Visualization of the type III secretion mediated Salmonella–host cell interface using cryo-electron tomography
Figures
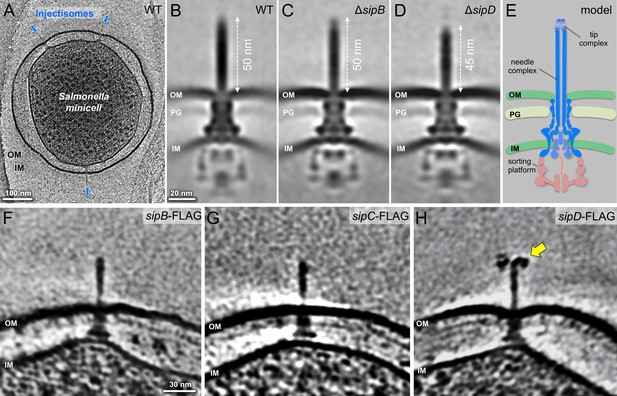
In situ structures of host-free S. Typhimurium T3SS injectisome in wild-type (WT), ΔsipB, and ΔsipD minicells.
(A) A central section of a tomogram showing S. Typhimurium minicell containing multiple injectisomes. (B–D) Central sections of sub-tomogram averages showing injectisomes of WT, ΔsipB, and ΔsipD, respectively. (E) A schematic of the injectisome. Outer membrane (OM), peptidoglycan (PG), sorting platform, and inner membrane (IM) of S. Typhimurium are annotated. (F–H) Central sections of tomograms showing injectisomes from strains expressing epitope-tagged (FLAG) SipB, SipC, and SipD, respectively. Yellow arrow indicates antibody bound to the epitope-tag.
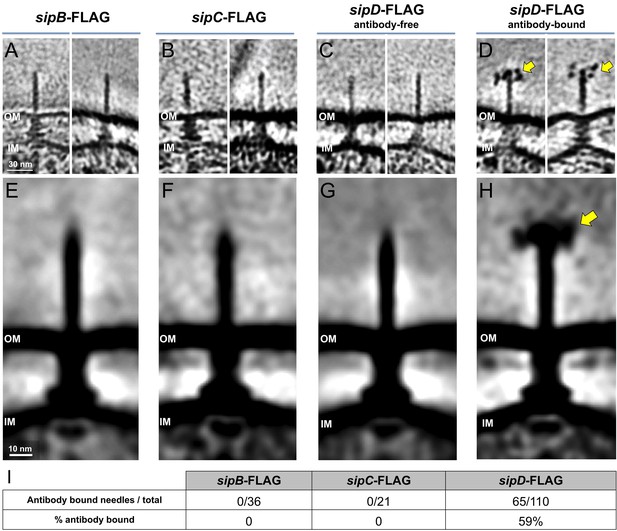
Detection of FLAG-epitope-tagged SipB, SipC, and SipD Central slices from representative tomograms showing.
(A) sipB-FLAG, (B) sipC-FLAG, (C) antibody-free sipD-FLAG, and (D) antibody-bound sipD-FLAG needles. (E-H) Sub-tomogram averages of FLAG-epitope-tagged S. Typhimurium strains shown in panels (A-D), respectively. Yellow arrows indicate anti-FLAG antibodies bound to the epitope-tag. (I) Quantification of anti-FLAG antibody bound needles.
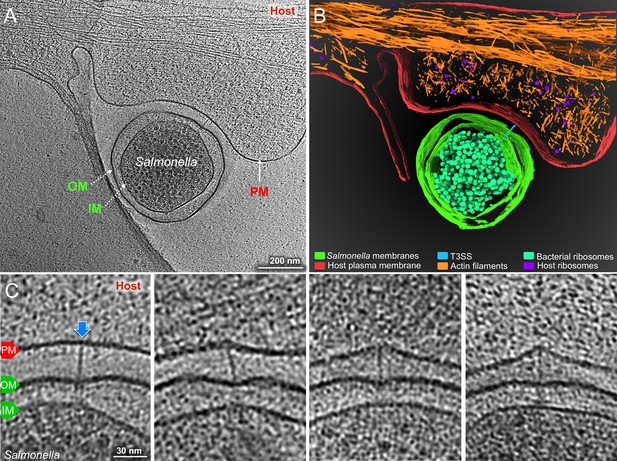
Visualization of the T3SS mediated Salmonella-Host interactions.
