Neuropeptide B mediates female sexual receptivity in medaka fish, acting in a female-specific but reversible manner
Figures
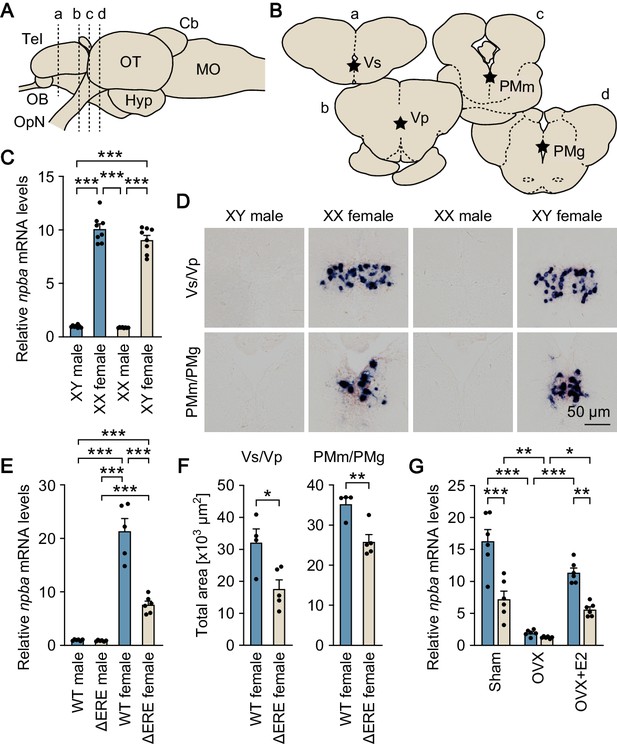
Sexually dimorphic npba expression is independent of sex chromosome complement but dependent on direct transcriptional activation by estrogen.
(A) Lateral view (anterior to left) of the medaka brain showing approximate levels of sections in panel B. For abbreviations of brain regions, see Supplementary file 1. (B) Coronal sections showing the location of Vs/Vp and PMm/PMg. (C) Levels of npba expression in the whole brain of artificially sex-reversed XX males/XY females, and wild-type XY males/XX females as determined by real-time PCR (n = 8 per group). ***, p<0.001 (Bonferroni’s post hoc test). (D) Representative micrographs of npba expression in Vs/Vp and PMm/PMg of sex-reversed XX males/XY females and wild-type XY males/XX females (n = 5 per group). Scale bars represent 50 μm. (E) Levels of npba expression in the whole brain of male and female mutants lacking the estrogen-responsive element in the npba promoter (ΔERE), and wild-type (WT) males and females as determined by real-time PCR (n = 6 per group, except WT females, where n = 5). ***, p<0.001 (Bonferroni’s post hoc test). (F) Total area of npba expression in Vs/Vp and PMm/PMg of ΔERE (n = 5) and WT (n = 4) females. *, p<0.05; **, p<0.01 (unpaired t-test). (G) Levels of npba expression in the whole brain of ΔERE (beige columns) and WT (blue columns) females that were sham-operated (Sham), ovariectomized (OVX), or ovariectomized and treated with estradiol-17β (OVX +E2) as determined by real-time PCR (n = 6 per group). There were significant main effects of genotype (F (1, 30)=45.03, p<0.0001) and treatment (F (2, 30)=60.19, p<0.0001) and a significant interaction between genotype and treatment (F (2, 30)=9.944, p=0.0005). *, p<0.05; **, p<0.01; ***, p<0.001 (Bonferroni’s post hoc test). See also Figure 1—figure supplement 1 and Figure 1—figure supplement 2.
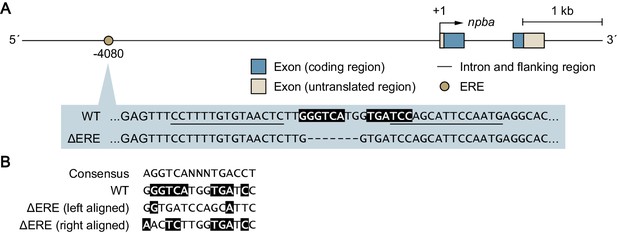
Genetic scheme for the ΔERE mutant medaka.
Mutant medaka in which an estrogen-responsive element (ERE) in the npba promoter was deleted (designated ΔERE mutant medaka) were generated by transcription activator-like effector nuclease (TALEN)-mediated genome editing. (A) Structure of the npba locus showing the position of the deleted ERE, which is expanded below to indicate the nucleotide sequences of the wild-type (WT) and ΔERE alleles. ERE is indicated by white letters on a black background. TALEN binding sites are underlined and deleted nucleotides are indicated by dashes. (B) Comparison of the nucleotide sequences of WT ERE and ΔERE, the deletion of which is both left- and right-aligned. Nucleotides identical to the consensus ERE sequence are indicated by white letters on a black background.
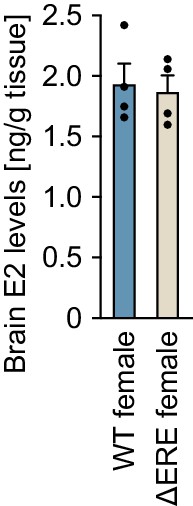
Levels of estradiol-17β (E2) in the brains of ∆ERE mutant and wild-type (WT) females.
n = 4 per group.
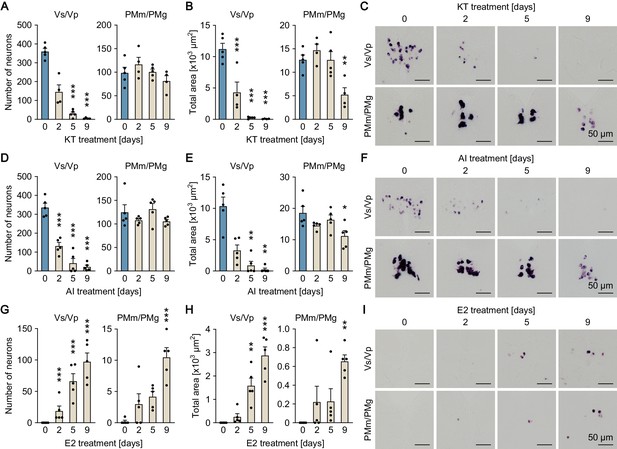
Sexually dimorphic npba expression can be reversed by altering the sex steroid milieu.
