Reversal of ApoE4-induced recycling block as a novel prevention approach for Alzheimer’s disease
Figures
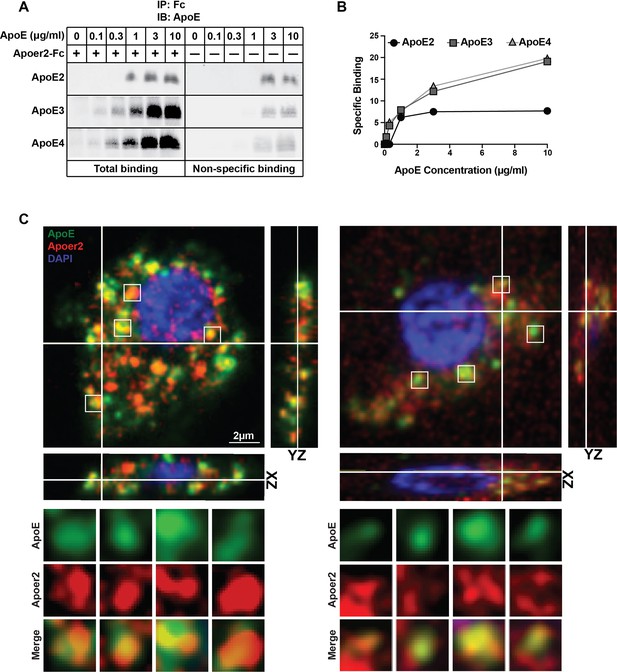
Binding of ApoE Isoforms to Apoer2.
(A and B) ApoE isoforms interact with ApoE receptor 2 (Apoer2) as tested by co-immunoprecipitation. ApoE3 and ApoE4 bind Apoer2 with similar affinity, whereas ApoE2 binding to Apoer2 is poor. ApoE-conditioned media (0, 0.1, 0.3, 1, 3 and 10 µg/ml ApoE) were incubated with Apoer2-Fc (secreted Apoer2 ectodomain fused to Fc) bound to protein-G beads and pulled down to perform immunoblotting for ApoE. Representative immunoblot images (A) and quantification (B) are shown. (C) Apoer2 co-localizes with ApoE in primary neurons. Primary cortical neurons were infected with lentiviral mCherry-Apoer2 (red) and subsequently treated with ApoE3-GFP-conditioned media (green). A single plane of a z-stack is shown with the orthogonal xz- and yz-views as indicated. White lines indicate the vertical and horizontal cuts. Boxed vesicles are shown enlarged in the panels below labeled ApoE, Apoer2 and Merge. Additionally 3D movies of the cells are provided online (Videos 1 and 2).
-
Figure 1—source data 1
Binding of ApoE Isoforms to Apoer2.
- https://doi.org/10.7554/eLife.40048.004
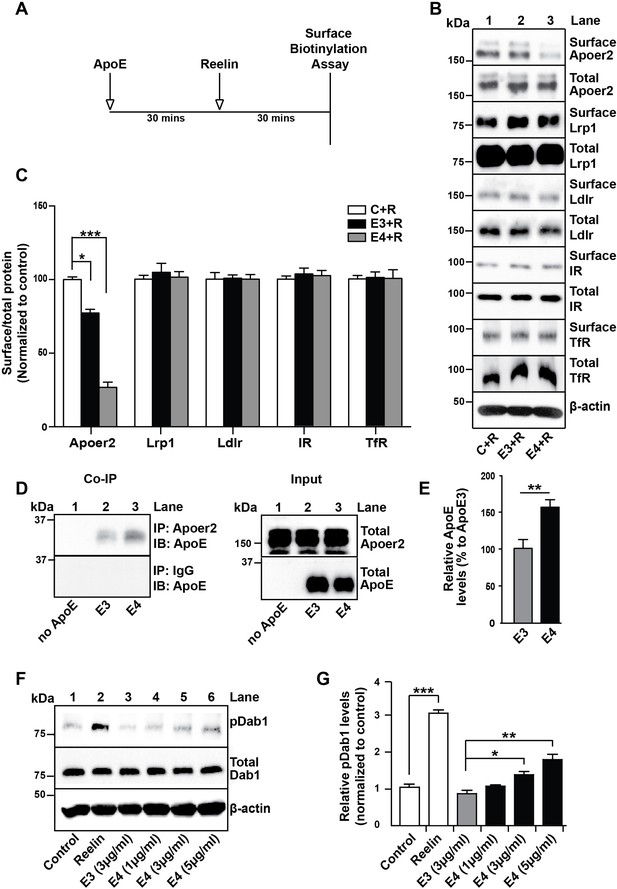
ApoE4 Impairs Recycling of the Reelin Receptor Apoer2.
