Circular dichroism spectra of purified GpsBSa (black) or GpsBSa-L35S (red). (E) Total extracts (T) of S. aureus cells producing GpsB-GFP or GpsBL35S-GFP were prepared and fractionated into soluble supernatant (S) and insoluble pellet (P) fractions by ultracentrifugation. GpsB-GFP was detected using rabbit anti-GpsB antiserum raised against purified GFP mut2. (F) Immunoblot of cell extracts from S. aureus cells harboring an inducible copy of gpsB antisense RNA (left) or empty vector (right) grown in the presence of inducer using antisera recognizing GpsB or SigA.
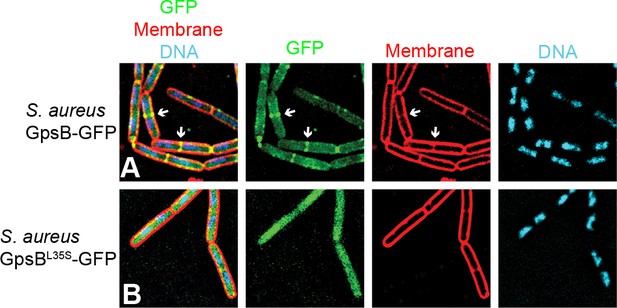
GpsBSa-GFP localizes to division septa in B.subtilis.
Localization of (A) GpsBSa-GFP or (B) GpsBSa-L35S-GFP in B. subtilis. First panel: overlay of GFP, membrane visualized using FM4-64, and DNA visualized using DAPI; second panel: GFP fluorescence; third panel: membrane; fourth panel: DNA. Arrows indicate sites of cell division.
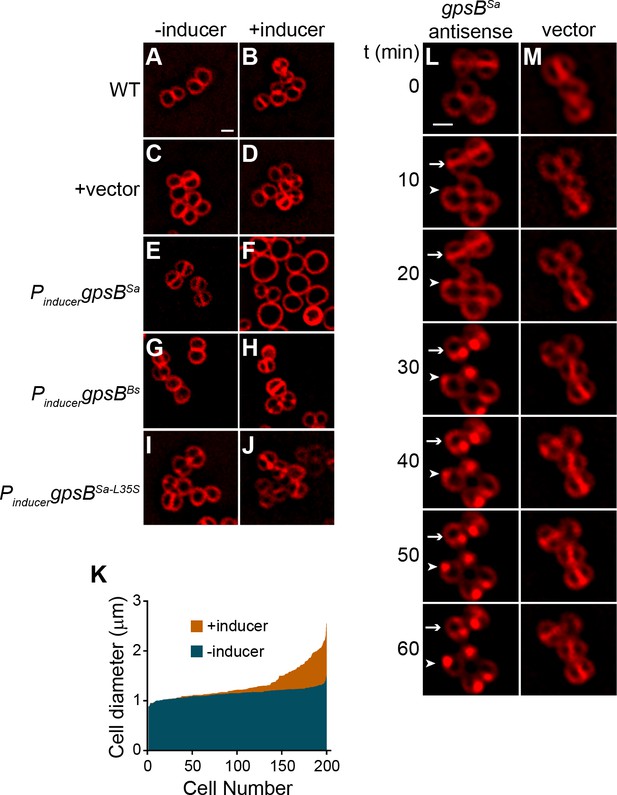
Overexpression and depletion phenotypes of gpsB in S.aureus.
(A–J) Morphology of wild type S. aureus cells (A-B; strain SH1000) or S. aureus strains harboring a plasmid encoding an inducible copy of gpsBSa (E–F; plasmid pPE45), vector backbone alone (C–D; pCL15), gpsBBs (G–H; pPE83), or gpsBSa-L35S (I–J; pPE79) in the absence (A, C, E, G, I) or presence (B, D, F, H, J) of inducer. (K) Histogram of cell diameters of 200 individual S. aureus cells harboring an inducible copy of gpsBSa grown in the absence (blue) or presence (orange) of inducer IPTG. (L–M) Morphology of S. aureus cells harboring a plasmid encoding an inducible copy of antisense RNA against gpsB (L; SH1000 pGG59) or empty vector (M) at times (min) indicated to the left after induction. Arrow indicates a cell that has already initiated cell division when GpsB depletion was initiated; arrowhead indicates a cell that had not initiated cell division at the time of GpsB depletion. Membranes visualized using FM4-64. Scale bar: 1 μm.
