Dynein-2 intermediate chains play crucial but distinct roles in primary cilia formation and function
Figures
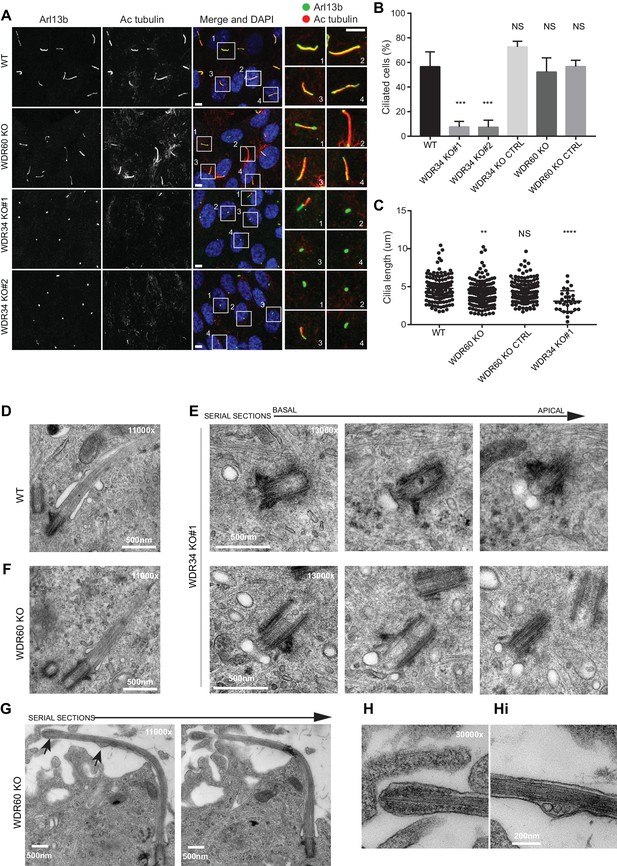
Role of dynein-2 intermediate chains WDR34 and WDR60 in ciliogenesis.
(A) Cilia stained with the markers Arl13b (green) and acetylated tubulin (AcTub, red) in RPE1 WDR60 and WDR34 KO cell lines. Panels on the right show enlargements of the boxed regions. Scale bars 5 μm. (B) Percentage of ciliated cells (n = 3; 656 WT, 430 WDR34 KO#1, 296 WDR34 KO#2, 397 WDR34 KO CTRL, 343 WDR60 KO and 195 WDR60 KO CTRL cells quantified). (C) Cilium length in WDR60 and WDR34 KO compared with WT cells and CRISPR control cells lines (n = 3; 120 WT, 158 WDR60 KO, 138 WDR60 KO CTRL and 30 WDR34#1 cells quantified). Mann-Whitney test was used, p-value: ****=<0.0001. (D–Hi) Representative 70 nm thick EM sections of (D) WT, (E) WDR34 KO and (F, G– Hi) WDR60 KO cilia. (E) Six serial sections through a WDR34 KO cilium showing no axoneme extension. (G) Two serial sections through a WDR60 KO cilium showing complete cilium. Arrows point to the bulbous ciliary tip and to a membrane protrusion containing membrane vesicles; enlargements are shown to the right (H and Hi). Scale bar length and magnification is indicated on the images.
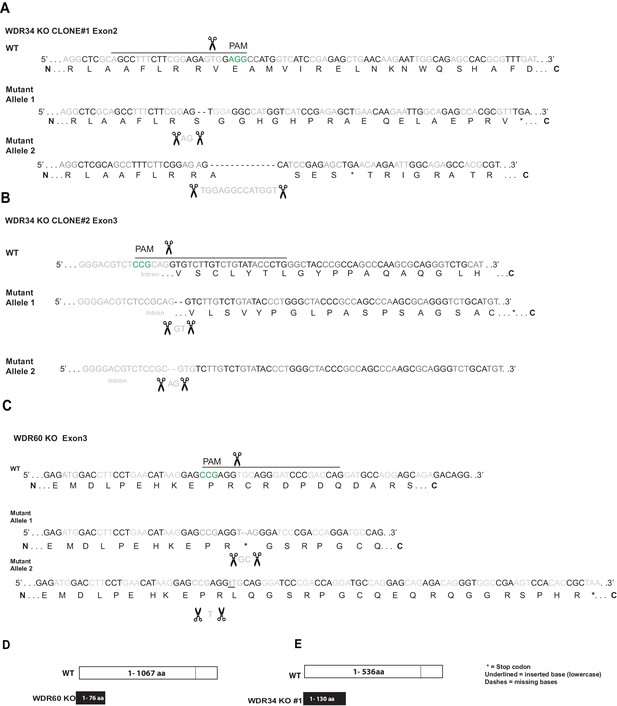
Generation of a WDR60 and WDR34 KO cell line.
(A and B) Genomic sequence for CRISPR-Cas9 target sites in the two WDR34 KO cell lines (identified as #1 and #2) and (C) WDR60 KO cells. Black lines and scissors depict gRNA binding and cut sites. Nucleotides in green show the CRISPR PAM site. Amino acid translation shown underneath; with the alternative translation in KO cells. Asterisk indicates a premature stop codon. In WDR34 KO cell clone #2 the mutation in the second allele affects the acceptor site of the intron. (D and E) Diagrams of WT type WDR60 and WDR34 and CRISPR mutant proteins. WDR60 is a protein of 1067 aa, in WDR60 KO the stop codon is introduced in exon 2 (76 aa). WDR34 is a protein of 536aa, in WDR34 KO #1 cells a stop codon is introduced in exon 2 (130aa).