Temporal changes in npba expression in Vs/Vp and PMm/PMg of 11-ketotestosterone (KT)-treated females (A, B, C), aromatase inhibitor (AI)-treated females (D, E, F), and estradiol-17β (E2)-treated males (G, H, I) (n = 5 per treatment and sampling day). (A, D, G) Number of npba-expressing neurons in Vs/Vp and PMm/PMg. ***, p<0.001 (versus day 0, Dunnett’s post hoc test). (B, E, H) Total area of npba expression in Vs/Vp and PMm/PMg. *, p<0.05; **, p<0.01; ***, p<0.001 (versus day 0, Dunnett’s post hoc test). (C, F, I) Representative micrographs showing npba expression in Vs/Vp and PMm/PMg. Scale bars represent 50 μm. See also Figure 2—figure supplement 1 and Figure 2—figure supplement 2.
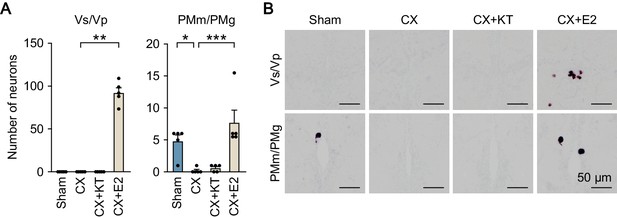
Effects of castration and sex steroid supplementation on npba expression in Vs/Vp and PMm/PMg of males.
(A) Number of npba-expressing neurons in Vs/Vp and PMm/PMg of sham-operated males (Sham) and castrated males that were exposed to vehicle alone (CX), 11-ketotestosterone (CX + KT), or estradiol-17β (CX + E2) (n = 5 per group). *, p<0.05; **, p<0.01; ***, p<0.001 (Bonferroni’s post hoc test). (B) Representative micrographs of npba expression in Vs/Vp and PMm/PMg of Sham, CX, CX + KT, and CX + E2 treated males. Scale bars represent 50 μm.
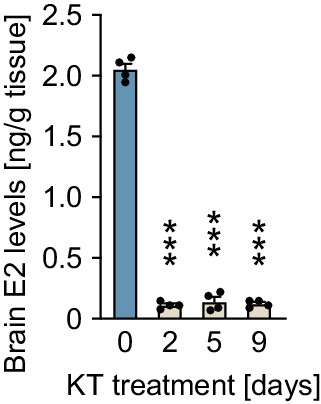
Levels of estradiol-17β (E2) in the brains of females treated with 11-ketotestosterone (KT).
n = 4 per sampling day. ***, p<0.001 (versus day 0, Dunnett’s post hoc test).
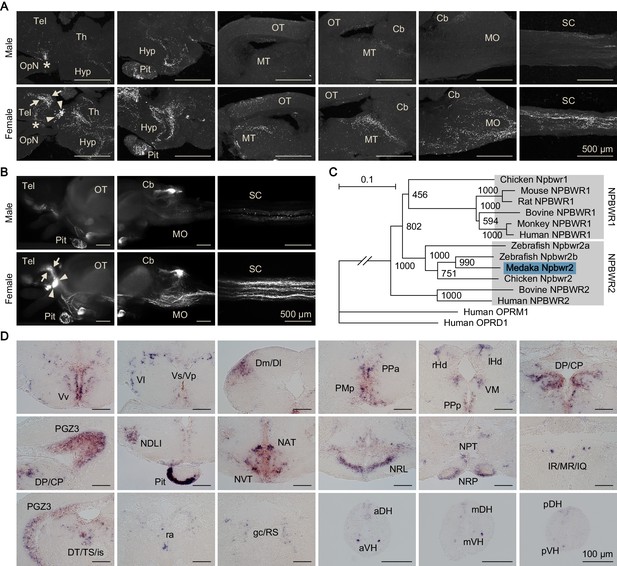
Female-specific Npba likely acts widely in the brain and spinal cord.
(A) Comparison of the distribution of Npba-immunoreactive axons in the brain, pituitary, and spinal cord between males and females. All images are sagittal sections with anterior to the left. Arrows and arrowheads indicate female-specific Npba-immunoreactive neuronal cell bodies in Vs/Vp and PMm/PMg, respectively. Asterisks indicate Npba-immunoreactive neuronal cell bodies in Pbl occurring in both sexes. Scale bars represent 500 μm. For abbreviations of brain regions, see Supplementary file 1. (B) Comparison of the distribution of GFP-labeled axons in the brain, pituitary, and spinal cord between npba-GFP transgenic males and females. Images in the left and middle panels are lateral views with anterior to the left; images in the right panels are horizontal views with anterior to the left. Arrows and arrowheads indicate female-specific GFP-labeled neuronal cell bodies in Vs/Vp and PMm/PMg, respectively. Scale bars represent 500 μm. (C) Phylogenetic tree of NPBWR1 and NPBWR2. The number at each node indicates bootstrap values for 1000 replicates. Human opioid receptors μ1 (OPRM1) and δ1 (OPRD1) were used as the outgroup for tree reconstruction. Scale bar represents 0.1 substitution per site. For species names and GenBank accession numbers, see Supplementary file 3. (D) Distribution of npbwr2 expression in the brain, pituitary, and spinal cord. All images are coronal sections. Images of only males are presented, because there were no obvious sex differences in the distribution of expression (n = 5 per sex). Scale bars represent 100 μm. For abbreviations of brain and spinal cord regions and brain nuclei, see Supplementary file 1. See also Figure 3—figure supplement 1, Figure 3—figure supplement 2, Figure 3—figure supplement 3, and Figure 3—figure supplement 4.