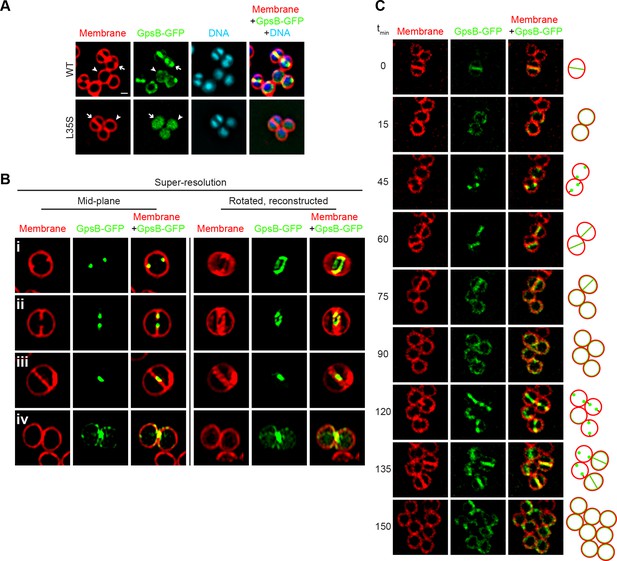
Dynamic redistribution of GpsB to mid-cell and periphery of S.aureus during the cell cycle.
(A) Localization of GpsB-GFP (top, SH1000 pPE46) or GpsBL35S-GFP (bottom, SH1000 pPE80) to mid-cell in actively dividing cells (arrow) and to the periphery of cells that are not dividing (arrowhead). First panel: membranes visualized using FM4-64; second panel: GFP fluorescence; third panel: chromosomes visualized using DAPI; fourth panel: overlay of membrane, GFP, and DNA. (B) GpsB-GFP localization in S. aureus cells at various stages of division (i–iv) using structured illumination microscopy (SIM). First column: membranes visualized using FM4-64; second column: GpsB-GFP fluorescence; third column: overlay, membrane and GpsB-GFP. Columns 4–6: reconstruction of deconvolved Z-stacks and rotation of the cells in columns 1–3, respectively, around the vertical axis. (C) Time-lapse fluorescence micrographs of a dividing S. aureus cell taken at the time intervals indicated at the left. Left panels: membranes visualized using FM4-64; middle panels: GpsB-GFP fluorescence; right panels: overlay, membrane and GpsB-GFP. Depictions of GpsB-GFP localization patterns are to the right of the panels. Scale bar: 1 μm.
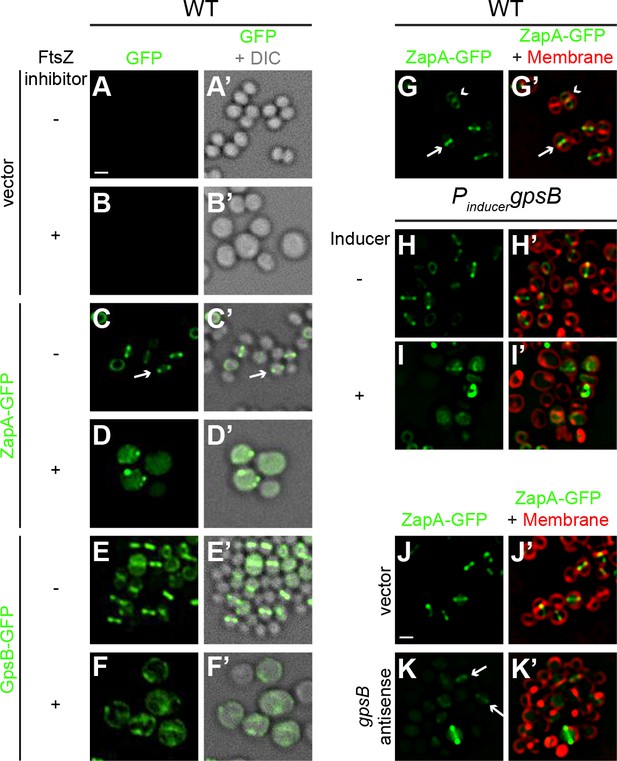
FtsZ and GpsB reciprocally influence each other’s subcellular localization pattern.
