Cryo-EM structure of respiratory complex I at work
Figures
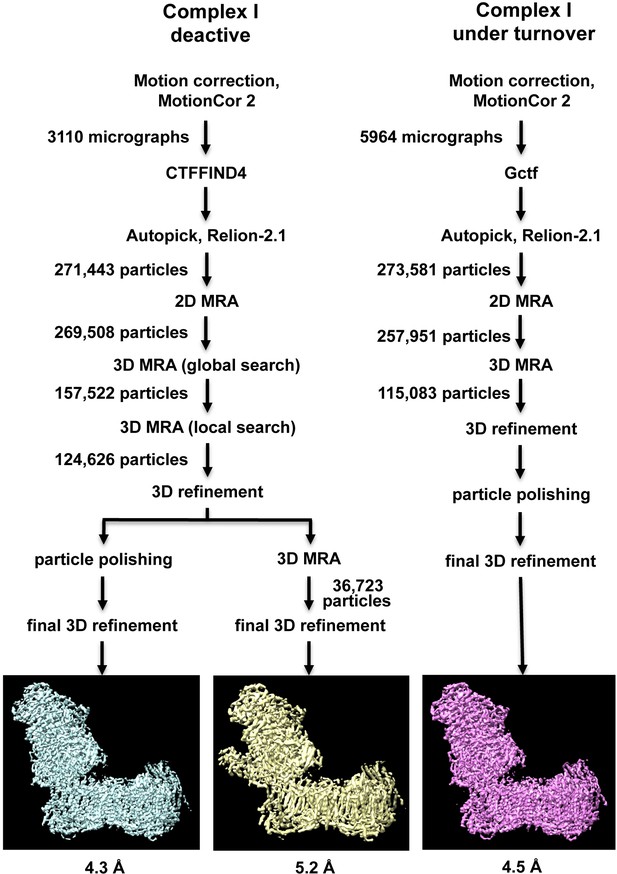
Image processing and two-dimensional classification of particle images.
Electron densities of complex I in the deactive state and under turnover conditions are light blue and magenta. About 30% of the particles belong to a subclass (yellow) which contains the accessory sulfur transferase subunit ST1 known to be bound substoichiometrically to Y. lipolytica complex I (D'Imprima et al., 2016).
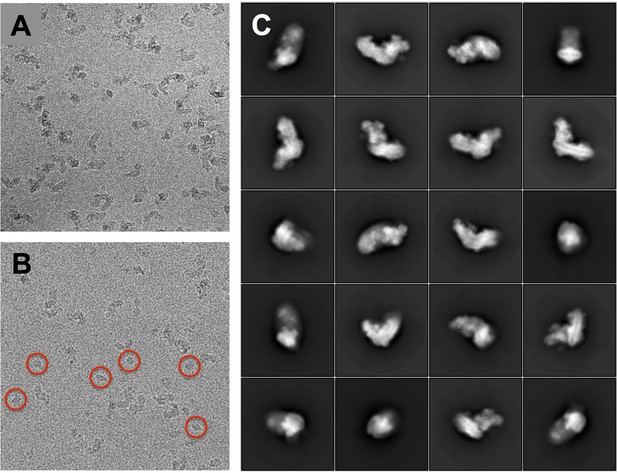
Raw micrographs and class averages.
Raw micrographs for (A) deactive complex I; (B) complex I under turnover (red circles highlight oxidase molecules). (C) Selection of 2D reference-free class averages of inactive complex I sorted by RELION2.1 in different orientations.
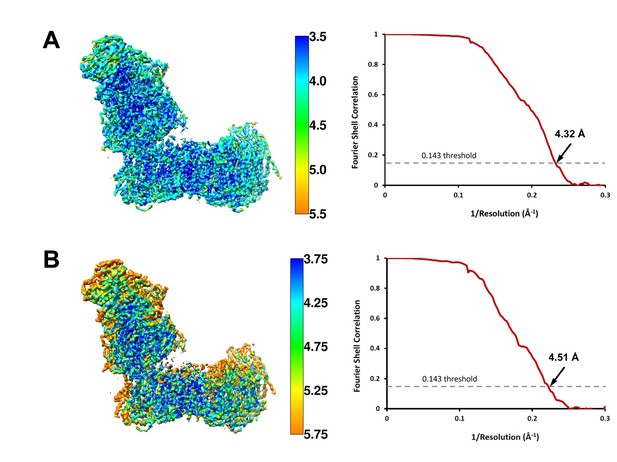
Local resolution and Fourier shell correlation (FSC) curves of (A) deactive complex I and (B) complex I under steady-state turnover conditions.
Left: cryo-EM maps of complex I analysed by ResMap (Kucukelbir et al., 2014) coloured according to local resolution. Right: FSC plots of final masked and refined cryo-EM maps. The map resolution is indicated by the point where the curve crosses the 0.143 threshold (Rosenthal and Henderson, 2003).