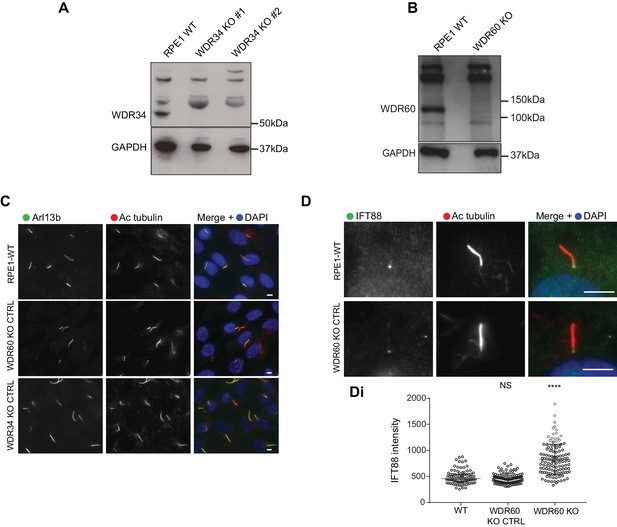
CRISPR control cells lines show no defect in ciliogenesis.
(A and B) Immunoblotting for WDR34 and WDR60 in WT and KO cells. (A) A 52 kDa band, corresponding to WDR34 is lost in WDR34 KO cells lysate. (B) A WDR60 band of 123 kDa is lost in the WDR60 KO cells lysate. (C) WDR34 KO CTRL and WDR60 KO CTRL cells stained with Arl13b (green) and AcTub (red). (D) WDR60 KO CTRL cells stained with IFT88 (green). (Di) IFT88 intensity quantification in WT and WDR60 KO CTRL cells (n = 3, 98 WT, 102 WDR60 KO CTRL, and 122 WDR60 KO cells quantified). Mann-Whitney test was used, p-value: *=<0.05 as above. Scale bars 5 μm.
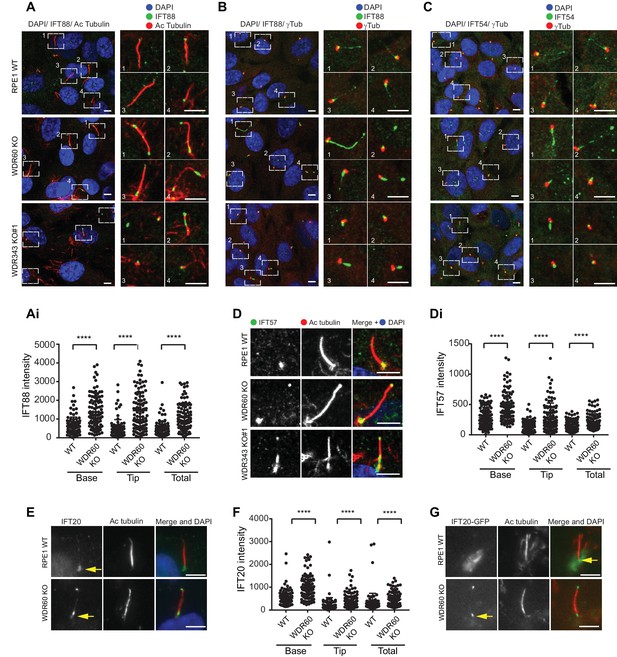
WDR34 and WDR60 are essential for IFT-B trafficking in primary cilia.
(A) Localization of IFT88 in WT, WDR60 KO, and WDR34 KO#1 cells stained with the ciliary marker Acetylated tubulin. (Ai) Quantification of IFT88 localization within cilia in WT and WDR60 KO cells (102 WT and 101 WDR60 cells quantified). (B and C) Localization of IFT88 (B) and IFT54 (C) in WT, WDR60 KO, and WDR34 KO#1 cells stained with the centrosome marker gamma-tubulin. (D) Localization of IFT57 in WT, WDR60 KO, and WDR34 KO#1 cells stained with acetylated tubulin. (Di) Quantification of IFT57 within cilia in WDR60 KO and WT cells (IFT57 106 WT and 98 WDR60 KO cells quantified; n = 3 independent experiments). Mann-Whitney test was used, p-value: ****=<0.0001. (E) Endogenous IFT20 accumulates at the ciliary tip in WDR60 KO cells. (F) Quantification of IFT20 localization within cilia in WDR60 KO cells (n = 3 independent experiments, 102 WT and 100 WDR60 KO cells quantified). (G) Localization of IFT20-GFP in WT and WDR60 KO fixed cells. Scale bar, all panels = 5 µm. Arrows point to the ciliary base.