(A) A central slice showing a S. Typhimurium minicell interacting with a host. Plasma membrane (PM) of HeLa cell, outer membrane (OM) and inner membrane (IM) of S. Typhimurium are annotated. (B) 3D rendering of the tomogram shown in (A). (C) Tomographic slices showing injectisomes interacting with the host PM. Blue arrows indicate needles attached to the host PM. Direction of the arrow represents the angle of needle perpendicular to the host PM.
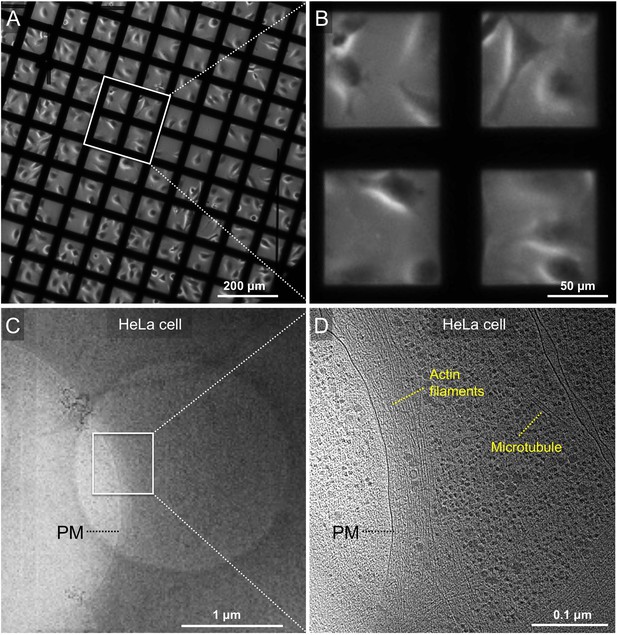
Cultivation of mammalian cells (HeLa) on EM grid for cryo-ET.
(A) Phase contrast microscopy image of HeLa cells grown on a gold Quantifoil grid. (B) A zoom-in view of the boxed area in panel a. (C) A snapshot of HeLa cell edge in a low-magnification montage. (D) Tomographic slice of the boxed area in panel C showing cellular features such as actin filaments and microtubules.
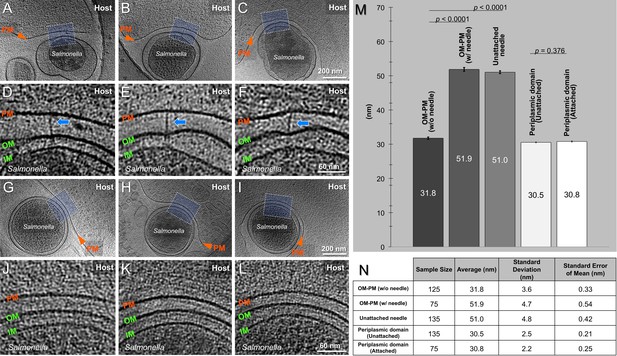
Inter-membrane spacing between outer membrane and plasma membrane.
(A-C) Central slices of tomograms showing different Salmonella - host cell contacts in the presence of T3SS injectisomes. (D-F) The zoom-in views of the boxed regions in the tomographic slices from panels (A-C) respectively. (G-I) Central slices of tomograms showing different Salmonella - host cell contacts without the presence of T3SS injectisomes. (J-L) The zoom-in views of the boxed regions in the tomographic slices from panels (G-I), respectively. Plasma membrane (PM) of HeLa cell, outer membrane (OM) and cytoplasmic membrane (CM) of S. Typhimurium are annotated. (M) Average membrane spacing between S. Typhimurium minicells and HeLa cells at different positions as indicated across the bottom of the bar graph. Error bars indicate s.e.m. Data were compared using an unpaired t test. (N) A summary of statistical measures including average, standard deviation, and standard error of mean.