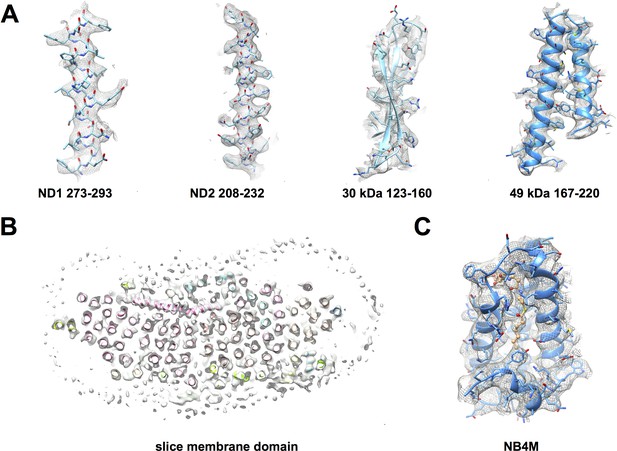
Cryo-EM map of deactive complex I with fitted models.
(A) Selected regions of the matrix domain. (B) A horizontal cross-section through the membrane arm shows TMH fits. (C) A region of the accessory LYR protein subunit NB4M (Angerer et al., 2017); the acyl chain appended to the phosphopantetheine group of the adjacent acyl carrier protein ACPM1 inserts into the interior of NB4M. Cofactor and acyl chain are drawn as ball-and-stick model.
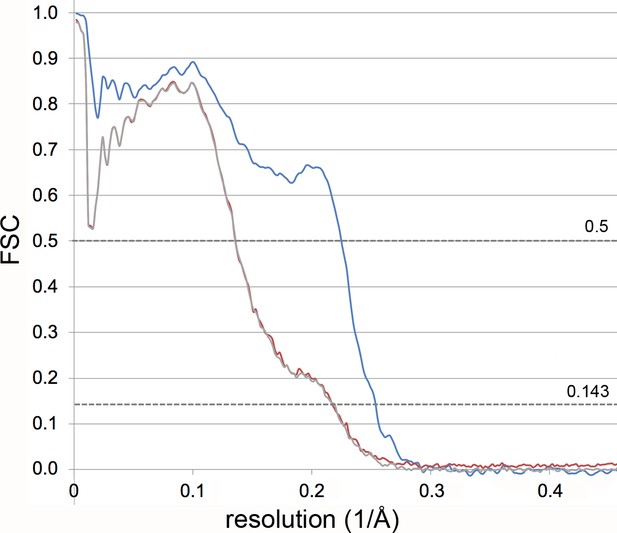
Validation of model refinement.
The FSC curve between the final refined model and the reconstruction from all particles of the deactive data set (blue) indicates a resolution of 4.5 Å. The model was scrambled by random displacement of the atoms up to 0.5 Å and refined against half map 1 to 4.3 Å. The FSC between the refined model and half map 1 (FSCwork, red), and between this model and the reconstruction from the other half of the particles (FSCfree, grey) both indicate the same resolution (4.6 Å) and are similar over the full resolution range, showing that no overfitting of the model to the map has taken place.
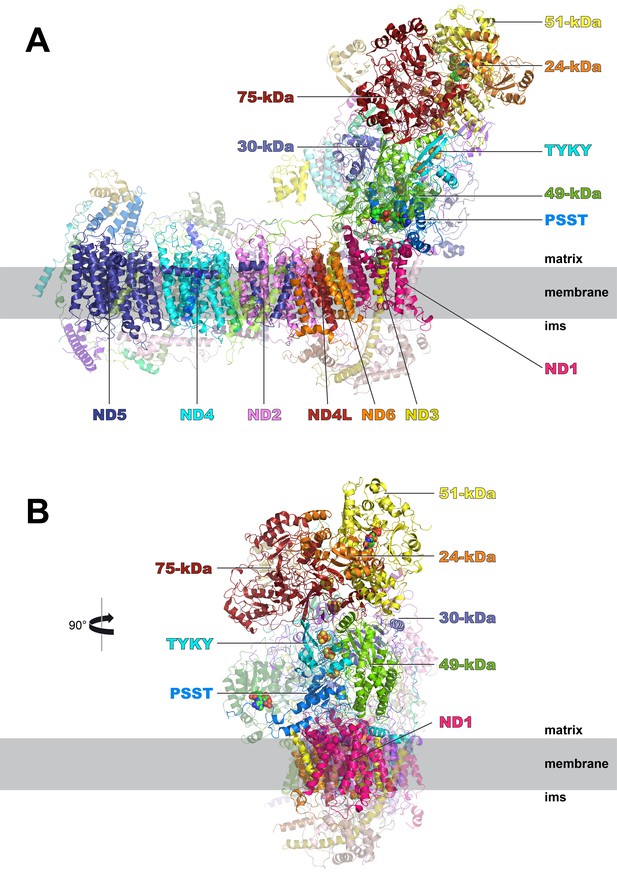
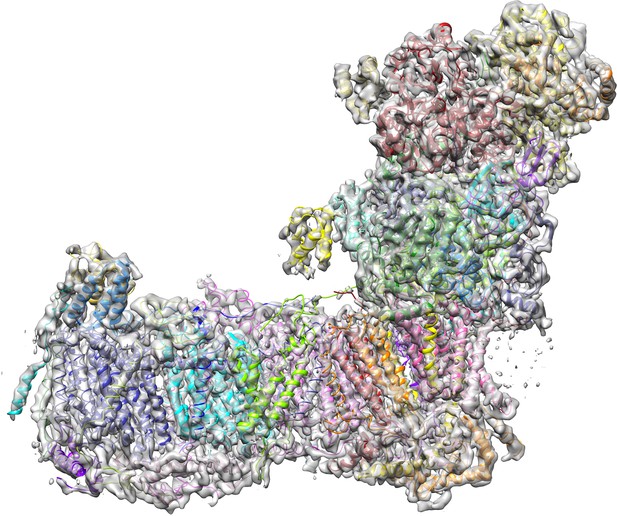
Cryo-EM structure of respiratory complex I from Y. lipolytica.