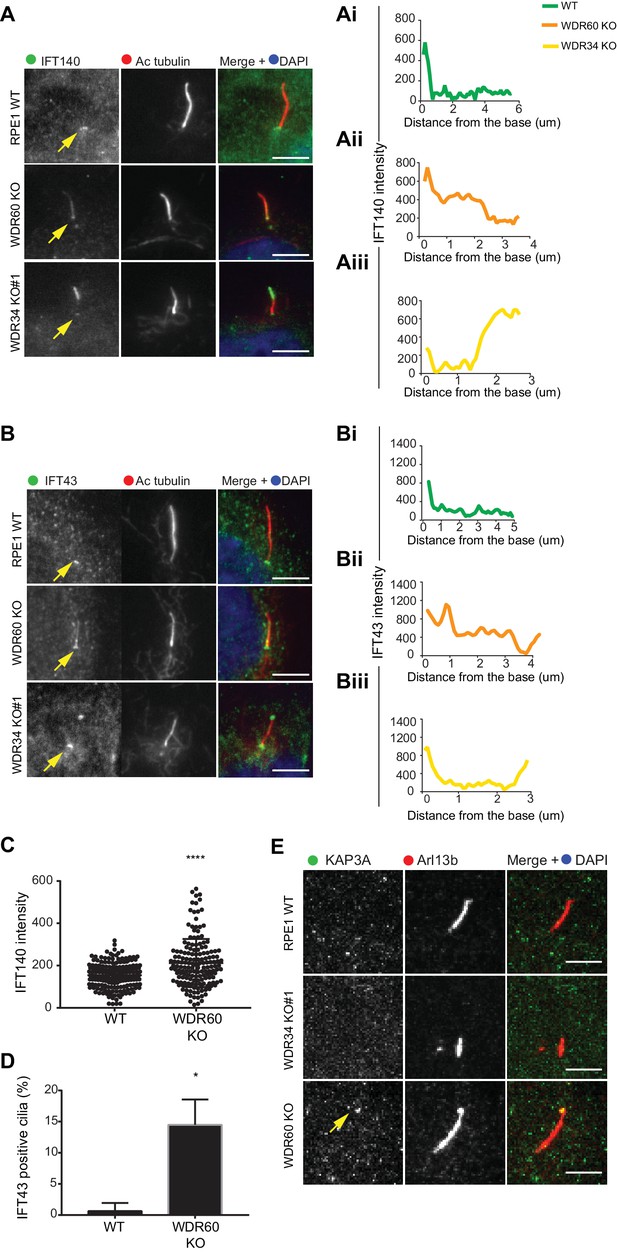
IFT-A trafficking defects in absence of WDR34 and WDR60.
(A–B) Localization of IFT-A proteins (A) IFT140 and (B) IFT43 in WT, WDR60 KO, and WDR34 KO#1 cells. Line graphs show lines scans of IFT intensity along the length of a representative cilium from WT (Ai and Bi, green), WDR60 KO (Aii and Bii, orange) and WDR34 KO (Aiii and Biii, yellow) cells. (C) Quantification of IFT140 intensity within cilia in WT and WDR60 KO cells (n = 3, 186 WT and 166 WDR60 KO cells quantified). (D) Quantification of the number of IFT43 positive cilia from WT and WDR60 KO cells (n = 3, 271 WT and 203 WDR60 KO cells quantified). (C–D) Mann-Whitney test was used, p-value: ****=<0.0001. (E) Localization of KAP3A in WT, WDR34 KO, and WDR60 KO cells. Scale bars = 5 μm. Arrows point to the ciliary base.
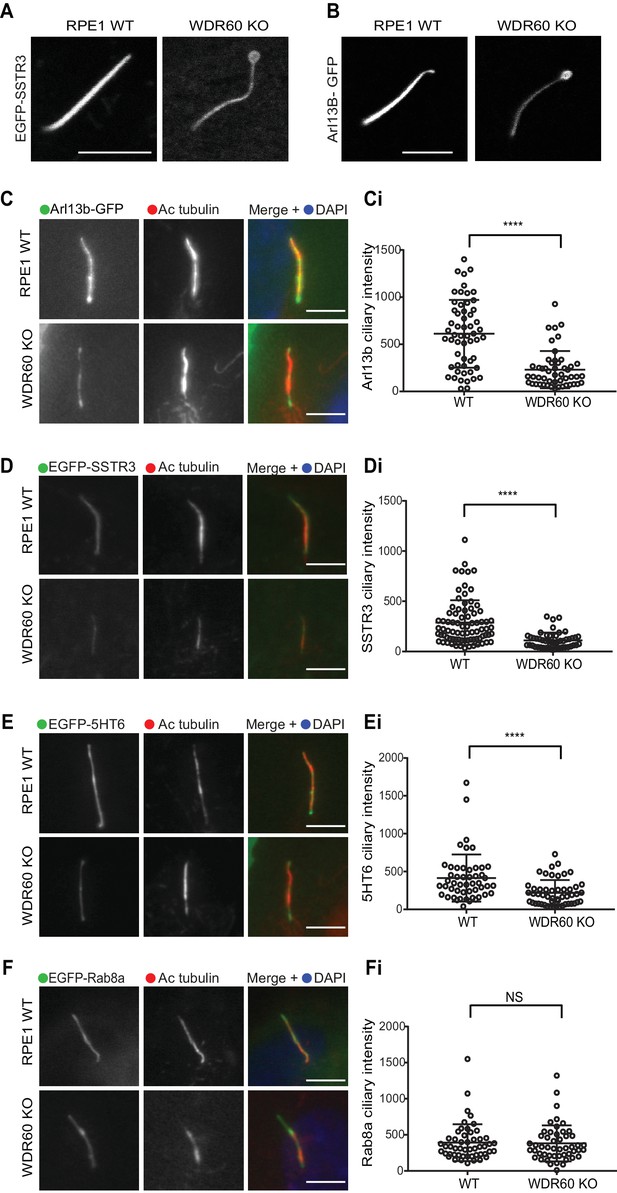
WDR60 is crucial for the composition of ciliary membrane proteins.
(A and B) Single frame images taken from live imaging movies of WT and WDR60 KO cells overexpressing (A) EGFP-SSTR3 and (B) GFP-Arl13b. (C–F) Fixed cell staining of overexpressed (C) Arl13b-GFP, (D) EGFP-SSTR3, (E) EGFP-5HT6, and (F) EGFP-Rab8a in WT and WDR60 KO cells. (Ci-Fi) Intensity quantification of the overexpressed protein indicated (Arl13b-GFP n = 3, 56 WT, and 50 WDR60 KO cells quantified; EGFP-SSTR3 n = 3, 80 WT and 50 WDR60 KO cells quantified; EGFP-5HT6 n = 3, 50 WT, and 51 WDR60 KO cells quantified; EGFP-Rab8a n = 3, 51 WT, and WDR60 50 KO cells quantified). Mann-Whitney test was used, p-value: ****=<0.0001. Scale bars 5 μm.