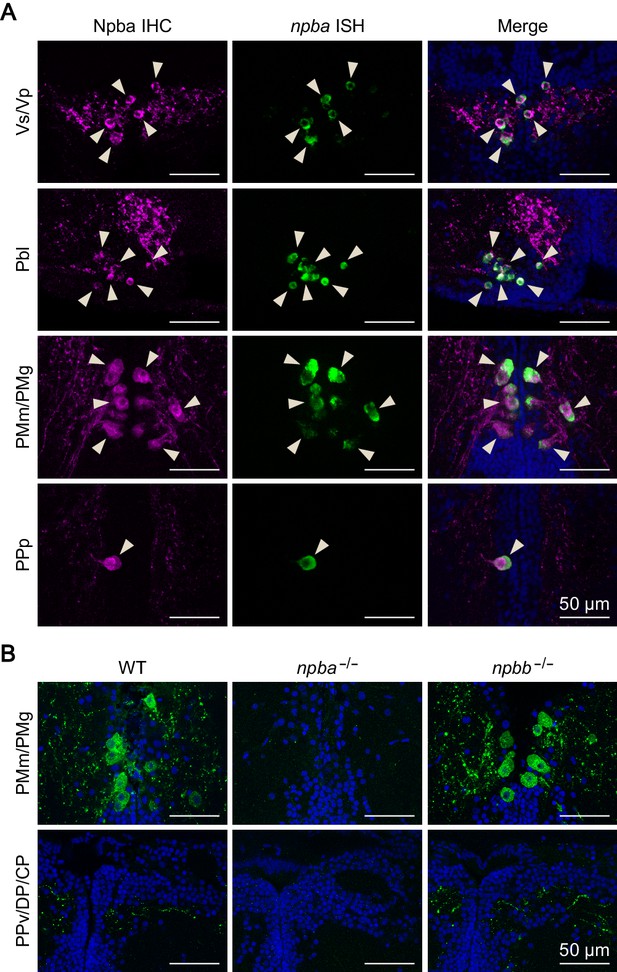
Verification of the specificity of the anti-Npba antibody.
(A) The specificity of the anti-Npba antibody was verified by a pattern of labeling consistent with npba-expressing neurons detected by in situ hybridization in the medaka brain. Left and middle panels show images of immunohistochemistry (IHC) using the anti-Npba antibody (green) and in situ hybridization (ISH) detecting npba expression (magenta), respectively, in the same sections; right panels show the merged images with nuclear counterstaining (blue). Arrowheads indicate representative neuronal cell bodies labeled by both IHC and ISH. The IHC signals that do not overlap with the ISH signals most likely represent the axons of Npba-expressing neurons (but not the cell bodies of other neurons), given their relatively small size and typical varicosity-like structures. Scale bars represent 50 μm. (B) The lack of cross-reactivity of the anti-Npba antibody with Npbb was confirmed by the observation that labeled cell bodies and axons (green) were present in npbb knockout (npbb-/-) as well as wild-type (WT) females, npbb-/- but totally absent in npba knockout (npba-/-) females (n = 15, 9, and eight for wild-type, npba-/-, and npbb-/- females). Representative micrographs of PMm/PMg, which contains neurons expressing both npba and npbb, and PPv/DP/CP, which contains hundreds of npbb-expressing neurons but no npba-expressing neurons, are shown. Scale bars represent 50 μm. For abbreviations of brain nuclei, see Supplementary file 1.
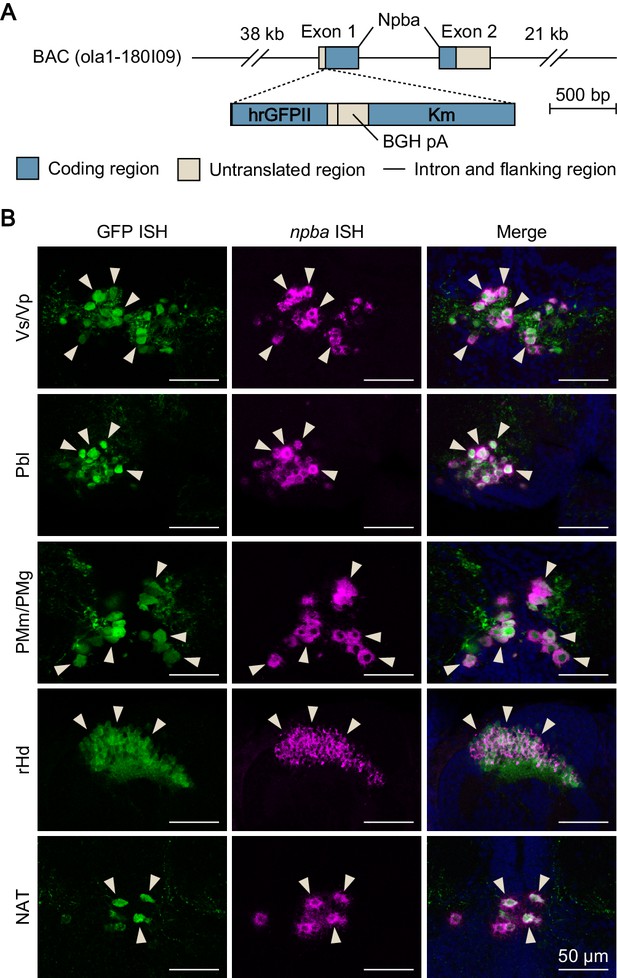
Generation and verification of npba-GFP transgenic medaka.