(A) Timeline for experiment shown in B and C. (B and C) Apolipoprotein E (ApoE) isoforms reduce surface expression of Apoer2. ApoE-conditioned media treatment reduces the surface expression of Apoer2 in presence of Reelin in primary neurons. Apoer2 surface levels show a higher reduction with ApoE4 than ApoE3. Other ApoE receptors, such as low-density lipoprotein receptor-related protein 1 (Lrp1) and low-density lipoprotein receptor (Ldlr), as well as the endocytic receptor for transferrin (TfR) and insulin receptor (IR) exhibit comparable surface levels in the presence of ApoE3 or ApoE4. Levels of surface proteins and total proteins were analyzed by immunoblotting using antibodies raised against Apoer2, Lrp1, Ldlr, IR and TfR. Quantitative analysis of the ratio of surface and total receptor levels is shown (C). (D and E) Proteins from primary neurons incubated with ApoE-conditioned media were immunoprecipitated with anti-Apoer2 or control rabbit IgG and immunoblotted with anti-ApoE antibody. Input is shown in the right panel of (D) and quantification in (E). (F and G) ApoE4, but not ApoE3, induces phosphorylation of Dab1 independent of Reelin. Primary neurons were incubated with ApoE-conditioned media or Reelin and tested for phospho-Dab1 and total Dab1. Quantitative analysis is shown (G). All data are expressed as mean ± SEM from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. Statistical analysis was performed using one-way ANOVA and Dunnett’s post-hoc test (C and G) or Student’s t-test (E).
-
Figure 2—source data 1
ApoE4 Impairs Recycling of the Reelin Receptor Apoer2.
- https://doi.org/10.7554/eLife.40048.008
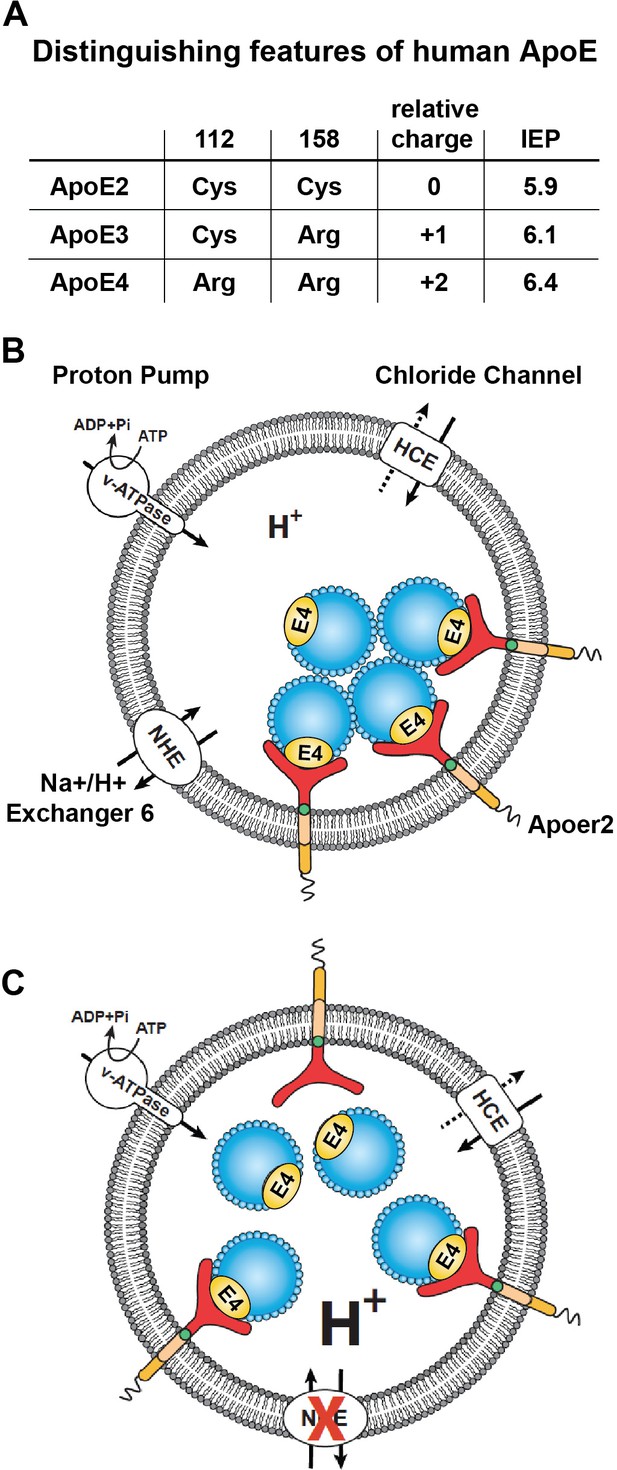
Working model illustrating the hypthetical mechanism of the vesicular trafficking defect incurred by human Apoe4.