Morphology of wild type S. aureus cells harboring empty vector (strain SH1000 pCL15) in the absence (A–A’) or presence (B–B’) of FtsZ inhibitor PC190723 as visualized in the GFP fluorescence channel (A–B) or by differential interference contrast (DIC; A’–B’). Localization of ZapA-GFP (proxy for FtsZ localization; strain SH1000 pRB42; C–D’) or GpsB-GFP (E-F’; SH1000 pPE46) in the absence (C-C'; E-E') or presence (D-D'; F-F') of S. aureus FtsZ GTPase activity inhibitor PC190723. (G–I’) Localization of ZapA-GFP in wild type (G-G’; SH1000 pRB42), or in cells harboring an IPTG-inducible copy of gpsB in the absence (H-H’; SH1000 pPE45 pRB42) or presence (I–I’) of IPTG. Localization of ZapA-GFP in cells harboring vector only (J-J’; SH1000 pRB42 pEPSA5) or vector producing gpsB antisense RNA (K-K’; SH1000 pRB42 pGG59). A-K: GFP fluorescence; A’-F’: overlay of GFP fluorescence and DIC; G’-K’: overlay of GFP fluorescence and membrane. Arrows indicate site of cell division; arrowheads indicate ZapA-GFP localization at the subsequent plane of cell division. Scale bar: 1 μm.
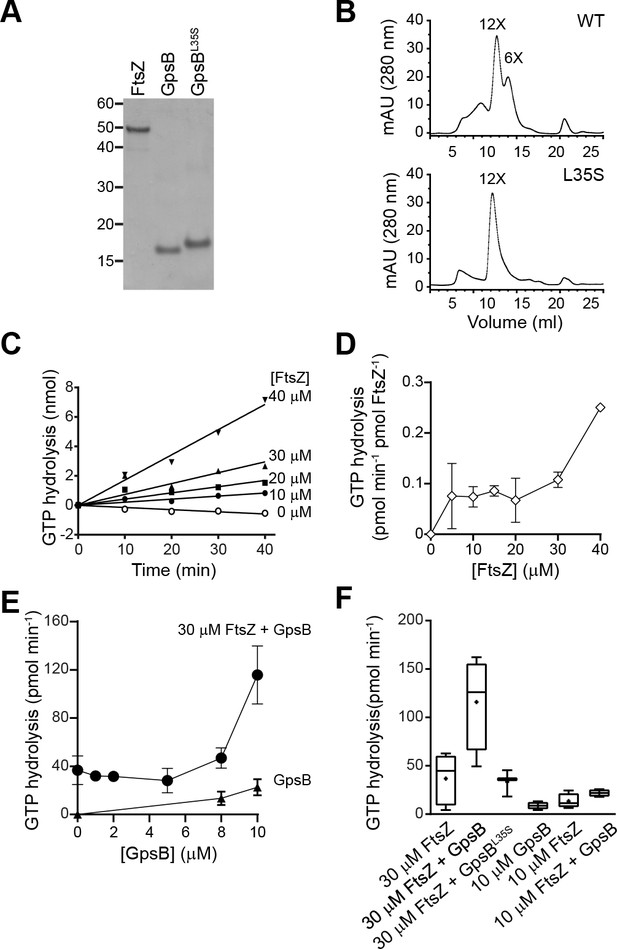
GpsB stimulates GTPase activity of FtsZ in vitro.