(A) Side view; (B) view from peripheral arm; central subunits (labelled, solid) and accessory subunits (transparent, compare Figure 8) are shown.
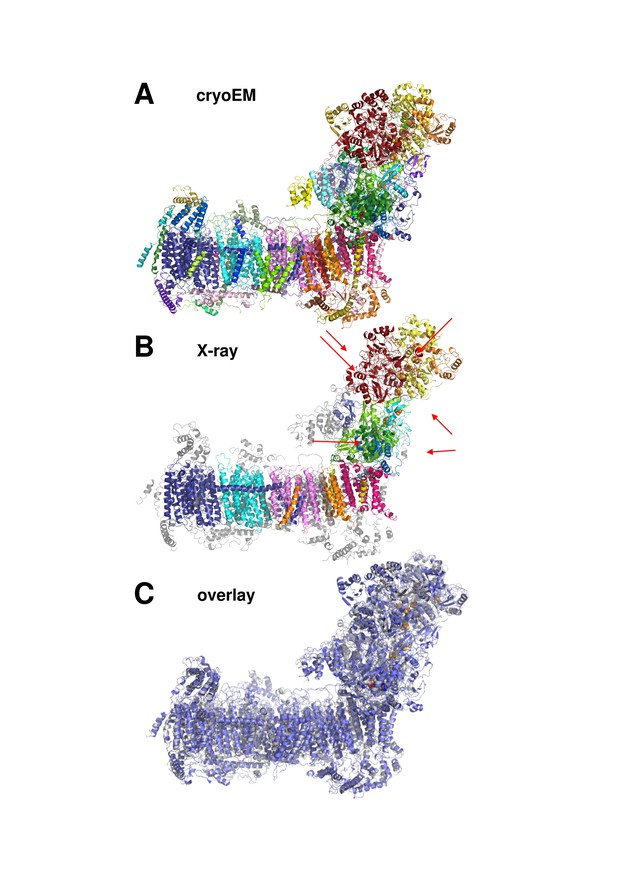
Cryo-EM and X-ray structures of deactive complex I are consistent.
(A) With 42 assigned subunits (colour coded as in Figures 4 and 8) and 7515 residues the cryo-EM structure is significantly more complete than the X-ray structure (B) with 15 assigned subunits and 4979 residues (colour coded as in (A). Unassigned parts of the model are grey. Red arrows indicate subunits NI8M, NUYM, NUZM, N7BM, NUMM, cofactors FMN (51 kDa subunit) and NADPH (NUEM subunit) that are missing or incomplete in the X-ray structure. (C) X-ray structure of deactive complex I from Y. lipolytica (Zickermann et al., 2015) (grey) overlaid with the cryo-EM structure of deactive Y. lipolytica complex I (blue).
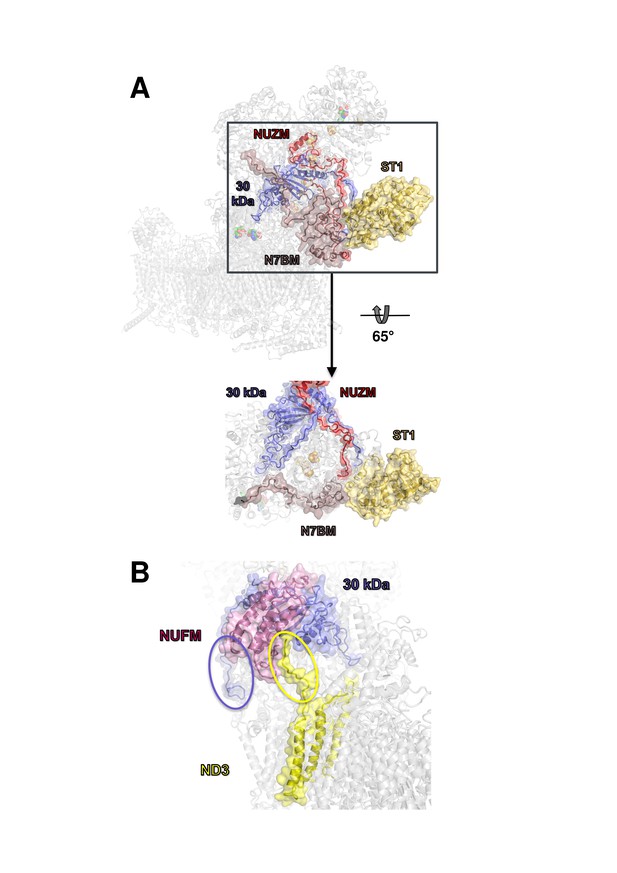
Docking site of accessory subunit ST1 and extensions of the 30 kDa and ND3 subunits in Y. lipolytica complex I.