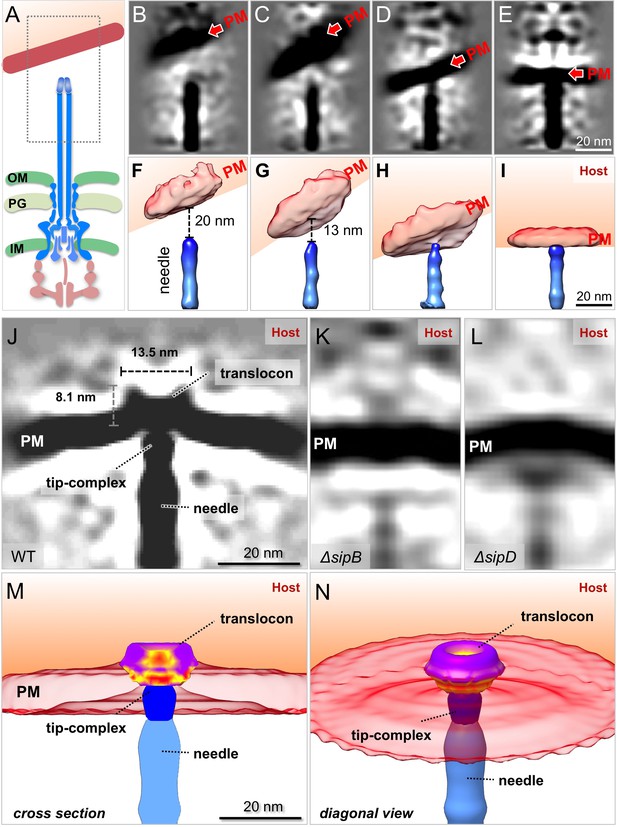
In situ structural analysis of the interface between the T3SS needle and the host membrane reveals a novel structure of the intact translocon.
(A) A schematic representation of the S. Typhimurium injectisome with a box highlighting the area used for alignment and classification. (B–E) Central sections and (F–I) 3-D surface views of class averages showing different spacings between the needle and the plasma membrane (PM). (J–L) Central sections of the sub-tomogram averages of the interface between the host PM and the needle of WT, ΔsipB, and ΔsipD, respectively. (M) Cross-section and (N) diagonal view of the surface rendering of the translocon in panel (J).
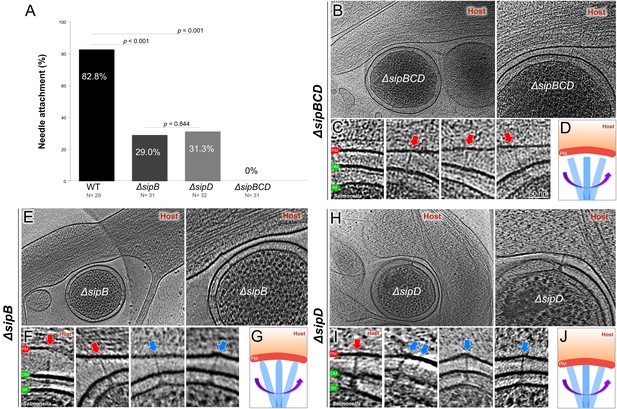
Deletion of the protein translocases disrupts the T3SS-dependent intimate attachment to the host PM, and the formation of the translocon.