(A) Cysteines/arginines at residues 112 and 158 account for the difference in relative charge and isoelectric point (IEP) of human ApoE isoforms. (B) Endosomal ApoE4/Apoer2 aggregates form upon acidification. Endosomal pH is regulated by the vacuolar-type H+-ATPase (vATPase, proton pump) and organellar Na+/H+ exchangers (NHEs, proton leak). After binding to ApoE4, Apoer2 undergoes endocytosis, is sequestered in endosomes and recycling is delayed. (C) Endosomal ApoE4/Apoer2 resolve when the pH is lowered further. Accelerated acidification through NHE6 inhibition activity promotes dissociation of ApoE4 and Apoer2, resulting in the efficient recycling of Apoer2 back to cell plasma membrane.
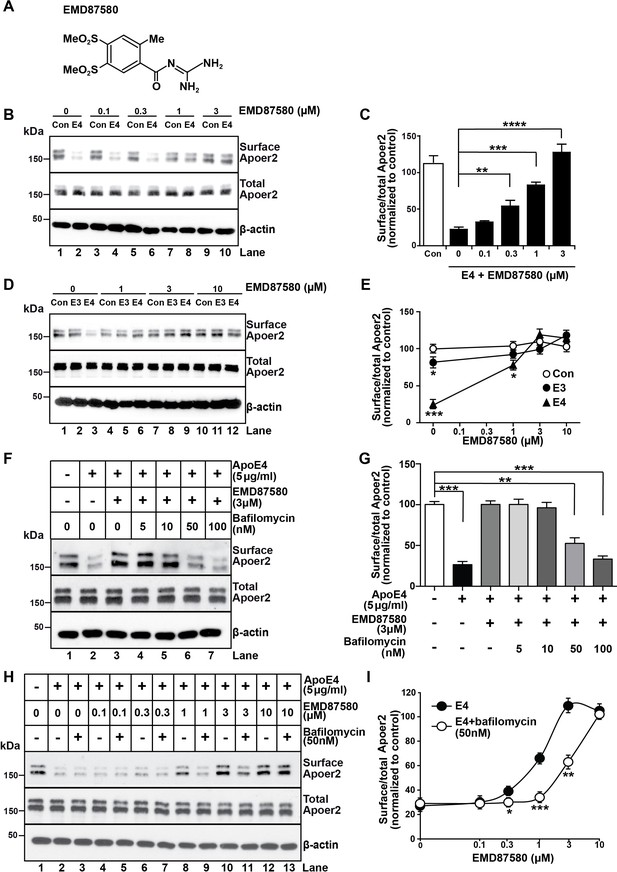
The NHE inhibitor EMD87580 prevents the intracellular trapping of ApoE4 and its receptor Apoer2.
(A) Chemical structure of the NHE inhibitor EMD87580. (B and C) EMD87580 increases the Reelin-induced surface expression of Apoer2 in ApoE4-treated neurons in a dose-dependent manner. Primary neurons were pre-treated with EMD87580 at the indicated concentrations and then incubated with Reelin with or without cell-derived ApoE4. Surface and total Apoer2 levels were analyzed by immunoblotting. (D and E) The effect of EMD87580 on Reelin-induced Apoer2 trafficking in the presence of ApoE3 or ApoE4. Primary neuronal cells were treated with EMD87580, Reelin and either ApoE3- or ApoE4-conditioned media. (F and G) Bafilomycin, a proton pump inhibitor, counteracts the effect of EMD87580 on Apoer2 recycling in a dose-dependent manner. Primary neurons were pre-treated with or without bafilomycin in the presence or absence of EMD87580 and subsequently incubated with ApoE4 and Reelin. Surface and total Apoer2 levels were analyzed by immunoblotting. (H and I) Bafilomycin shifts the EMD87580 dose response curve of Apoer2 surface expression. All values are expressed as mean ±SEM from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. Statistical analysis was performed using one-way ANOVA and Dunnett’s post-hoc test (C, E and G) or Student’s t-test (I).