(A) Coomassie-stained gel of purified FtsZ, GpsB, and GpsBL35S. (B) Size exclusion chromatograms of purified GpsB (top) or GpsBL35S (bottom). Predicted multimerization states of the purified protein, based on migration of MW standards, is indicated above peaks (12X, dodecamer; 6X, hexamer). Shown is a representative example of at least 3 independent purifications. (C) Initial velocities of GTP hydrolysis by FtsZ as a function of time at various FtsZ concentrations. (D) GTP hydrolysis turnover rate of FtsZ as a function of FtsZ concentration. (E) GTP hydrolysis of increasing concentrations of GpsB alone (▲) or 30 μM FtsZ in the presence of increasing GpsB concentrations (●). Error bars represent SEM (n = 3). (F) Median GTP hydrolysis rates of 30 μM FtsZ and 10 µM FtsZ in the absence and presence of 10 μM GpsB or GpsBL35S. The ends of the boxes represent the first and third quartile of measurements; bars represent the entire range of measurements; line indicates median value; ‘+' indicates mean value (n = 3).
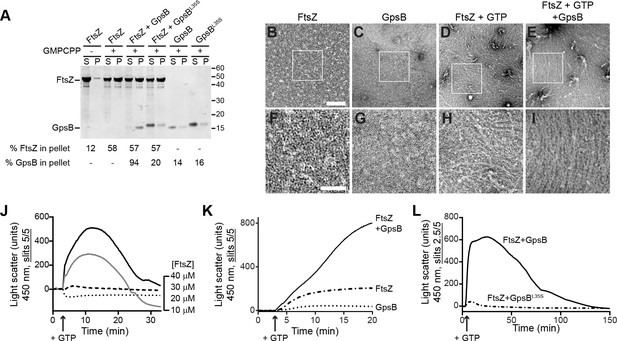
GpsB bundles FtsZ polymers in vitro.
(A) Co-sedimentation of GpsB with polymerized FtsZ in vitro. 30 μM FtsZ were incubated in the presence or absence of GMPCPP, and 10 μM GpsB or GpsBL35S as indicated. Polymerized FtsZ was collected by ultracentrifugation and proteins in the resulting supernatant (S) and pellet (P) were separated by SDS-PAGE and detected by Coomassie staining. Percentage of total FtsZ or GpsB in the pellet are indicated below. Migration of MW markers are indicated to the right. Shown is a representative gel of 3 independent replicates. (B–E) Morphology of (B) purified FtsZ, (C) purified GpsB, (D) polymerized FtsZ incubated with GTP, or (E) polymerized FtsZ incubated with GTP and GpsB visualized using negative stain transmission electron microscopy. Scale bar: 200 nm. (F–I) magnified views of the areas indicated in (B–E), respectively. Scale bar: 100 nm. (J) Assembly of 10 µM (dotted trace), 20 µM (dashed), 30 µM (gray) or 40 µM (black) FtsZ measured using 90° angle light scattering. (K) FtsZ assembly in the presence (solid) or absence (dash-dot) of GpsB, or GpsB alone (dotted), measured by 90° angle light scattering. (L) Assembly and disassembly of FtsZ in the presence of limiting amount of GTP monitored by 90° angle light scattering. Time of GTP addition is indicated with an arrow. Note the difference in slit width in (L). Shown are representative traces of at least 3 independent experiments.