(A) Structure of the transgene in npba-GFP transgenic medaka. A 22 bp sequence containing the translation initiation site of npba in a medaka bacterial artificial chromosome (BAC) clone (clone ID: 180_I09) was replaced by a 2136 bp DNA cassette containing the humanized Renilla reniformis GFP II (hrGFPII)-coding sequence, bovine growth hormone polyadenylation signal (BGH pA), and kanamycin resistance gene (Km). This BAC clone contains the whole transcriptional unit of npba together with 38 kb of 5′-flanking and 21 kb of 3′-flanking sequence. (B) The specificity of GFP expression was verified by double in situ hybridization (ISH) of GFP and npba in the brain of npba-GFP transgenic medaka. Left and middle panels show images of GFP (green) and npba expression (magenta), respectively, in the same sections; right panels show the merged images with nuclear counterstaining (blue). Note that GFP fluorescence remained visible after processing for ISH. Arrowheads indicate representative neuronal cell bodies labeled by both GFP and npba ISH. Scale bars represent 50 μm. For abbreviations of brain nuclei, see Supplementary file 1.
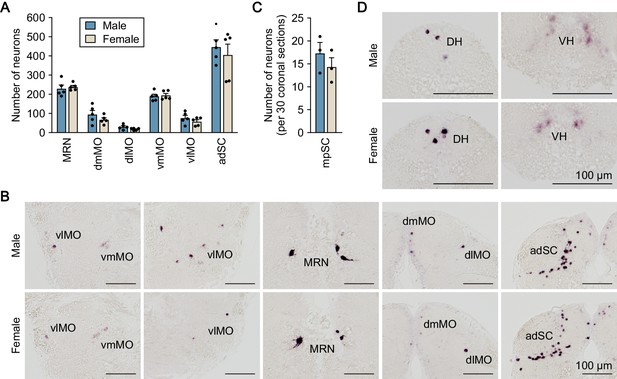
Distribution of npba-expressing neurons in the medaka medulla oblongata and spinal cord.
(A) Distribution of npba-expressing neurons in the medulla oblongata and the anterior part of the spinal cord of males (blue columns) and females (beige columns) (n = 5 per sex). (B) Representative micrographs showing npba-expressing neurons in the respective regions of the medulla oblongata and anterior part of the spinal cord. Scale bars represent 100 μm. (C) Number of npba-expressing neurons in the middle to posterior part of the spinal cord (mpSC) of males (blue columns) and females (beige columns) (n = 3 per sex). The number of neurons per 30 coronal sections of 10 μm thickness was counted. (D) Representative micrographs showing npba-expressing neurons in mpSC. Scale bars represent 100 μm. For abbreviations of brain and spinal cord regions and brain nuclei, see Supplementary file 1.
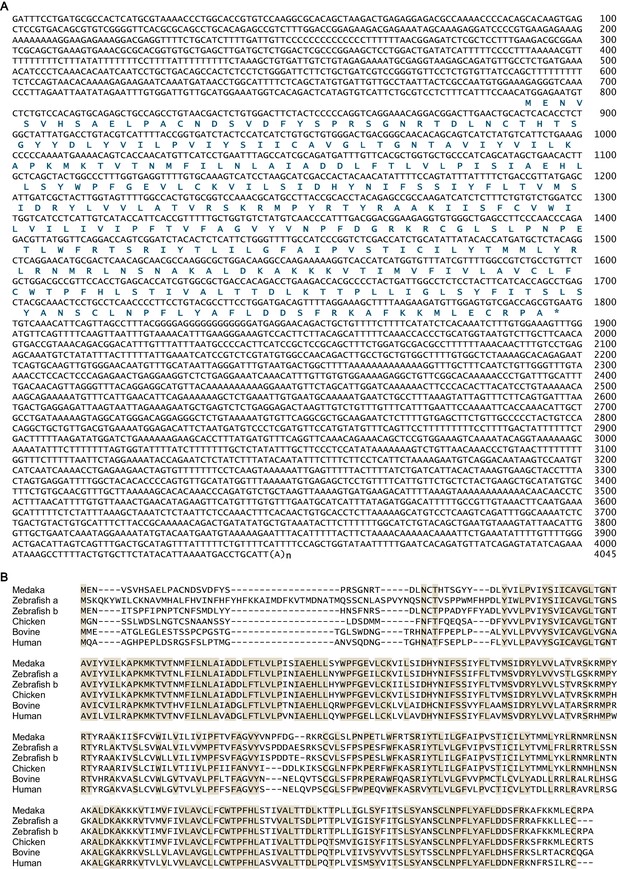
Sequence information for medaka npbwr2.
(A) Nucleotide and deduced amino acid sequences of the medaka npbwr2 cDNA. Asterisk indicates the stop codon. Nucleotide numbers are shown at the right of each sequence line. (B) Alignment of deduced amino acid sequences of Npbwr2 from medaka and other vertebrates. Identical amino acids in all sequences are shaded in beige. For species names and GenBank accession numbers, see Supplementary file 3.
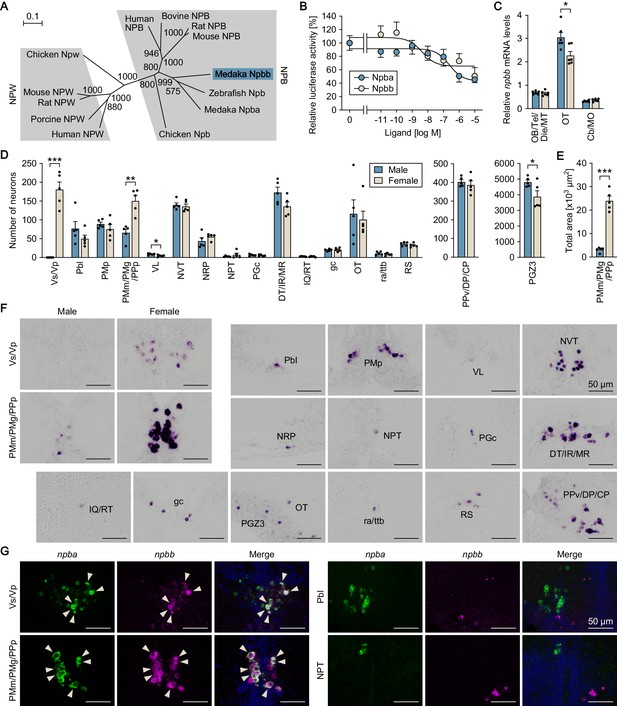
Medaka possess an additional NPB gene, designated npbb.