(A) ST1 (yellow) binds to N7BM (violet), NUZM (red) and the extended N-terminus of the 30 kDa subunit (blue); (B) N-terminal extension of the 30 kDa subunit (blue oval) and interaction of the C-terminal extension of subunit ND3 with NUFM (yellow oval).
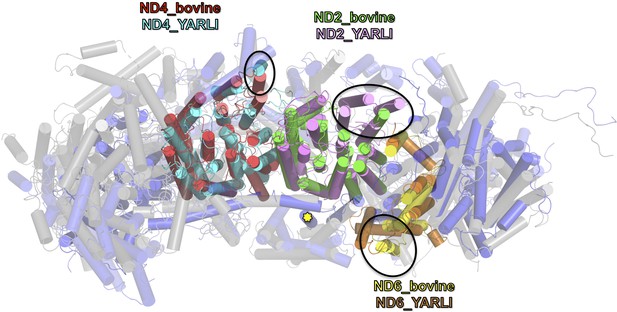
Overlay of membrane arm subunits of complex I from Y. lipolytica and B. taurus.
Top view of membrane arm with subunits of the peripheral arm removed for clarity (Y. lipolytica blue, B. taurus, grey; selected subunits are coloured as indicated). The first three helices of ND2 are missing in bovine complex I and the position of TMH 4 of ND6 is different. These changes result in an incision of the membrane arm of mammalian complex I at the position of ND2. Subunit ND5 of complex I from Y. lipolytica has an extra C-terminal TMH (yellow asterisk [Zickermann et al., 2015]), and TMH 1 of ND4 is oriented differently in the membrane.
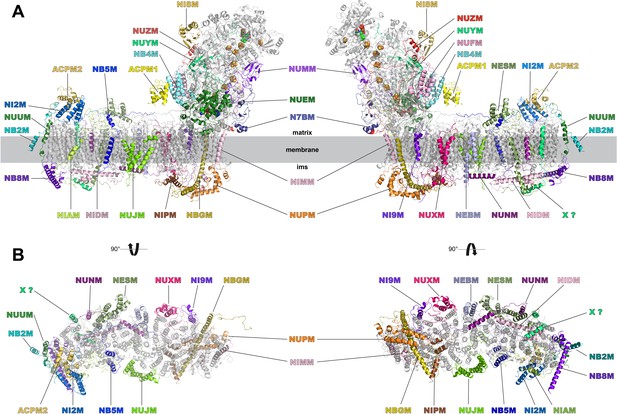
Accessory subunits of complex I.
Central subunits (see Figure 4) are shown in grey, accessory subunits are labelled and coloured. (A) Side views, (B) view from the matrix (left) and from the intermembrane space (right) with peripheral arm subunits removed for clarity.
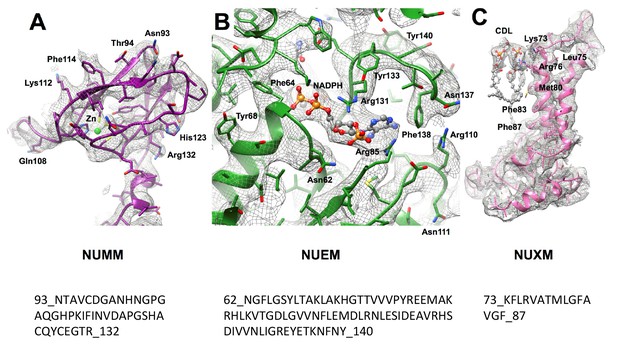
Assignment of accessory subunits.
Example densities with corresponding sequence stretches are shown for the Zn binding domain of NUMM and for the NADPH binding domain of NUEM. In agreement with secondary structure predictions subunit NUXM has four αhelices, two of which are transmembrane. Side chains were modelled for short sequence stretch in this subunit.
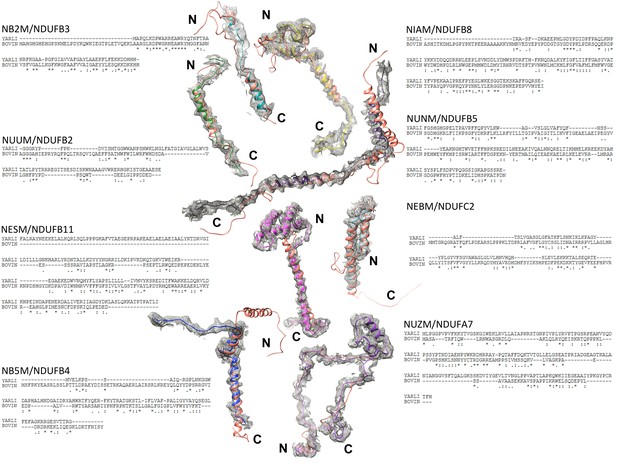
Assignment of accessory subunits.
Sequence and structural alignments with bovine complex I (PDB ID 5O31 (Blaza et al., 2018), bovine subunits, orange) are shown for eight subunits that were modelled largely or entirely as poly-alanine.