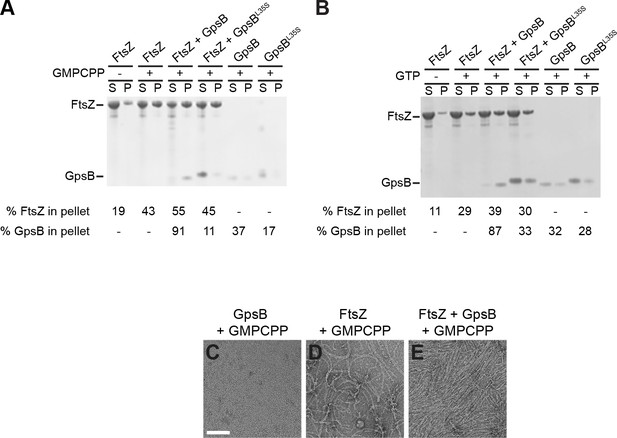
GTP hydrolysis is not required for GpsB-mediated FtsZ bundling.
(A–B) Co-sedimentation of GpsB with polymerized FtsZ in vitro. 30 μM FtsZ were incubated in the presence or absence of (A) GMPCPP or (B) GTP, with 10 μM GpsB or GpsBL35S as indicated. Polymerized FtsZ was collected by ultracentrifugation at (A) 20k × g or (B) 90k × g, and proteins in the resulting supernatant (S) and pellet (P) were separated by SDS-PAGE and detected by Coomassie staining. Percentage of total FtsZ or GpsB in the pellet are indicated below. Shown are representative gels of 3 independent replicates. (C–E) Morphology of (C) purified GpsB incubated with GMPCPP, (D) polymerized FtsZ incubated with GMPCPP, or (E) polymerized FtsZ incubated with GMPCPP and GpsB visualized using negative stain transmission electron microscopy. Scale bar: 100 nm.
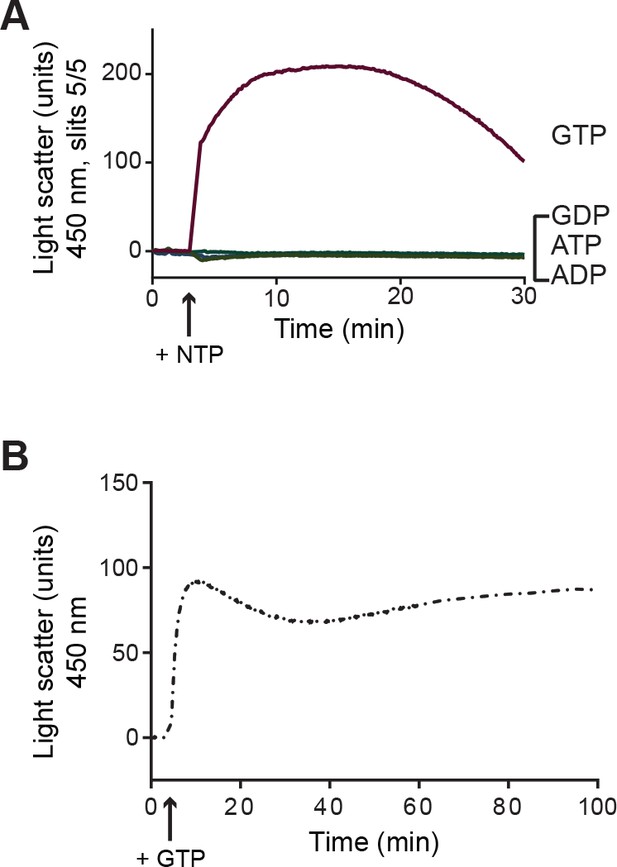
Assembly of 30 µM FtsZ measured using 90° angle light scattering in the presence of GTP (red trace), GDP (blue), ATP (light green), or ADP (dark green).
(A) Time of nucleotide addition (NTP) is indicated with an arrow. (B) Assembly of 30 µM FtsZ in the presence of GTP and a nucleotide regeneration system measured using 90° angle light scattering.