(A) Phylogenetic tree of NPB and NPW. The number at each node indicates bootstrap values for 1000 replicates. Scale bar represents 0.1 substitution per site. For species names and GenBank accession numbers, see Supplementary file 3. (B) Ability of medaka Npba and Npbb to activate medaka Npbwr2. Receptor activation was assessed by measuring cAMP-responsive element-driven luciferase activity in cells transfected with Npbwr2. The x-axis shows the concentration of Npba and Npbb, and the y-axis shows luciferase activity as a percentage of that observed in the absence of Npba/Npbb. (C) Overall levels of npbb expression in the male (blue columns) and female (beige columns) brain dissected into three parts as determined by real-time PCR (n = 6 per sex). *, p<0.05 (unpaired t-test). For abbreviations of brain regions, see Supplementary file 1. (D) Distribution of npbb-expressing neurons in the male (blue columns) and female (beige columns) brain (n = 5 per sex). The data are split into three graphs for clarity. *, p<0.05; **, p<0.01; ***, p<0.001 (unpaired t-test). For abbreviations of brain nuclei, see Supplementary file 1. (E) Total area of npbb expression in PMm/PMg/PPp in males (blue column) and females (beige column) (n = 5 per sex). ***, p<0.001 (unpaired t-test). (F) Representative micrographs showing npbb-expressing neurons in different brain nuclei. Micrographs of both sexes are shown for Vs/Vp and PMm/PMg/PPp, where npbb expression is confined or almost confined to females. Micrographs of males only are shown for other nuclei, where sex differences were not detected or, if present, were not sufficiently demonstrated by photographs. (G) Coexpression of npba and npbb in the same neurons in Vs/Vp and PMm/PMg, but not in other brain nuclei. In each set of panels, the left and middle ones show images of npba (green) and npbb (magenta) expression, respectively, in the same sections; the right ones show the merged images with nuclear counterstaining (blue). Arrowheads indicate representative neurons coexpressing npba and npbb. Scale bars represent 50 μm. For abbreviations of brain nuclei, see Supplementary file 1. See also Figure 4—figure supplement 1 and Figure 4—figure supplement 2.

Sequence information for medaka npbb.
(A) Nucleotide and deduced amino acid sequences of the medaka npbb cDNA. The predicted signal peptide is underlined and the mature Npbb polypeptide is boxed. Asterisk indicates the stop codon. Nucleotide numbers are shown at the right of each sequence line. (B) Comparison of mature NPB polypeptide sequences from medaka and other vertebrate species. Identical amino acids in all sequences are shaded in beige. For species names and GenBank accession numbers, see Supplementary file 3.
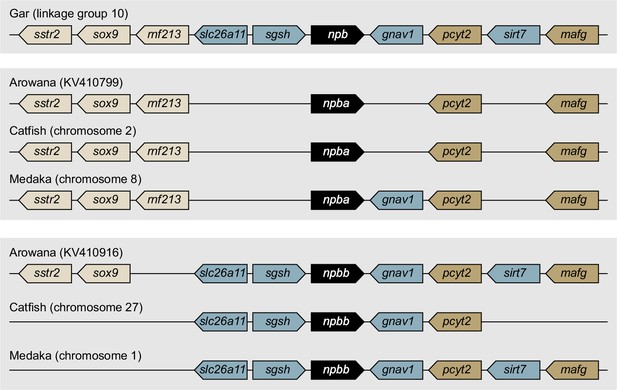
Comparison of syntenic relationships of genes in the vicinity of gar, arowana, catfish, and medaka NPB genes.
Genes are represented by arrows denoting the direction of transcription. gnav1, guanine nucleotide-binding protein (G protein) α v1; mafg, MAF bZIP transcription factor G; pcyt2, phosphate cytidylyltransferase 2, ethanolamine; rnf213, ring finger protein 213; sgsh, N-sulfoglucosamine sulfohydrolase; sirt7, sirtuin 7; slc26a11, solute carrier family 26 member 11; sox9, SRY (sex determining region Y)-box 9; sstr2, somatostatin receptor 2.
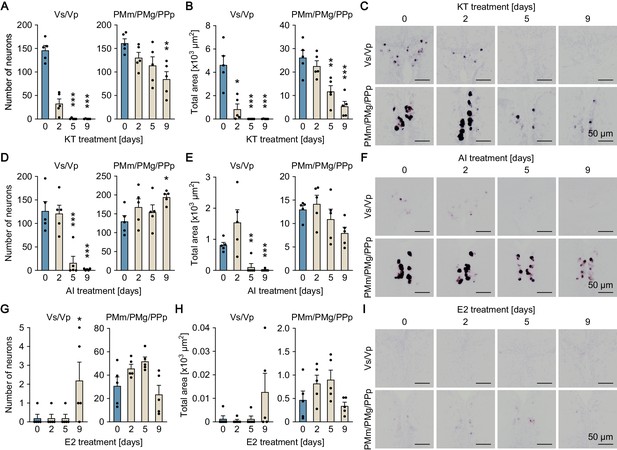
Sexually dimorphic npbb expression can also be reversed by altering the sex steroid milieu.
Temporal changes in npbb expression in Vs/Vp and PMm/PMg of 11-ketotestosterone (KT)-treated females (A, B, C), aromatase inhibitor (AI)-treated females (D, E, F), and estradiol-17β (E2)-treated males (G, H, I) (n = 5 per treatment and sampling day). (A, D, G) Number of npbb-expressing neurons in Vs/Vp and PMm/PMg. *, p<0.05; **, p<0.01; ***, p<0.001 (versus day 0, Dunnett’s post hoc test). (B, E, H) Total area of npbb expression in Vs/Vp and PMm/PMg. *, p<0.05; **, p<0.01; ***, p<0.001 (versus day 0, Dunnett’s post hoc test). (C, F, I) Representative micrographs showing npbb expression in Vs/Vp and PMm/PMg. Scale bars represent 50 μm. See also Figure 5—figure supplement 1 and Figure 5—figure supplement 2.