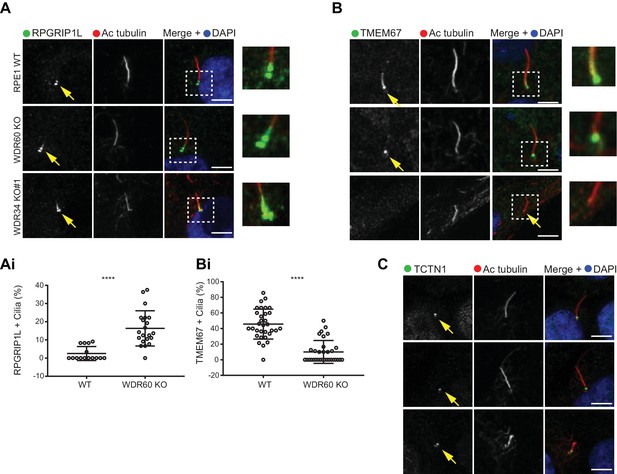
Dynein-2 is important for transition zone assembly.
(A–C) Localization of (A) RPGRIP1L, (B) TMEM67, and (C) TCTN1 in WT and KO cells. Enlargements from the box regions are shown on the right. (Ai and Bi) Percentage of RPGRIP1L and TMEM67 positive cilia (RPGRIP1L n = 3, 188 WT, and 272 WDR60 KO cells quantified; TMEM67 n = 3, 359 WT, and 243 WDR60 KO cells quantified). Mann-Whitney test was used p-value: ****=<0.0001. Scale bars 5 μm. Arrows point to the ciliary base.
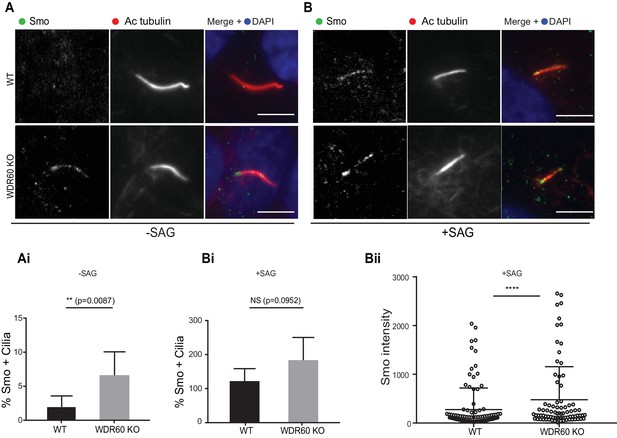
Loss of WDR60 affects Smo localization in the cilia.
(A and B) Immunofluorescence of WT and WDR60 KO cells in presence or absence of SAG and stained for Smo (green), AcTub (red), and DAPI (blue). (Ai and Bi) Percentage of Smo positive cilia in SAG untreated (n = 3, 148 WT and 120 WDR60 KO cells quantified) and treated cells (n = 3, 670 WT and 580 WDR60 KO cells quantified). (Bii) Quantification of the total intensity of ciliary Smo labeling in cells treated with SAG (n = 3, 102 WT and 82 WDR60 KO cells quantified). Mann-Whitney test was used, p-value: ****=<0.0001. Scale bars 5 μm.
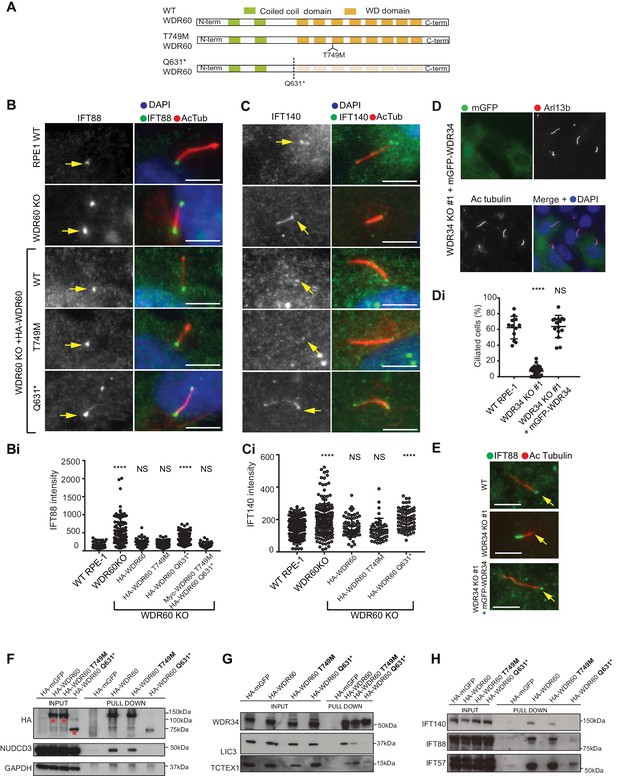
WDR34 and WDR60 KO rescue experiments.