(A) Percentage of minicells attached to the host membrane via needle-membrane contact. Data were compared using a chi-squared test. (B, C) Central slices from tomograms showing the ΔsipBCD injectisomes interacting with the host PM. (E, F) Central slices from tomograms showing the ΔsipB injectisomes interacting with the host PM. (H, I) Central slices from tomograms showing the ΔsipD injectisomes interacting with the host PM. Blue arrows indicate needles attached to the host PM. Red arrows indicate unattached needles. (D, G, J) Schematic models depicting needle-attachment patterns from three mutants, respectively.
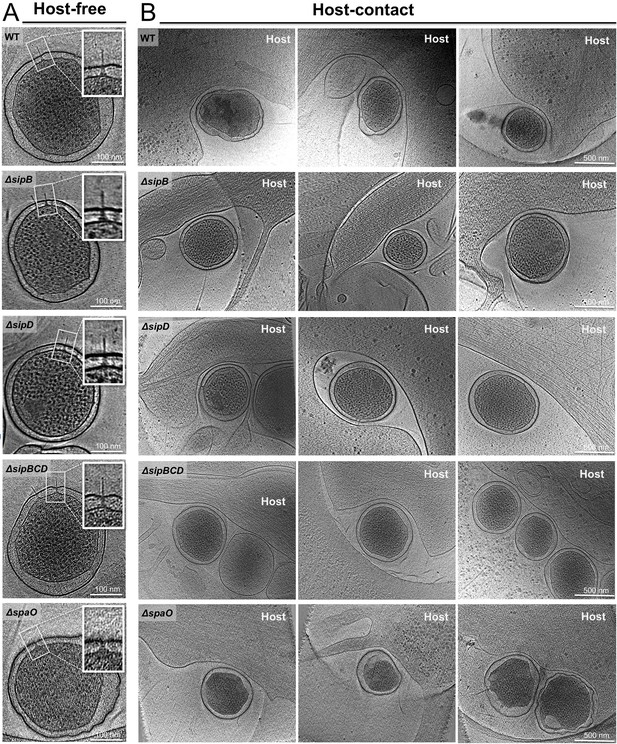
Gallery of snapshots from host-free and host-interacting S. Typhimurium minicells.
Central slices from representative tomograms showing (A) host-free and (B) host-interacting S. Typhimurium minicells from WT, ΔsipB, ΔsipD, ΔsipBCD, and ΔspaO strain.
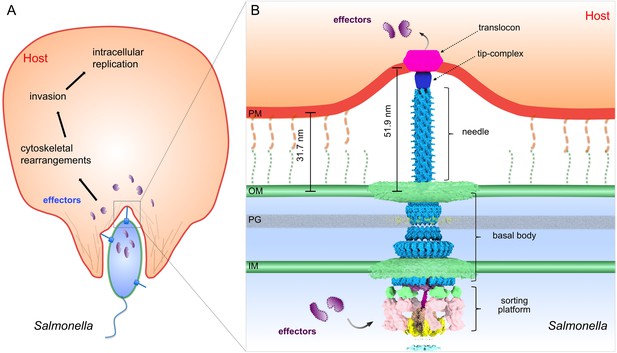
Model of the S. Typhimurium injectisome interacting with the host cell membrane.
(A) A schematic diagram of S. Typhimurium interacting with the host cell. (B) Molecular model of the T3SS injectisome at the Salmonella-host cell interface.
Videos
A typical reconstruction shows the detailed interaction between the T3SS machines and the target cell and the membrane remodeling.
https://doi.org/10.7554/eLife.39514.008An animation shows the T3SS mediated Salmonella-host interaction and a plausible pathway of effector translocation.
https://doi.org/10.7554/eLife.39514.013Tables
Needle lengths of S. Typhimurium WT, ΔsipB, ΔsipD and ΔsipBCD cells.