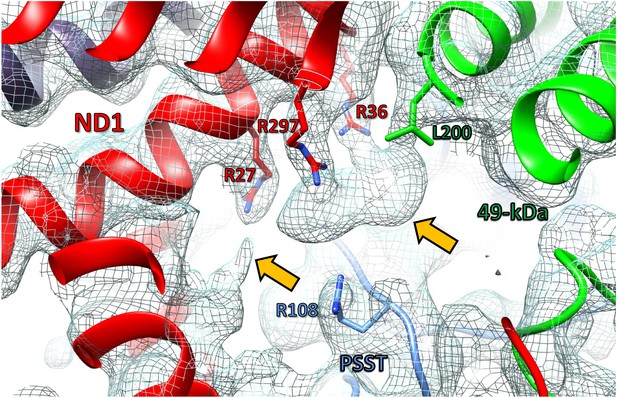
Unassigned density (orange arrows) at the interface of subunits ND1, 49 kDa, and PSST.
Slice of interface region of membrane and peripheral arm of complex I in the deactive state (model, cartoon representation; selected residues, stick representation; cryo-EM map, mesh). Note that the density is also present in the maps of complex I under steady-state turnover conditions.
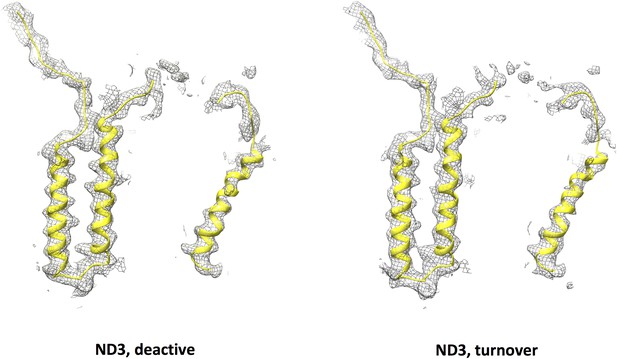
Model for subunit ND3 (yellow) and cryo-EM density (grey mesh) of subunit ND3 in the deactive state (left) and under turnover conditions (right).
The central part of the long loop connecting TMH1 and 2 is disordered.
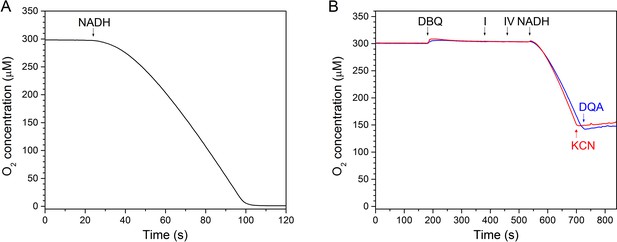
In vitro assay of a minimal respiratory chain of complex I and bo3-type ubiquinol oxidase.
(A) Assay conducted at the substrate concentration used for cryo-EM sample preparation (2 μM complex I, 1 μM oxidase, 2 mM NADH and 200 μM DBQ at 18°C. The reaction was started by addition of NADH. (B) Inhibition of complex I by DQA (blue) and of the Vitreoscilla oxidase by CN- (red).
Complex I in the deactive state and under turnover conditions.
The model for complex I in the deactive state (colour) is overlaid with the cryo-EM density (grey) for complex I under turnover conditions. There are no differences in overall structure, so there is no indication that the matrix arm moves relative to the membrane arm during turnover. Occupation and conformational changes of the substrate binding sites are shown in Figure 13.
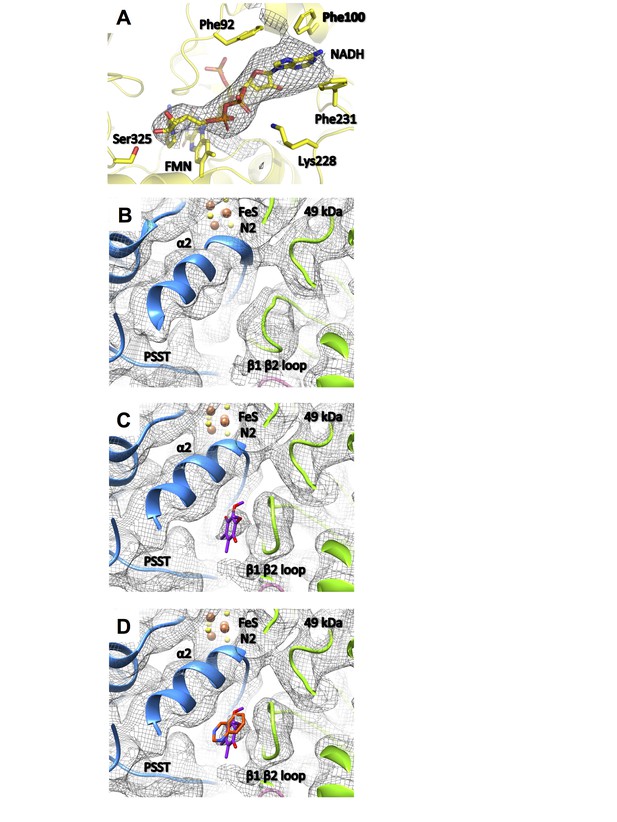
Substrate binding sites of complex I.
(A) Under steady-state turnover conditions NADH (mesh, cryo-EM density) binds to the FMN cofactor and residues of the 51 kDa subunit; (B) ubiquinone binding site in the deactive state (mesh, cryo-EM density; 49 kDa subunit, green; PSST subunit, blue) and under steady-state turnover (C). The ubiquinone headgroup (purple) binds between the β1-β2 loop of the 49 kDa subunit and helix α2 of PSST. (D) This binding site overlaps with the position of the toxophore of decyl-quinazolineamine (orange) that was modelled based on anomalous diffraction of brominated inhibitor derivatives in the X-ray structure of Y. lipolytica complex I (Zickermann et al., 2015).