(A) Diagrams of WT and WDR60 mutant proteins structures. (B and C) IFT88 or IFT140 staining (green) with AcTub labeling (red) of the stable cell lines shown in Figure 7—figure supplement 1A, as well as WT cells. (Bi) Total intensity quantification of IFT88 labeling across the length of primary cilia in each cell line (n = 3; 97 WT, 125 WDR60 KO, 202 WDR60 KO+ HA-WDR60, 119 WDR60 KO+ HA-WDR60 [T749M], 150 WDR60 KO+ HA-WDR60 [Q631*] cells quantified). (Ci) Total intensity quantification of IFT140 labeling across the length of primary cilia in each cell line (n = 3; 168 WT, 164 WDR60 KO, 63 WDR60 KO+ HA-WDR60, 56 WDR60 KO+ HA-WDR60 [T749M], 71 WDR60 KO+ HA-WDR60 [Q631*] cells quantified). (D) WDR34 KO#1 cells stably expressing mGFP-tagged WT and mutant WDR34. Primary cilia staining with Arl13b (red) and AcTub (blue) (Di) Percentage of ciliated cells in WT, WDR34 KO#1 cells and WDR34 KO#1 cells stably expressing mGFP-WDR34 (n = 3; 357 WT, 430 WDR34 KO, 399 WDR34 KO+ mGFP-WDR34 cells quantified). (E) IFT88 staining in WT, WDR34 KO#1 cells, and WDR34 KO#1 cells expressing GFP-tagged WDR34. One-way ANOVA followed by Kruskal-Wallis test was used p-value: ****=<0.0001. Scale bars 5 μm. Arrows point to the ciliary base. (F– H) Immunoprecipitation of HA-tagged GFP, WT WDR60, WDR60 [T749M] and WDR60 [Q631*] mutant proteins followed by immunoblot for (F) HA, NudCD3, and GAPDH; red asterisks show expression of full-lengths WDR60 and truncated Q631*, (G) WDR34, LIC3/DYNC2LI1, and TCTEX1/DYNLT1 and (H) IFT140, IFT88, and IFT57.
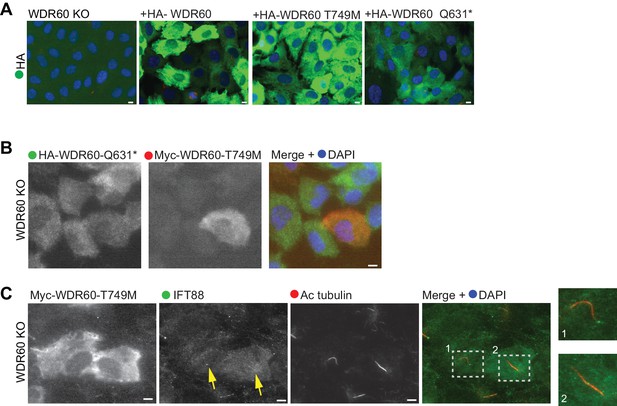
Co-expression of disease-associated mutations do not affect IFT88 localization.
(A) Immunofluorescence staining of HA (green) in WDR60 KO cell lines stably expressing HA-tagged WT and mutant WDR60 constructs. (B and C) WDR60 KO stable cell line co-expressing both WDR60 [T749M] and [Q631*] mutants stained for (B) Myc or HA tag or (C) IFT88 and AcTub. Enlargements from the box region are shown on the right. Arrows point to the ciliary base. Scale bars 5 μm.
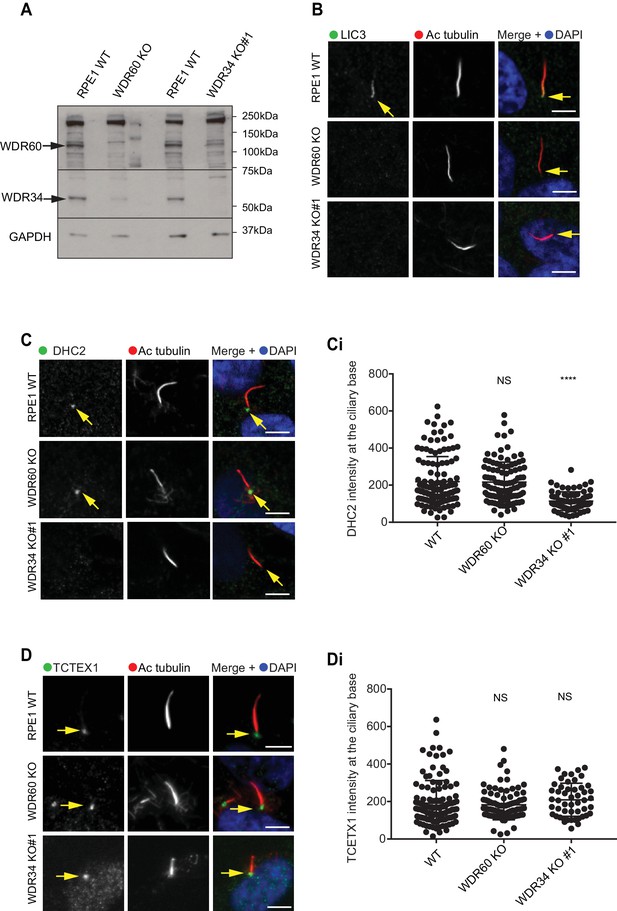
Dynein-2 assembly in primary cilium.
(A) Immunoblotting for WDR60 and WDR34 in WT, WDR34 KO#1 and WDR60 KO cells. Arrows indicate WDR34 and WDR60 proteins. (B) LIC3/DYNC2LI1 localization in the cilia of WT, WDR34 KO#1 and WDR60 KO cells. (C) DHC2/DYNC2H1 localization at the ciliary base in WT and KO cells. (Ci) Intensity quantification shows a reduction of DHC2/DYNC2H1 at the ciliary base in WDR34 KO#1 cells (n = 3, 120 WT, 106 WDR60 KO, and 71 WDR34 KO #1 cells quantified). (D) TCTEX1/DYNLT1 localizes at the ciliary base in WT and KO cells. (Di) Intensity quantification of TCTEX1/DYNLT1 at the ciliary base (n = 3 115 WT, 85 WDR60 KO, and 50 WDR34 KO#1 cells quantified). Mann-Whitney test, p-value: ****=<0.0001. Scale bars 5 μm. Arrows point to the ciliary base.