-
Figure 4—source data 1
The NHE inhibitor EMD87580 prevents the intracellular trapping of ApoE4 and its receptor Apoer2.
- https://doi.org/10.7554/eLife.40048.011
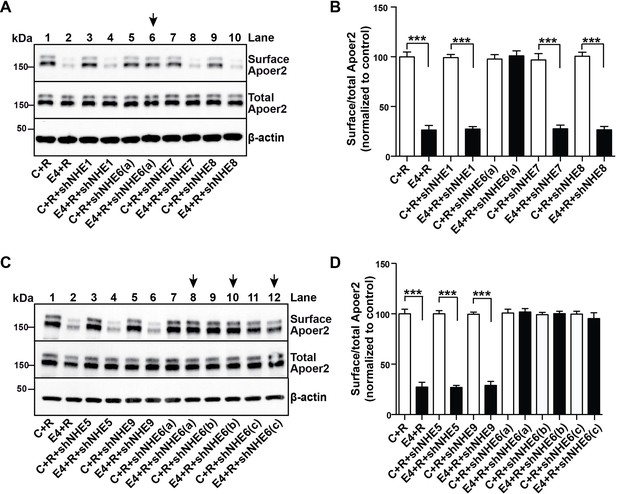
A Specific Role for NHE6 in Apoer2 trafficking.
(A and C) shRNA knockdown of NHE6, but not other NHEs (NHE1, 5, 7, 8, 9) restores ApoE4-impaired Apoer2 recycling. Lentivirus-mediated shRNAs targeting NHE1, 5, 6, 7, 8, or 9 were applied to primary neurons. Cells were then treated with ApoE4-conditioned media and Reelin, and cell surface and total Apoer2 were determined by immunoblotting. Arrows indicate conditions with restored Apoer2 surface levels. Three different shRNA constructs against NHE6 showed significant attenuation of Apoer2 cell surface levels (shNHE6 a, b, c). (B and D) Quantitative analysis of (A) and (C). All data are expressed as mean ±SEM of three independent experiments. ***p<0.001. Statistical analysis was performed using Student’s t-test (B and D).
-
Figure 5—source data 1
A Specific Role for NHE6 in Apoer2 trafficking.
- https://doi.org/10.7554/eLife.40048.013
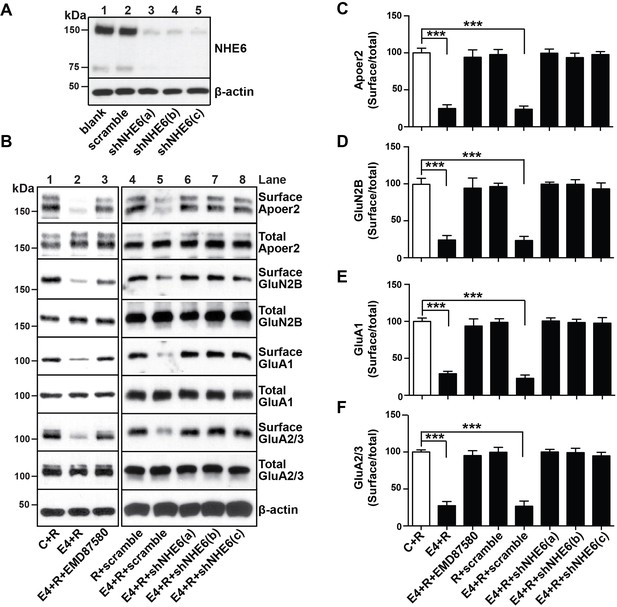
NHE6 knockdown alleviates surface trafficking deficits induced by ApoE4.
(A) Lentiviral shRNA knockdown efficiency of NHE6 protein expression in primary rat cortical neurons. (B) Lentiviral shRNA directed against NHE6 restores the ApoE4-induced trafficking deficits of surface receptors. Primary cortical neurons were infected with three different lentiviral shRNAs directed against NHE6 (shNHE6 a, b and c; lanes 6–7) or scrambled shRNA control (lanes 4, 5). Infected cultures were treated without (lanes 1 and 4) or with (lanes 2, 3, 5–8) cell-derived ApoE4 and Reelin (all lanes) and the cell surface biotinylation assay was performed for Apoer2, GluN2B, GluA1 and GluA2/3. (D–F) Quantitative analysis of immunoblot signal from (B). All data are expressed as mean ±SEM from three independent experiments. *p<0.05, **p<0.01, ***p<0.001. Statistical analysis was performed using one-way ANOVA and Dunnett’s post-hoc test (C–F).