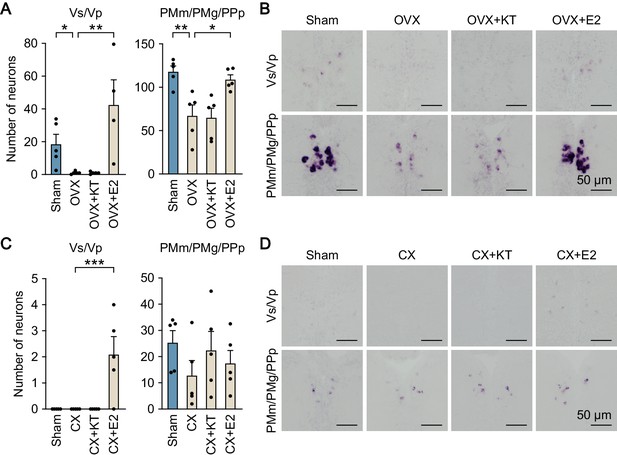
Effects of gonadectomy and sex steroid supplementation on npbb expression in Vs/Vp and PMm/PMg.
(A) Number of npbb-expressing neurons in Vs/Vp and PMm/PMg/PPp of sham-operated females (Sham) and ovariectomized females that were exposed to vehicle alone (OVX), 11-ketotestosterone (OVX + KT), or estradiol-17β (OVX + E2) (n = 5 per group). *, p<0.05; **, p<0.01 (Bonferroni’s post hoc test). (B) Representative micrographs of npbb expression in Vs/Vp and PMm/PMg/PPp of Sham, OVX, OVX + KT, and OVX + E2 females. Scale bars represent 50 μm. (C) Number of npbb-expressing neurons in Vs/Vp and PMm/PMg/PPp of Sham males and castrated males that were exposed to vehicle alone (CX), 11-ketotestosterone (CX + KT), or estradiol-17β (CX + E2) (n = 5 per group). ***, p<0.001 (Bonferroni’s post hoc test). (D) Representative micrographs of npbb expression in Vs/Vp and PMm/PMg/PPp of Sham, CX, CX + KT, and CX + E2 males. Scale bars represent 50 μm.
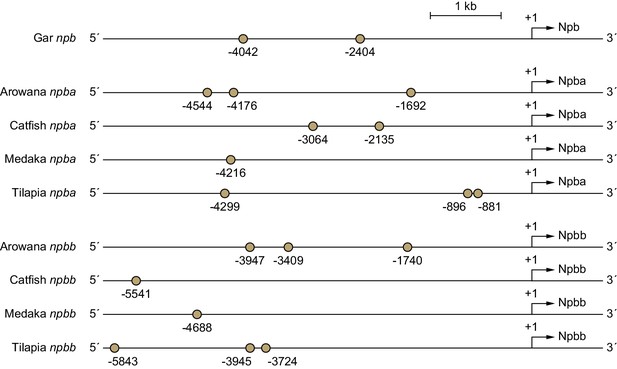
Putative estrogen-responsive elements (EREs) predicted in the proximal promoter regions of gar, arowana, catfish, medaka, and tilapia NPB genes.
Ocher circles represent putative EREs. Numbers indicate nucleotide positions relative to the translation initiation site (+1).
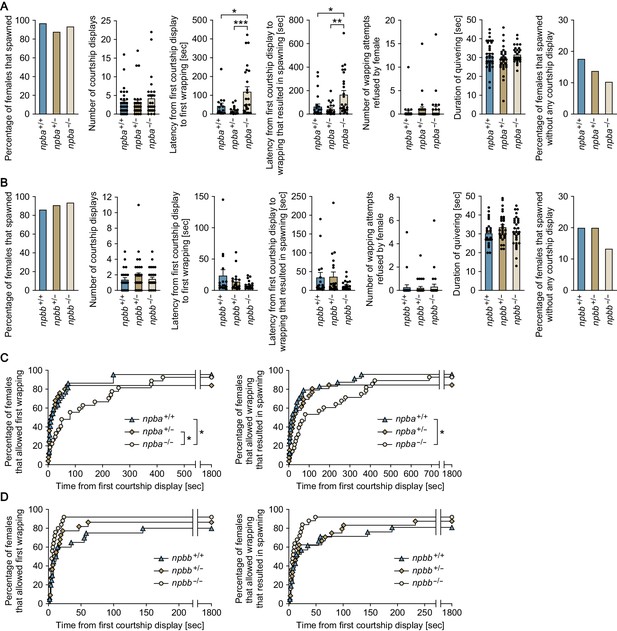
npba is involved in female sexual receptivity.
(A, B) Various parameters in the mating behavior of npba (A) and npbb (B) single knockout females were measured and compared with wild-type females (n = 34, 33, and 31 for npba+/+, npba+/-, and npba-/- females, respectively; n = 29, 33, and 32 for npbb+/+, npbb+/-, and npbb-/- females, respectively). Blue, ocher, and beige columns represent wild-type, heterozygous knockout, and homozygous knockout females, respectively. *, p<0.05; **, p<0.01; ***, p<0.001 (Bonferroni’s post hoc test). (C, D) The latency data for npba (C) and npbb (D) single knockouts were further analyzed using Kaplan-Meier plots. Blue triangles, ocher diamonds, and beige circles represent wild-type, heterozygous knockout, and homozygous knockout females, respectively. *, p<0.05 (Gehan-Breslow-Wilcoxon test with Bonferroni’s correction). See also Figure 6—figure supplement 1.