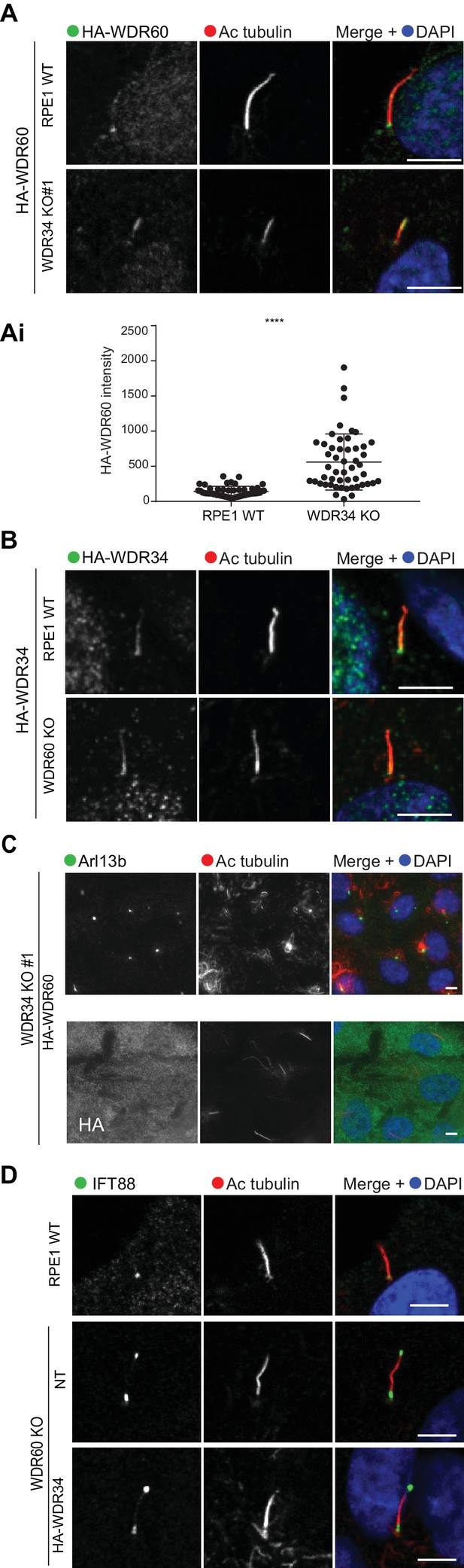
Overexpression of WDR34 cannot rescue WDR60 KO phenotype and vice versa.
(A) HA-WDR60 localization in WT and WDR34 KO cells. (Ai) Intensity quantification of HA-WDR60 in primary cilia (n = 3, 50 WT and 50 WDR34 KO cells quantified). Mann-Whitney test was used, p-value: ****=<0.0001. (B) HA-WDR34 localization in WT and WDR60 cells. For HA immunolabeling in Fig. A and B cells were treated with cytoskeletal buffer as described in methods. (C) Arl13b staining of WDR34 KO cells expressing HA-WDR60. HA labeling in green shows stable expression of HA-WDR60. (D) IFT88 localizes predominantly at the ciliary base in WT cells whereas WDR60 KO cells stably expressing HA-WDR34 accumulate IFT88 at the ciliary tip, similar to untransfected WDR60 KO cells. Scale bars 5 μm.
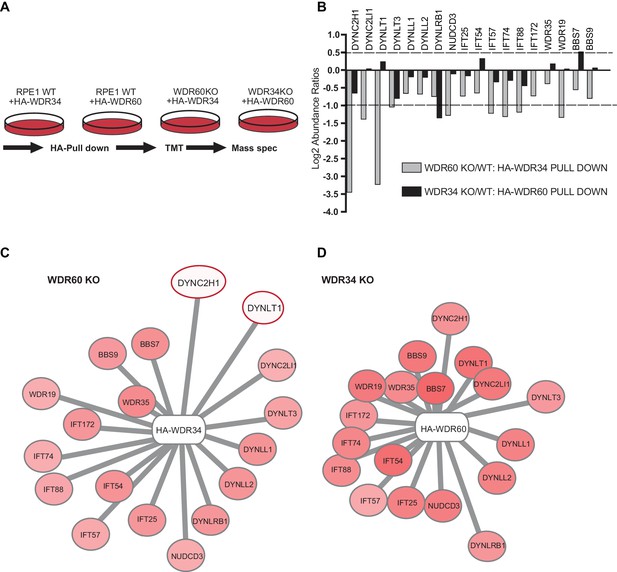
Proteomics of HA-WDR34 and HA-WDR60 interactomes in knockout cell lines.
(A) Schematic representation of the HA pull downs and TMT proteomic methodology used. (B) The graph shows the log2-transformed ratios of the abundances between WDR60 KO/WT cells (grey) and WDR34KO/WT cells (black). Proteomic data were obtained from two independent experiments. The table shows raw data from one experiment. Similar results were obtained by normalizing the data with respect to the overexpressed protein abundance. (C and D) Schematic representations of HA-WDR34 and HA-WDR60 interactors with a red- white color scale. Red values correspond to an increased protein interaction and white to a reduced protein interaction with HA-WDR34 or HA-WDR60. The red circles indicate the two interactions most affected by the loss of WDR60. Distances represent the strength of protein-protein interaction in KO compared to WT cells. Protein more distant from the center present a decreased interaction with HA-WDR34/WDR60. Protein closer to the center present unchanged or increased interaction.