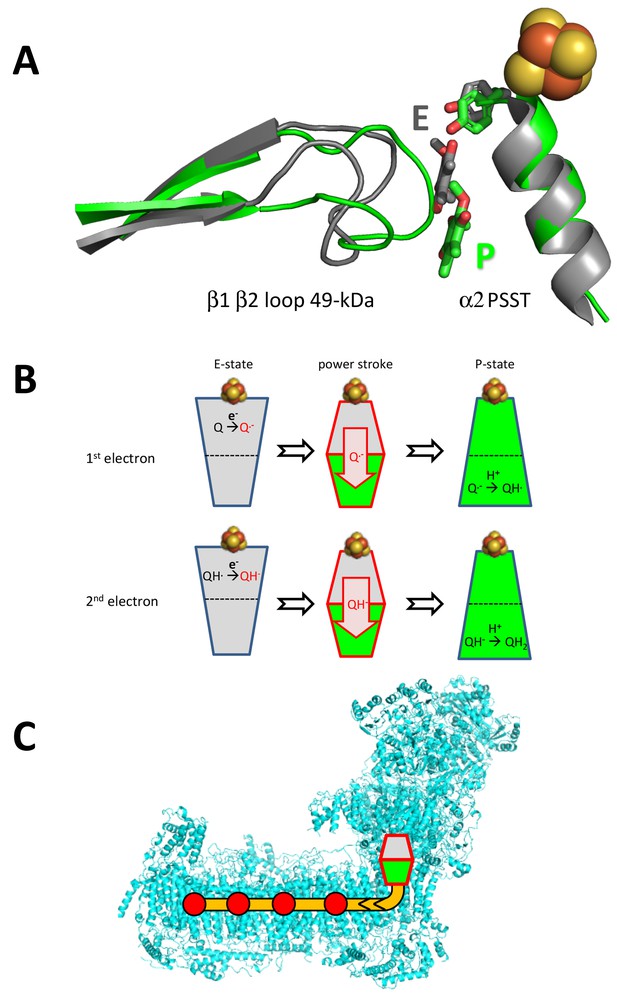
Alternating binding positions of ubiquinone support a two-state stabilization change mechanism for respiratory complex I (Brandt, 2011).
(A) ubiquinone and Tyr144 of the 49 kDa subunit (stick representation) and the β1-β2 loop of the 49 kDa and helix α2 and FeS cluster N2 of the PSST subunit in the ubiquinone binding pocket of Y. lipolytica complex I (green) were superimposed on ubiquinone and the corresponding structures in T. thermophilus (grey). The position of ubiquinone in T. thermophilus (PDB ID: 4HEA) was fitted according to Figure 4 in (Baradaran et al., 2013). The position of ubiquinone in T. thermophilus is assigned to the E-state (E) while the position of ubiquinone determined in our study is assigned to the P-state (P). (B) Electron transfer from iron-sulfur cluster N2 occurs in the E-state (grey), while ubiquinone intermediates are protonated in the P-state (green). The stabilization of negatively charged redox intermediates of ubiquinone drive the transition from the E- to the P-state, changing the binding site for the ubiquinone headgroup. This would create conformational and electrostatic strain in the loops lining the ubiquinone binding pocket. (C) The strain provides the energy for a power stroke transmitted through a chain of titratable residues (orange) into the membrane arm, where it drives the proton pump modules (red dots) (Zickermann et al., 2015).

Density for ubiquinone headgroup.
View from the membrane arm into the ubiquinone binding pocket. Overlay of density maps (deactive, blue; turnover conditions, red; model for complex I under turnover conditions). Note that the blue map has slightly higher resolution and that the b1 b2 loop changes between the two states.
Tables
Data collection, refinement and model statistics.
https://doi.org/10.7554/eLife.39213.009| Deactive | steady-state turnover | |
|---|---|---|
| Data collection Microscope | FEI Tecnai Polara | FEI Titan Krios |
| Camera | Gatan K2 Summit | Falcon III |
| Voltage (kV) Nominal magnification Calibrated pixel size (Å) | 300 200,000x 1.09 | 300 75,000x 1.053 |
| Total exposure (e-/Å2) | 60.5 | 30.7–40.7 |
| Exposure rate (e-/pixel/s) Number of frames | 9 40 | 0.4 81 |
| Defocus range (μm) | 1.5–3.0 | 1.5–4.5 |
| Image processing Motion correction software | MotionCor2 | MotionCor2 |
| CTF estimation software | CTFFIND4 | Gctf |
| Particle selection software | EMAN boxer and RELION2.1 | RELION2.1 |
| Initial/final micrographs | 3,110/3,110 | 5,964/5,650 |
| Particles selected | 271,443 | 273,581 |
| Applied B-factor (Å2) | −142 | −215 |
| Final resolution (Å) | 4.3 | 4.5 |
| Refinement statistics Modeling software | COOT, PHENIX | |
| Number of residues | 7515 | |
| Map CC (whole unit cell) | 0.786 | |
| RMS deviations Bond-lengths (Å) | 0.0099 | |
| Bond-angles (°) | 1.52 | |
| Av. B-factor (Å2) | 151.46 | |
| Ramachandran plot Outliers (%) | 0.74 | |
| Allowed (%) | 13.43 | |
| Favoured (%) | 85.83 | |
| Rotamer outliers (%) | 0.71 | |
| Molprobity score | 2.11 | |
| All-atom clashscore | 8.55 | |
| PDB ID | 6GCS |
Composition of subunits.