-
Figure 6—source data 1
NHE6 knockdown alleviates surface trafficking deficits induced by ApoE4.
- https://doi.org/10.7554/eLife.40048.015
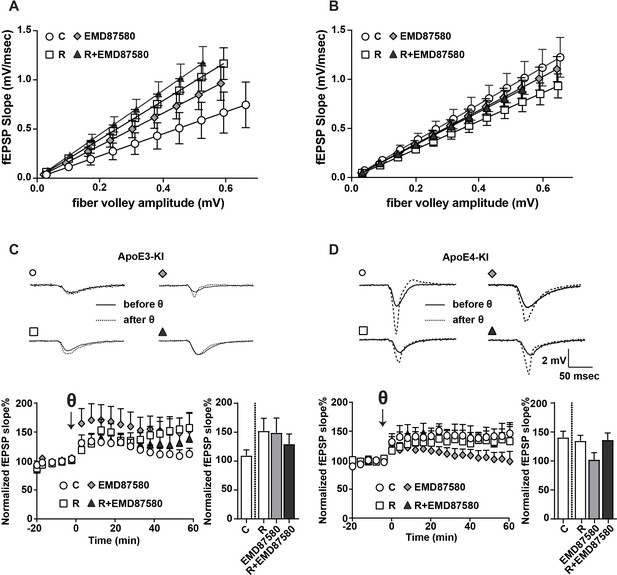
EMD87580 treatment differentially alters synaptic plasticity in ApoE3-KI and ApoE4-KI mice.
Mice were pre-treated with EMD87580 in vivo and acute hippocampal slices were subsequently analyzed by recording extracellular field potentials (A and B) Input-output curves are shown for ApoE3-KI (A) and ApoE4-KI (B). (A) ApoE3-KI slices treated with Reelin or treated with EMD87580 and Reelin exhibited increased I/O slopes compared to control (Ctrl: 1.127 ± 0.18; EMD87580: 1.653 ± 0.15; Reelin: 1.97 ± 0.14; Reelin and EMD87580: 2.23 ± 0.16; F = 9.567, p<0.05). (B) I/O curves were increased in ApoE4-KI slices at baseline (1.86 ± 0.16) compared to ApoE3-KI control (1.127 ± 0.18). Neither EMD87580 nor Reelin significantly affected the I/O slopes in ApoE4-KI slices (EMD: 1.705 ± 0.10; Reelin: 1.43 ± 0.09; Reelin and EMD: 1.67 ± 0.09; F = 1.8, p=0.14). (C and D) Results from LTP recordings in ApoE3-KI (C) and ApoE4-KI (D). Representative traces before (solid line) and 40 min after (dashed line) theta-burst stimulation (TBS) for each treatment paradigm are shown in the top panels. Bottom panels depict LTP recordings and quantification of average LTP responses between 40 and 60 min after TBS (bar graphs). (C) ApoE3-KI slices treated with Reelin (152.4% ± 21.69, n = 5) and EMD87580 (149.30 ± 25.29, n = 7) had increased LTP compared to control slices (109.7% ± 9.7, n = 6). Combined Reelin treatment with EMD87580 increased LTP (129.4 ± 17.6, n = 7) compared to control. (D) Untreated ApoE4-KI slices showed enhanced LTP (140.80% ± 10.5, n = 12) when compared to untreated ApoE3-KI slices (109.7% ± 9.7, n = 6). ApoE4-KI slices treated with Reelin did not further potentiate LTP (134.70% ± 9.63, n = 14), whereas ApoE4-KI slices treated with EMD87580 exhibited reduced LTP (102 ± 11.9, n = 14). ApoE4-KI slices with EMD87580 show increased LTP when treated with Reelin (136.30 ± 11.87, n = 15) as compared to EMD87580 treatment alone. Open circles: no additions; Open squares: Reelin alone; Gray diamonds: EMD87580 alone; Filled triangles: Reelin and EMD87580 treated.
-
Figure 7—source data 1
EMD87580 treatment differentially alters synaptic plasticity in ApoE3-KI and ApoE4-KI mice.