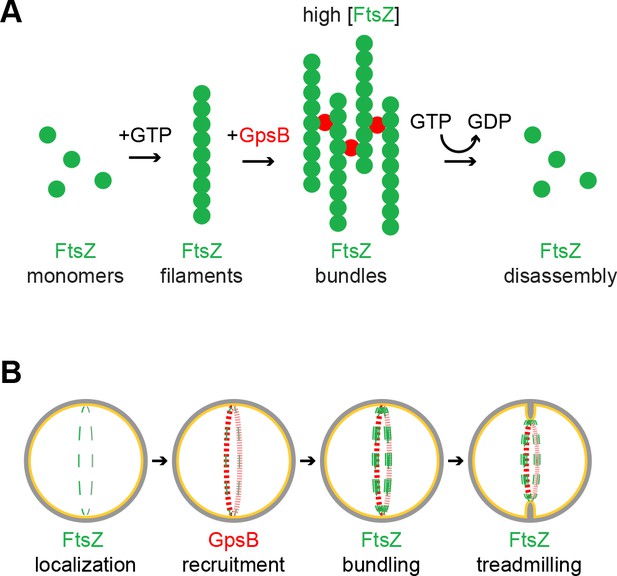
Model of GpsB remodeling of FtsZ in S.aureus.
(A) Molecular model. FtsZ (green) filaments, which form upon GTP binding, directly interact with GpsB (red). Filament-bound GpsB molecules promote lateral interactions between FtsZ filaments, thereby raising the local concentration of FtsZ, which drives GTP hydrolysis that leads to FtsZ disassembly. (B) Cellular model. FtsZ (green) ring localizes at mid-cell and recruits GpsB (red), which initially drives lateral interactions between FtsZ filaments to promote Z-ring stabilization at that position. Subsequent stimulation of FtsZ GTP hydrolysis, caused by a local increase in FtsZ concentration, stimulates FtsZ treadmilling which is linked to membrane constriction and concurrent peptidoglycan synthesis at mid-cell.
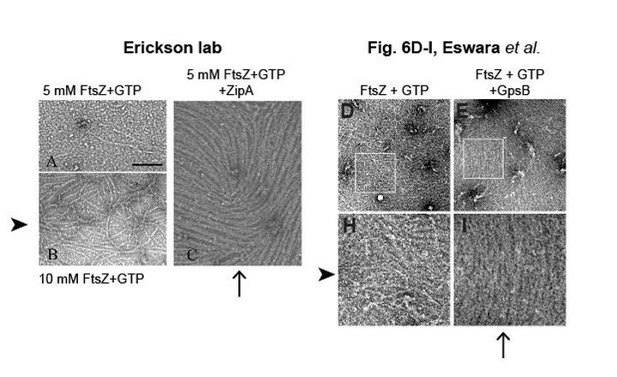
Bundling of E. coli FtsZ filaments by ZipA, compared to bundling of S. aureus FtsZ filaments by GpsB.
https://doi.org/10.7554/eLife.38856.016Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Bacillus subtilis) | PY79 | Youngman et al. (1984) | Wild type | |
| Strain, strain background (Bacillus subtilis) | GG7 | This paper | amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | GG8 | This paper | amyE::Phyperspank-gpsBSa-gfp spec | |
| Strain, strain background (Bacillus subtilis) | GG35 | This paper, derived from FG345, Gueiros-Filho and Losick (2002) | ΔezrA::spec::erm amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | CS26 | This paper, derived from BKE22320 (BGSC) | ΔponA::erm amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | CS24 | This paper, derived from BKE15770 (BGSC) | ΔprkC::erm amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | CS40 | This paper | ΔgpsB::tet amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | CS94 | This paper, derived from KR546, Ramamurthi and Losick (2009) | ΔdivIVA::erm amyE::Phyperspank-gpsBSa spec | |
| Strain, strain background (Bacillus subtilis) | GG9 | This paper, derived from AD3007, Eswaramoorthy et al. (2011) | amyE::Phyperspank-gpsBSaspec ftsAZΩftsAZ-gfp erm | |
| Strain, strain background (Bacillus subtilis) | GG18 | This paper | amyE::Phyperspank-gpsBBs spec | |
| Strain, strain background (Bacillus subtilis) | GG19 | This paper | amyE::Phyperspank-gpsBBs-gfp spec | |
| Strain, strain background (Bacillus subtilis) | PE448 | This paper | amyE::Phyperspank-gpsBSa-L35S-gfp spec | |