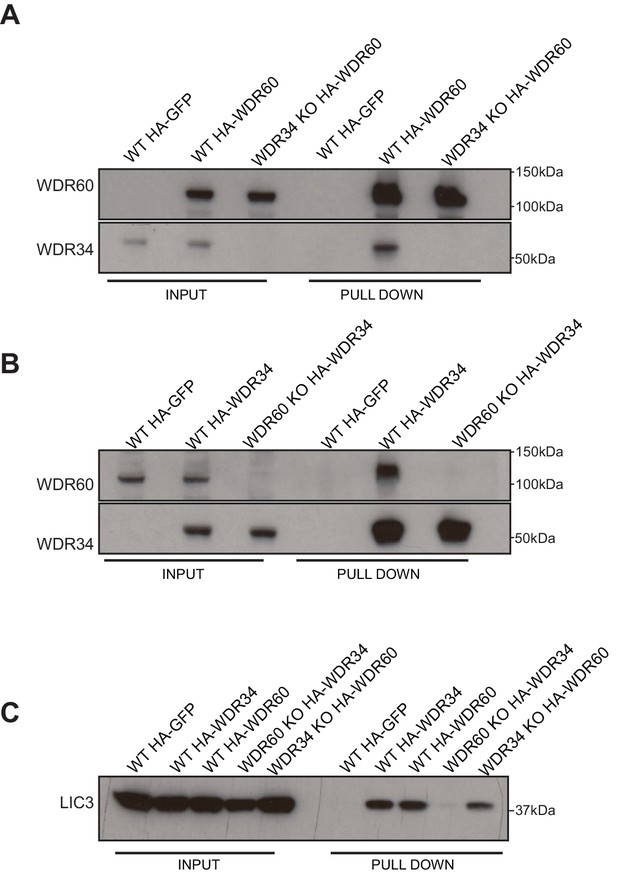
HA-WDR34 and HA-WDR60 expression in WT and KO cells.
(A) Pull down of HA-WDR60 in WT and WDR34 KO cells. HA-WDR60 pulls down WDR34 in WT but not in KO cells (B) Pull down of HA-WDR34 in WT and WDR60 KO cells. HA-WDR34 pulls down WDR60 in WT but not in KO cells. (C) Immunoprecipitation of HA-tagged GFP, WDR60, and WDR34 followed by immunoblot for LIC3.
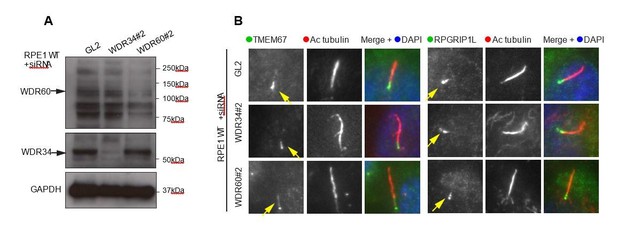
(A) RNAi of WDR34 and WDR60 validated by immunoblotting with GAPODH as a loading control.
(B) Immunofluorescence of TMEM67 and RPGRIP1L in WDR34 and WDR60 depleted cells. TMEM67 is seen at the base and within the cilium proximal to this in both control and depleted cells, RPGRIP1L is more tightly restricted to the base of the cilium in all cases. These are representative images, we have analysed multiple fields of view in each case using z-stacks and cannot identify any difference.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293T | ATCC CRL-3216 | RRID:CVCL_0063 | Purchased from ATCC, not verified further |
| Cell line (Homo sapiens) | hTERT- RPE-1 | ATCC CRL-4000 | RRID:CVCL_4388 | Purchased from ATCC, not verified further |
| Antibody | Acetylated tubulin | Sigma-Aldrich | Cat# T6793 RRID:AB_477585 | 1:2000 for IF |
| Antibody | HA | Cell Signaling Technology | Cat# 3724 RRID:AB_1549585 | 1:2000 WB, 1:1000 IF |
| Antibody | IFT88 | Proteintech Group | Cat# 13967–1-AP RRID:AB_2121979 | 1:200 WB, 1:300 IF |
| Antibody | IFT140 | Proteintech Group | Cat# 17460–1-AP RRID:AB_2295648 | 1:200, WB 1:100 IF |
| Antibody | IFT43 | Proteintech Group | Cat# 24338–1-AP RRID:AB_2749824 | 1:50 IF |
| Antibody | IFT20 | Proteintech Group | Cat# 13615–1-AP RRID:AB_2280001 | 1:200 IF |
| Antibody | TMEM67 | Proteintech Group | Cat# 13975–1-AP RRID:AB_10638441 | 1:50 IF |
| Antibody | RPGRIP1L | Proteintech Group | Cat# 55160–1-AP RRID:AB_10860269 | 1:100 IF |
| Antibody | DYNC2H1 | Proteintech Group | Cat# 55473–1-AP AB_2749823 | 1:100 IF |
| Antibody | DYNC2LI1/LIC3 | Proteintech Group | Cat# 15949–1-AP RRID:AB_2093643 | 1:250 WB, 1:100 IF |
| Antibody | TCTEX1 | Santa Cruz Biotechnology | Cat# sc-28537 RRID:AB_2093671 | 1:200 WB, 1:100 IF |
| Antibody | Arl13B | Proteintech Group | Cat# 17711–1-AP RRID:AB_2060867 | 1:1000 IF |
| Antibody | TCTN1 | Proteintech Group | Cat# 15004–1-AP RRID:AB_10644442 | 1:100 IF |
| Antibody | Smoothened | Abcam | Cat# ab38686 RRID:AB_882615 | 1:100 IF |
| Antibody | Myc | gift from Harry Mellor | PMID:20233848 | 1:1000 IF |
| Antibody | WDR60 | Sigma-Aldrich | Cat# HPA021316 RRID:AB_2215577 | 1:300 WB in Figure 1—figure supplement 2 and Figure 9—figure supplement 1 |
| Antibody | WDR60 | Novus | Cat# NBP1-90437 RRID:AB_11023602 | WB in Figure 9 |
| Antibody | WDR34 | Novus | Cat# NBP1-88805 RRID:AB_11006071 | 1:300 WB |
| Antibody | GAPDH | Abcam | Cat# ab9484 RRID:AB_307274 | 1:1000 WB |
| Antibody | p150glued | BD Labs | Cat# 612709 RRID:AB_399948 | 1:1000 WB |
| Antibody | LIS1 | Bethyl | Cat# A300-409A RRID:AB_2159907 | 1:1000 WB |
| Antibody | dic74 | Millipore | Cat# MAB1618 RRID:AB_2246059 | 1:1000 WB |
| Antibody | NUDCD3 | Atlas Antibodies | Cat# HPA019136 RRID:AB_1852370 | 1:350 WB |
| Antibody | Gamma-Tubulin | Sigma-Aldrich | Cat# T5326, RRID:AB_532292 | |