https://doi.org/10.7554/eLife.39213.010| Subunit | human/bovine | Chain | Total residues/ range built | Modelled with side chains | Modelled as poly-alanine | [%] residues modelled | [%] with side chains | [%] unknown |
|---|---|---|---|---|---|---|---|---|
| central subunits | ||||||||
| NUAM | NDUFS1/ 75 kDa | A | 694/ 1–691 | 1–691 | 0 | 99 | 99 | 1 |
| NUBM | NDUFV1/ 51 kDa | B | 470/ 15–457 | 15–437 | 437–457 | 95 | 90 | 6 |
| NUCM | NDUFS2/ 49 kDa | C | 444/ 28–443 | 58–67 77–443 | 28–57 68–76 | 93 | 85 | 7 |
| NUGM | NDUFS3/ 30 kDa | G | 251/ 1–232 | 30–189 | 1–29 190–232 | 92 | 64 | 8 |
| NUHM | NDUFV2/ 24 kDa | H | 215/ 3–187 | 23–187 | 3–22 | 86 | 78 | 14 |
| NUIM | NDUFS8/ TYKY | I | 198/ 19–198 | 26–198 | 19–26 | 91 | 87 | 9 |
| NUKM | NDUFS7/ PSST | K | 183/ 15–183 | 15–183 | 0 | 92 | 92 | 8 |
| NU1M | NU1M/ ND1 | 1 | 341/ 1–340 | 1–179 184–205 217–251 268–340 | 180–183 206–216 252–267 | 100 | 91 | 0 |
| NU2M | NU2M/ ND2 | 2 | 469/ 1–85 99–465 | 1–25 53–85 99–415 | 26–52 416–465 | 96 | 80 | 4 |
| NU3M | NU3M/ ND3 | 3 | 128/ 1–34 49–124 | 1–34 49–118 | 119–124 | 81 | 77 | 19 |
| NU4M | NU4M/ ND4 | 4 | 486/ 7–481 | 85–189 201–434 | 7–84 190–200 435–481 | 98 | 70 | 2 |
| NU5M | NU5M/ ND5 | 5 | 655 5–479 489–652 | 28–436 457–474 568–592 614–652 | 5–27 437–456 475–479 489–567 593–613 | 98 | 75 | 2 |
| NU6M | NU6M/ ND6 | 6 | 185 2–184 | 2–77 160–184 | 78–159 | 99 | 55 | 1 |
| NULM | NULM/ ND4L | L | 89 1–86 | 1–86 | 0 | 97 | 97 | 3 |
| accessory subunits peripheral arm | ||||||||
| NUEM | NDUFA9/ 39 kDa | E | 355 17–334 | 17–334 | 0 | 82 | 82 | 18 |
| NUFM | NDUFA5/ B13 | F | 136 13–131 | 34–131 | 13–33 | 88 | 72 | 12 |
| NUMM | NDUFS6/ 13 kDa | M | 119 13–117 | 43–117 | 13–42 | 88 | 75 | 12 |
| NUYM | NDUFS4/ 18 kDa | Y | 137 19–133 | 19–133 | 0 | 84 | 84 | 16 |
| NUZM | NDUFA7/ B14.5a | Z | 182 30–166 | 0 | 30–166 | 75 | 0 | 25 |
| N7BM | NDUFA12/B17.2 | h | 137 6–135 | 6–135 | 0 | 95 | 95 | 5 |
| NB4M | NDUFA6/ B14 | P | 123 3–118 | 3–118 | 0 | 94 | 94 | 6 |
| ACPM1 | NDUFAB1/SDAP | O | 84 4–80 | 4–80 | 0 | 92 | 92 | 8 |
| NI8M | NDUFA2/ B8 | f | 86 5–84 | 5–84 | 0 | 93 | 93 | 7 |
| Accessory subunits PP module | ||||||||
| NUPM | Ndufa8/ pgiv | U | 171 10–167 | 16–142 | 10–15 143–167 | 92 | 74 | 8 |
| NUJM | NDUFA11/B14.7 | J | 197 18–157 | 18–133 | 134–157 | 71 | 59 | 29 |
| NB6M | NDUFA13/B16.6 | W | 122 3–120 | 14–120 | 3–13 | 97 | 88 | 3 |
| NIPM | NDUFS5/ 15 kDa | 9 | 88 8–66 | 14–66 | 08–13 | 67 | 60 | 33 |
| NUXM | -/- | X | 168 4–120 | 72–96 | 4–71 97–120 | 70 | 15 | 30 |
| NI9M | NDUFA3/ B9 | g | 66 3–62 | 3–62 | 91 | 91 | 9 | |
| NIMM | NDUFA1/ MWFE | D | 86 1–80 | 1–58 | 59–80 | 93 | 67 | 7 |
| NEBM | NDUFC2/ B14.5b | b | 73 1–64 | 1–64 | 87 | 0 | 13 | |
| accessory subunits PD module | ||||||||
| NESM | NDUFB11/ESSS | S | 204 29–187 | 29–187 | 78 | 0 | 22 | |
| NIAM | NDUFB8/ ASHI | a | 125 11–110 | 11–110 | 80 | 0 | 20 | |
| NUNM | NDUFB5/ SGDH | n | 119 23–115 | 23–115 | 78 | 0 | 22 | |
| NB2M | NDUFB3/ B12 | c | 59 8–49 | 31–49 | 8–30 | 71 | 32 | 29 |
| NB5M | NDUFB4/ B15 | j | 92 3–75 | 16–41 | 3–15 42–75 | 82 | 27 | 18 |
| NB8M | NDUFB7/ B18 | 8 | 98 3–84 | 3–80 | 81–84 | 84 | 80 | 16 |
| ACPM2 | NDUFAB1/SDAP | Q | 88 3–87 | 3–87 | 0 | 97 | 97 | 3 |