A summary of statistical measures including needle length average, standard deviation, and standard error of mean. Data were compared using an unpaired t test.
| Sample size | Average (nm) | Standard Deviation | Standard Error of Mean(nm) | P value campared to WT | |
|---|---|---|---|---|---|
| WT | 135 | 51.0 | 4.8 | 0.42 | |
| ∆sipB | 46 | 50.6 | 4.0 | 0.59 | 0.62 |
| ∆sipD | 61 | 46.5 | 3.9 | 0.50 | <0.0001 |
| ∆sipBCD | 46 | 45.3 | 3.0 | 0.44 | <0.0001 |
| Reagent type (species) or resources | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | SB1780 (Salmonella enterica serovar Typhymurium SL1344) | PMID: 23481398 | minD::cat (wt) | Galán Laboratory (Yale University) |
| Strain, strain background | SB3542 | This study | ΔsipB minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3543 | This study | ΔsipD minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3141 | This study | ΔsipBCD minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3046 | PMID: 28283062 | ΔspaO minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3544 | This study | sipB3xFLAG minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3545 | This study | sipB3xFLAG minD::cat | Galán Laboratory (Yale University) |
| Strain, strain background | SB3546 | This study | sipB3xFLAG minD::cat | Galán Laboratory (Yale University) |
| Genetic reagent | pSB3292 (Plasmid) | PMID: 28283062 | hilA in pBAD24 | Galán Laboratory (Yale University) |
| Genetic reagent | minD::cat P22 (P22 bacteriophage lysate) | Galán Laboratory (Yale University) | P22 lysate from SB1780 S. Typhimurium strain | Source of minD:: cat allele |
| Cell line | HeLa | ATCC | Hela (ATCC CCL-2) | |
| Antibody | M2 | Sigma-Aldrich | F3165 | 1:1000 by volume |
| Chemical compound, drug | LB Broth | Fisher BioReagents | BP1426 | |
| Chemical compound, drug | LB Agar | Fisher BioReagents | BP1425 | |
| Chemical compound, drug | L-arabinose | VWR | 1B1473 | |
| Chemical compound, drug | Ampicillin Sodium Salt | Fisher BioReagents | BP1760-25 | |
| Other | Gold grid | Quantifoil | R 2/1 on Au 200 mesh | |
| Software, algorithm | SerialEM | PMID: 16182563 | http://bio3d.colorado.edu/SerialEM/ | Data acquisition |
| Software, algorithm | MotionCor2 | PMID: 28250466 | http://msg.ucsf.edu/em/software/motioncor2.html | Motion correction |
| Software, algorithm | Tomoauto | PMID: 26863591 | https://github.com/DustinMorado/tomoauto | Tomogram reconstruction |
| Software, algorithm | Tomo3D | PMID: 25528570 | https://sites.google.com/site/3demimageprocessing/tomo3d | Tomogram reconstruction |
| Software, algorithm | IMOD | PMID: 8742726 | http://bio3d.colorado.edu/imod/ | Tomogram reconstruction |
| Software, algorithm | I3 | PMID: 16973379 | http://www.electrontomography.org/ | Sub-tomogram averaging |
| Software, algorithm | UCSF Chimera | PMID: 15264254 | http://www.cgl.ucsf.edu/chimera/ | 3D rendering |
| Software, algorithm | UCSF ChimeraX | PMID: 28710774 | https://www.rbvi.ucsf.edu/chimerax/ | 3D rendering |
Number of tomograms collected and analyzed.
https://doi.org/10.7554/eLife.39514.014| Number of tomograms collected | |
|---|---|
| WT - HeLa | 458 |
| ΔsipB - HeLa | 46 |
| ΔsipD - HeLa | 52 |
| ΔsipBCD - HeLa | 86 |
| ΔspaO - HeLa | 84 |
| WT minicells | 85 |
| ΔsipB minicell | 115 |
| ΔsipD minicell | 142 |
| sipB-FLAG - HeLa | 9 |
| sipC-FLAG - HeLa | 8 |
| sipD-FLAG - HeLa | 11 |
| sipB-FLAG | 5 |
| sipB-FLAG | 7 |
| sipB-FLAG | 13 |
| Total | 1051 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39514.015