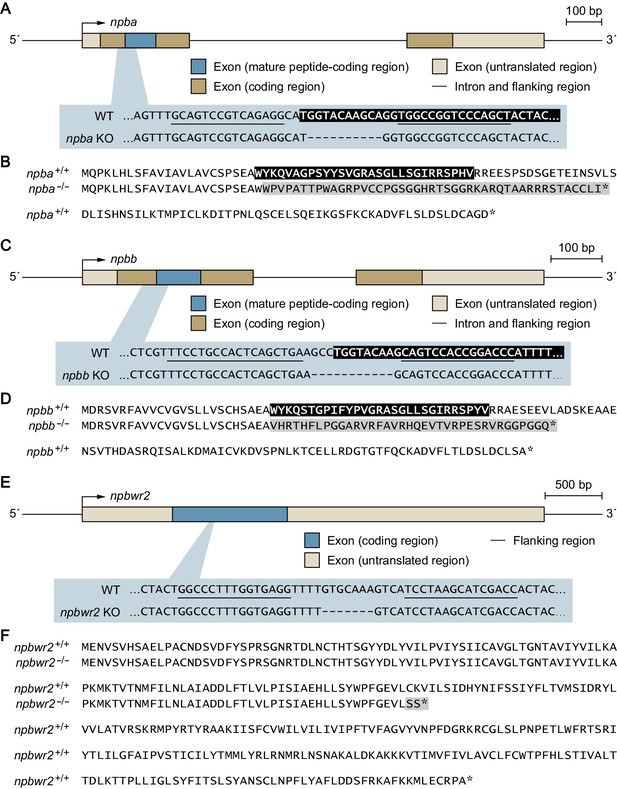
Genetic scheme for the npba-/-, npbb-/-, and npbwr2-/- mutant medaka.
Knockout medaka deficient for npba (A, B), npbb (C, D), and npbwr2 (E, F) were generated by transcription activator-like effector nuclease (TALEN)-mediated genome editing. (A) Gene structure of npba showing the location of the deletion target site, which is enlarged to show the nucleotide sequences of the npba+/+ and npba-/- alleles. The mature Npba polypeptide-coding sequence is indicated in white letters on a black background. TALEN binding sites are underlined, and deleted nucleotides are indicated by dashes. (B) Comparison of the deduced Npba precursor protein sequences of the npba+/+ and npba-/- alleles. The mature Npba polypeptide is indicated in white letters on a black background. The altered sequence caused by a frameshift is shaded in gray. Asterisks indicate stop codons. (C) Gene structure of npbb showing the location of the deletion target site, which is enlarged to show the nucleotide sequences of the npbb+/+ and npbb-/- alleles. The mature Npbb polypeptide-coding sequence is indicated in white letters on a black background. TALEN binding sites are underlined and deleted nucleotides are indicated by dashes. (D) Comparison of the deduced Npbb precursor protein sequences of the npbb+/+ and npbb-/- alleles. The mature Npbb polypeptide is indicated in white letters on a black background. The altered sequence caused by a frameshift is shaded in gray. Asterisks indicate stop codons. (E) Gene structure of npbwr2 showing the location of the deletion target site, which is enlarged to show the nucleotide sequences of the npbwr2+/+ and npbwr2-/- alleles. TALEN binding sites are underlined and deleted nucleotides are indicated by dashes. (F) Comparison of the deduced Npbwr2 protein sequences of the npbwr2+/+ and npbwr2-/- alleles. The altered sequence caused by a frameshift is shaded in gray. Asterisks indicate stop codons.
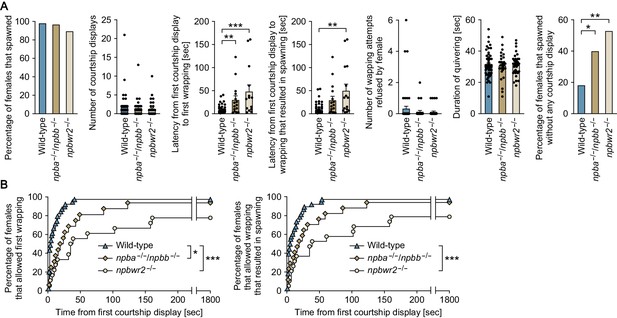
Npba/Npbb/Npbwr2 signaling is involved in female sexual receptivity.
(A) Various parameters in the mating behavior of npba/npbb double knockout (npba-/-/npbb-/-) females (n = 31) and npbwr2 knockout (npbwr2-/-) females (n = 38) were measured and compared with wild-type females (n = 55). Blue, ocher, and beige columns represent wild-type, npba/npbb double knockout, and npbwr2 knockout females, respectively. *, p<0.05; **, p<0.01; ***, p<0.001 (Dunnett’s post hoc test for data on number, latency, and duration; Fisher’s exact test for data on percentage). (B) The latency data were further analyzed using Kaplan-Meier plots. Blue triangles, ocher diamonds, and beige circles represent wild-type, npba/npbb double knockout, and npbwr2 knockout females, respectively. *, p<0.05; ***, p<0.001 (Gehan-Breslow-Wilcoxon test with Bonferroni’s correction). See Video 1, Video 2, and Video 3. Supplementary Data List.
Videos
A representative video showing the mating behavior of wild-type females.
https://doi.org/10.7554/eLife.39495.022A representative video showing the mating behavior of npba/npbb double knockout females.
https://doi.org/10.7554/eLife.39495.023A representative video showing the mating behavior of npbwr2 knockout females.