| NIDM | NDUFB10/PDSW | d | 91 3–91 | 3–91 | 0 | 98 | 98 | 5 |
| NI2M | NDUFB9/ B22 | R | 108 6–107 | 6–98 | 99–107 | 94 | 86 | 6 |
| NUUM | NDUFB2/ AGGG | e | 89 6–50 | 6–50 | 51 | 0 | 49 | |
Lipid content of typical complex I preparation
https://doi.org/10.7554/eLife.39213.017| Lipid | nmol lipid/nmol complex I |
|---|---|
| phosphatidylcholine | 19.3 |
| lyso-phosphatidylcholine | 0.1 |
| phosphatidylethanolamine | 13.3 |
| phosphatidylserine | 0.5 |
| phosphatidylinositol | 11.8 |
| cardiolipin | 21.7 |
| Σ | 66.7 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Y. lipolytica GB20) | ∆mus51, lys1-, leu2-, ura3-, 30Htg2, ndh2i | PMID 24706851 | ||
| Strain, strain background (E. coli CLY) | derived from C43(DE3), ∆cyoBCD::kan | PMID 17267395 | Prof. Robert B. Gennis, University of Illinois | |
| Genetic reagent (pET17b-V14) | cyoABCDE in pET 17b | this work | Dr. Hao Xie, MPI for Biophysics, Frankfurt | |
| Chemical compound, drug | n-Dodecyl β- maltoside | Glycon Biochemicals GmbH | Cat. # D97002-C | |
| Chemical compound, drug | Decylubiquinone | Sigma-Aldrich/Merck | Cat. # D7911 | |
| Chemical compound, drug | β-Nicotinamide adenine dinucleotide | Sigma-Aldrich/Merck | Cat. # N8129 | |
| Chemical compound, drug | Asolectin from soybean | Sigma-Aldrich/Merck | Cat. # 11145 | |
| Chemical compound, drug | CHAPS, Anagrade | Anatrace | Cat. # C316 | |
| Software, algorithm | Coot | PMID: 15572765 | RRID: SCR_014222 | |
| Software, algorithm | CTFFIND4 | PMID:26278980 | ||
| Software, algorithm | Gctf | PMID:26592709 | ||
| Software, algorithm | MolProbity | PMID: 20057044 | RRID: SCR_14226 | |
| Software, algorithm | MotionCor2 | PMID: 28250466 | ||
| Software, algorithm | Phenix | PMID: 20124702 | RRID: SCR_014224 | |
| Software, algorithm | PyMOL | Schrödinger, LLC | RRID: SCR_000305 | |
| Software, algorithm | RELION | PMID: 27845625 | RRID: SCR_016274 | |
| Software, algorithm | UCSF Chimera | PMID: 15264254 | RRID: SCR_004097 | |
| Software, algorithm | TMHMM Server | PMID: 11152613 | RRID: SCR_014935 |
Oligonucleotides used in this work.
https://doi.org/10.7554/eLife.39213.023| Oligonucleotides | Sequence (5’−3’) |
|---|---|
| Vbo3-NdeIa | GCGCATATGAAGCAGATGATTCAGGTC |
| Vbo3-HindIIIa | GGGAAGCTTTC AAAAATAAATATGCGGCAAC |
| Vbo3-10 | TAATCTATGTTAGGTAAACTCGATTGG |
| Vbo3-15 | ATTTCCTCCTGCAGCAGATGCAGCAAC |
| Vbo3-19b | GCTGCAGGAGGAAAT TAATAATCTATGTTAGGT |
| Vbo3-20b | ACCTAACATAGATTATTA ATTTCCTCCTGCAGC |
-
*Restriction enzymes sites are underlined.
‡The nucleotide sequences encoding the TEV cleavage site and the deca-histidine tag are shown in red and blue, respectively. The artificial intergenic region containing the Vitreoscilla ribosomal binding site is shown in magenta.
Additional files
-
Supplementary file 1
Coordinate file of complex I under steady-state turnover conditions with NADH and ubiquinone binding sites occupied (compare EMD-4385).
Please note that the position of the ubiquinone head group was identified but that the precise orientation of the molecule in the site remained ambiguous due to limited detail of the 4.5 Å resolution map.
- https://doi.org/10.7554/eLife.39213.024
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39213.025