| Antibody | Anti-HA-Agarose | Sigma-Aldrich | Cat# A2095 RRID:AB_257974 | |
| Antibody | Alexa- secondary antibodies | Invitrogen | 1:1000 | |
| Antibody | HRP-secondaries | Jackson ImmunoResearch | ||
| recombinant DNA reagent | pSpCas9(BB)−2A-GFP | Addgene | Cat# PX458 PMID:24157548 | |
| recombinant DNA reagent | pGEM T Easy vector | PROMEGA | Cat# A1360 | |
| recombinant DNA reagent | L13-Arl13b-GFP (plasmid) | gift from Tamara Caspary/Addgene | Cat# 40879 PMID:21976698 | |
| recombinant DNA reagent | IFT20-GFP (plasmid) | gift from Gregory Pazour/Addgene | Cat# 45608 PMID:16775004 | |
| recombinant DNA reagent | pEGFPN3-SSTR3 (plasmid) | gifts from Kirk Mykytyn/Addgene | Cat# 35624 PMID:18256283 | |
| recombinant DNA reagent | pEGFPN3-5HT6 (plasmid) | Addgene | Cat# 35623 PMID:18256283 | |
| recombinant DNA reagent | EGFP-Rab8 (plasmid) | gift from Johan Peränen | ||
| recombinant DNA reagent | pLVX-Puro-mGFP -WDR34 (plasmid) | PMID:25205765 | NM_052844.3, NP_443076 | |
| recombinant DNA reagent | WDR60 (cDNA) | Life Technologies | Uniprot: Q8WVS4 ENSG00000126870 | |
| recombinant DNA reagent | pLVX-Puro-HA- WDR60 (plasmid) | this paper | ||
| recombinant DNA reagent | pLVX-Puro-HA- WDR60 T749M (plasmid) | this paper | ||
| recombinant DNA reagent | pLVX-Puro-Myc- WDR60 T749M (plasmid) | this paper | ||
| recombinant DNA reagent | pLVX-Puro-HA- WDR60 Q631* (plasmid) | this paper | ||
| chemical compound, drug | SAG | Selleckchem | Cat# S7779 | |
| chemical compound, drug | DAPI | Life Technologies | Cat# D1306 | |
| chemical compound, drug | Amersham ECL | GE Healthcare | Cat# RPN2106 | |
| chemical compound, drug | DSP | Thermo Fisher Scientific | Cat# 22585 | |
| chemical compound, drug | Protease inhibitors | Millipore | Cat# 539137 | |
| chemical compound, drug | LDS sample buffer | Life Technologies | Cat# NP007 | |
| chemical compound, drug | Sample reducing agent | Life Technologies | Cat# NP007 | |
| chemical compound, drug | Lipofectamine 2000 | Life Technologies | Cat# 11668027 | |
| chemical compound, drug | Lenti-XTM HTX Packaging System | Clontech | Cat# 631275 | |
| Sequence-based reagent | WDR34 gRNA sequences (Exon2) | 5'-AGCCTTTCTTC GGAGAGTGG-3' | ||
| Sequence-based reagent | WDR34 gRNA sequences (Exon3) | 5'-CAGGTGTCTTGT CTGTATAC −3' | ||
| Sequence-based reagent | WDR60 gRNA sequences (Exon3) | 5'-AGGTGCAGGGA TCCCGACCA-3' | ||
| Software and Algorithms | FIJI/ImageJ | https://fiji.sc/ | PMID:22743772 PMID:22743772 PMID:22743772 | |
| Software and Algorithms | Prism 6 | http://www.graphpad.com | ||
| Software and Algorithms | Proteome Discoverer software v2.1 | Thermo Fisher Scientific |
Additional files
-
Supplementary file 1
Excel file listing proteomic results.
Page one shows results of HA-WDR60 pull down in WT and WDR34 KO cells. Page two shows results of HA-WDR34 pull down in WT and WDR60 KO cells. ‘Description’ refers to Homo sapiens RefSeq database. ‘Identified in # experiments’ shows protein presence in 1/2 or 2/2 separate analyses. Proteins are listed in order of abundance comparing WDR34 or WDR60 KO cells respect to WT.
- https://doi.org/10.7554/eLife.39655.017
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39655.018