| Strain, strain background (Staphylococcus aureus) | SH1000 (aka PL3055) | Horsburgh et al. (2002) | Wild type | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pCL15 | Luong and Lee (2006) | bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pPE45 | This paper | pCL15 backbone, Pspac-gpsBSa bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pPE83 | This paper | pCL15 backbone, Pspac-gpsBBs bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pPE79 | This paper | pCL15 backbone, Pspac-gpsBSa-L35S bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pPE46 | This paper | pCL15 backbone, Pspac-gpsBSa-gfp bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pPE80 | This paper | pCL15 backbone, Pspac-gpsBSa-L35S-gfp bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pEPSA5 | Forsyth et al. (2002) | bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pGG59 | This paper | pEPSA5 backbone,Pxyl-gpsBantisense bla cat | |
| Strain, strain background (Staphylococcus aureus) | SH1000 pRB42 | This paper | pJB67 backbone, PCd-zapASa-gfp bla erm | |
| Sequence-based reagent (oligonucleotide) | oP36 | This paper | AAAAAGCTTACATAA GGAGGAACTACTATGTCAGATGTTTCATTGAAATTATCAGCA | |
| Sequence-based reagent (oligonucleotide) | oP37 | This paper | AAAGCTAGCTTTACCA AATACAGCTTTTTCTAAGTTTGA | |
| Sequence-based reagent (oligonucleotide) | oP38 | This paper | AAAGCATGCTTATTTACCAAATACAGCTTTTTCTAAGTTTGA | |
| Sequence-based reagent (oligonucleotide) | oP46 | This paper | AAAGCTAGCATGAGTAAAGGAGAAGAACTTTTC | |
| Sequence-based reagent (oligonucleotide) | oP24 | This paper | GCCGCATGCTTATTTGTATAGTTCATCCATGCC | |
| Sequence-based reagent (oligonucleotide) | oP100 | This paper | AAAGTCGACACATA AGGAGGAACTACTATGCTTGCTGAT AAAGTAAAGCTTTCTGCG | |
| Sequence-based reagent (oligonucleotide) | oP101 | This paper | AAAGCTAGCATCA TAAAGCTTGCTGCCAAAAACGTG | |
| Sequence-based reagent (oligonucleotide) | oP102 | This paper | AAAGCTAGCTCAAT CATAAAGCTTGCTGCCAAAAACGTG | |
| Sequence-based reagent (oligonucleotide) | oP195 | This paper | AAAGGATCCTCAATCATAAAGCTTGCTGCCAAAAACGTG | |
| Sequence-based reagent (oligonucleotide) | oP187 | This paper | AAAGAATTCTTATTTACCAAATACAGCTTTTTCTAAGTTTGAAATACGTTTTAAAATATCTAC | |
| Sequence-based reagent (oligonucleotide) | oP188 | This paper | AAAGGATCCGAGG TGGAAAAAATGTCAGATGTTTCATTGAA ATTATCAGC | |
| Sequence-based reagent (oligonucleotide) | oP236 | This paper | AAAGTCGACTAATGAGGAGGAAAAAATGACACAGTTTAAAAACAAGGTAAATGTATCAATTAATGATCAG | |
| Sequence-based reagent (oligonucleotide) | oP237 | This paper | AAAGCTAGCCGCTGCTG CAATTTGTGAATTTGTTGTTTCAAACGT | |
| Sequence-based reagent (oligonucleotide) | oP47 | This paper | AAAGGATCCTTATTTGTATAGTTCATCCATGCC | |
| Antibody | anti-GpsB | This paper | Raised against purified GpsB-His6 | |
| Antibody | anti-SigA | Ramamurthi lab | Raised against purified B. subtilis SigA-His6 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38856.014