https://doi.org/10.7554/eLife.39495.024Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Oryzias latipes) | npba | DOI: 10.1210/en.2013-1806 | Genbank:NM_001308979 | |
| Gene (O. latipes) | npbb | NBRP Medaka | clone ID:olova57o22; clone ID:olova6h09; clone ID:olova8d12; clone ID:olova13m04; clone ID:olova28d23; clone ID:olova58i17 | |
| Gene (O. latipes) | npbwr2 | Genbank:LC375958 | ||
| Gene (O. latipes) | actb | Genbank:NM_001104808 | ||
| Strain, strain background (O. latipes) | d-rR | NBRP Medaka | strain ID:MT837 | maintained in a closed colony over 10 years in Okubo lab |
| Genetic reagent (O. latipes) | ΔERE mutant | this paper | generated and maintained in Okubo lab | |
| Genetic reagent (O. latipes) | npba-GFP transgenic | this paper | generated and maintained in Okubo lab | |
| Genetic reagent (O. latipes) | npba-/- | this paper | generated and maintained in Okubo lab | |
| Genetic reagent (O. latipes) | npbb-/- | this paper | generated and maintained in Okubo lab | |
| Genetic reagent (O. latipes) | npbwr2-/- | this paper | generated and maintained in Okubo lab | |
| Cell line (Homo sapiens) | HEK293T | Riken BRC Cell Bank | Cell number:RCB2202; RRID:CVCL_0063 | |
| Cell line (Escherichia coli) | DY380 | DOI: 10.1038/35093556; DOI: 10.1006/geno.2000.6451 | ||
| Transfected construct | pcDNA3.1/V5-His-TOPO | Thermo Fisher Scientific | Thermo Fisher Scientific:K480001 | |
| Transfected construct | pGL4.29 | Promega | Promega:E8471 | |
| Transfected construct | pGL4.74 | Promega | Promega:E6921 | |
| Antibody | anti-Npba antibody | this paper | RRID:AB_2810229 | rabbit polyclonal; against the entire Npba polypeptide (1:500 or 1:1000) |
| Antibody | Dylight 549-conjugated goat anti-rabbit IgG | Vector Laboratories | Vector Laboratories:DI-1549; RRID:AB_2336407 | (1:500 or 1:1000) |
| Antibody | Alexa Flour 488-conjugated goat anti-rabbit IgG | Thermo Fisher Scientific | Thermo Fisher Scientific:A-11070; RRID:AB_2534114 | (1:500) |
| Antibody | horseradish peroxidase-conjugated anti-fluorescein antibody | PerkinElmer | PerkinElmer:NEF710001EA; RRID:AB_2737388 | (1:500 or 1:1000) |
| Antibody | alkaline phosphatase-conjugated anti-DIG antibody | Roche Diagnostics | Roche Diagnostics:11093274910; RRID:AB_514497 | (1:500–1:10000) |
| Recombinant DNA reagent | medaka bacterial artificial chromosome (BAC) clone | NBRP Medaka | clone ID:180_I09 | |
| Recombinant DNA reagent | phrGFP II-1 mammalian expression vector | Agilent Technologies | Agilent Technologies:240143 | |
| Recombinant DNA reagent | pGEM-Teasy vector | Promega | Promega:A1360 | |
| Peptide, recombinant protein | Npba polypeptide | this paper | WYKQVAGPSYYSVGRASGLLSGIRRSPHV-NH2 | |
| Peptide, recombinant protein | Npbb polypeptide | this paper | WYKQSTGPIFYPVGRASGLLSGIRRSPYV-NH2 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Promega:E1910 | |
| Commercial assay or kit | RNeasy Lipid Tissue Mini Kit | Qiagen | Qiagen:74804 | |
| Commercial assay or kit | RNeasy Plus Universal Mini Kit | Qiagen | Qiagen:73404 | |
| Commercial assay or kit | Omniscript RT Kit | Qiagen | Qiagen:205111 | |
| Commercial assay or kit | SuperScript VILO cDNA Synthesis Kit | Thermo Fisher Scientific | Thermo Fisher Scientific:11754050 | |
| Commercial assay or kit | LightCycler 480 SYBR Green I Master | Roche Diagnostics | Roche Diagnostics:04887352001 | |
| Commercial assay or kit | Golden Gate TALEN and TAL Effector Kit 2.0 | Addgene | Addgene:1000000024 | |
| Commercial assay or kit | mMessage mMachine SP6 Kit | Thermo Fisher Scientific | Thermo Fisher Scientific:AM1340 | |
| Commercial assay or kit | Marathon cDNA Amplification Kit | Takara Bio | Takara Bio:634913 | |
| Commercial assay or kit | Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Thermo Fisher Scientific:4367659 | |
| Commercial assay or kit | TSA Plus Fluorescein System | PerkinElmer | PerkinElmer:NEL741001KT | |
| Chemical compound, drug | aromatase inhibitor (AI); Fadrozole | Sigma-Aldrich | Sigma-Aldrich:F3806-10MG | |
| Chemical compound, drug | estradiol-17β; E2 | Fujifilm Wako Pure Chemical Corporation | Fujifilm Wako Pure Chemical Corporation:058–04043 | |
| Chemical compound, drug | 11-ketotestosterone; KT | Cosmo Bio | Cosmo Bio:117 ST | |
| Software, algorithm | GraphPad Prism | GraphPad Software | RRID:SCR_002798 | |
| Software, algorithm | Adobe Photoshop | Adobe Systems | RRID:SCR_014199 | |
| Software, algorithm | ImageJ | http://rsbweb.nih.gov/ij/ | RRID:SCR_003070 | |
| Software, algorithm | InterProScan | http://www.ebi.ac.uk/interpro/search/sequence-search | RRID:SCR_005829 | |
| Software, algorithm | SignalP | http://www.cbs.dtu.dk/services/SignalP/ | RRID:SCR_015644 | |
| Software, algorithm | ClustalW | http://clustalw.ddbj.nig.ac.jp/index.php | RRID:SCR_017277 | |
| Software, algorithm | Jaspar | http://jaspar.genereg.net/ | RRID:SCR_003030 | |
| Other | DAPI stain | Thermo Fisher Scientific | Thermo Fisher Scientific:D1306; RRID:AB_2629482 | (1:1000) |
Additional files
-
Supplementary file 1
Abbreviations of brain and spinal cord regions and brain nuclei.
- https://doi.org/10.7554/eLife.39495.025
-
Supplementary file 2
Primers used in this study.
- https://doi.org/10.7554/eLife.39495.026
-
Supplementary file 3
Species names and GenBank accession numbers of the protein sequences used in this study.
- https://doi.org/10.7554/eLife.39495.027
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39495.028