- https://doi.org/10.7554/eLife.40048.017
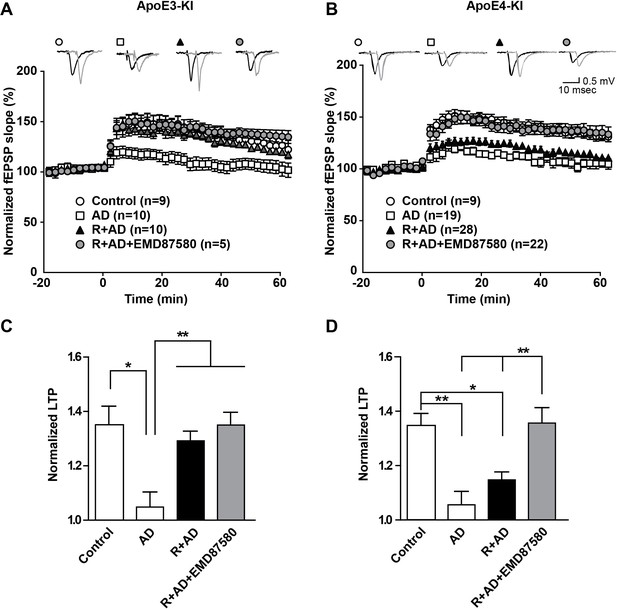
NHE inhibition counteracts Aβ-induced LTP suppression in ApoE4-KI mice.
(A–D) Treatment of hippocampal slices with AD brain extracts impairs long-term potentiation (LTP) in ApoE3-KI and ApoE4-KI mice. Reelin can attenuate the LTP deficits induced by AD extracts in ApoE3-KI, but not ApoE4-KI mice. Inhibition of NHE counteracts the LTP deficits induced by AD extract in ApoE4-KI mice. Hippocampal slices were prepared from 2- to 3-month-old ApoE3-KI and ApoE4-KI mice. Extracellular field recordings were performed in slices treated with AD brain extract, Reelin and/or EMD87580. Control slices were treated with control brain extract. Theta burst stimulation (TBS) was performed 20 min after stable baseline was attained. Representative traces are shown in each panel, before TBS induction (black) and 40 min after TBS (grey). (C, D) Quantitative analysis of normalized fEPSP slopes at 40–60 min post TBS for (A), respectively (B). All data are expressed as mean ±SEM. *p<0.05, **p<0.01. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post-hoc test (C, D).
-
Figure 8—source data 1
NHE inhibition counteracts Aβ-induced LTP suppression in ApoE4-KI mice.
- https://doi.org/10.7554/eLife.40048.019
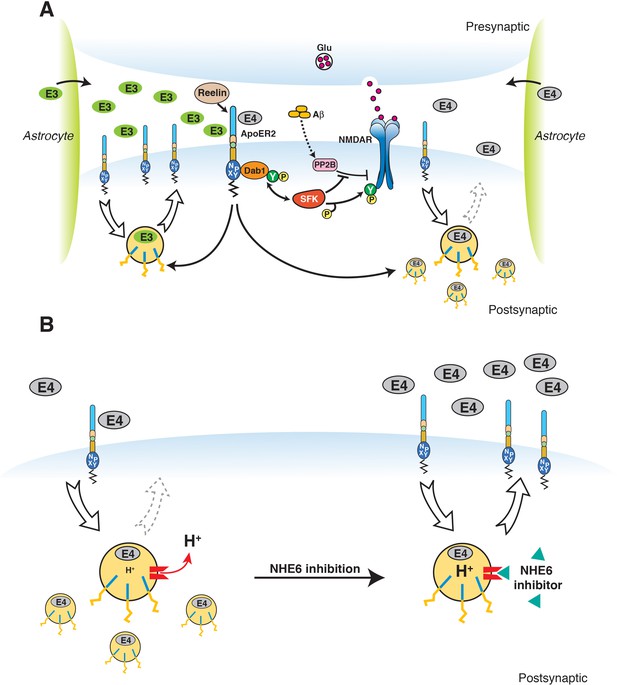
Restoration of vesicular trafficking and synaptic homeostasis by NHE6 inhibition in the presence of ApoE4.
(A) Effect of ApoE isoforms on ApoE receptor signaling at the synapse. Apoer2 induces NMDAR tyrosine phosphorylation by activating SFKs in response to Reelin in the postsynaptic neuron. Astrocyte-derived ApoE3 (green ovals) or ApoE4 (gray ovals) bind to Apoer2 and are constitutively but slowly internalized. Apoer2 undergoes accelerated endocytosis in response to Reelin signaling. ApoE4 sequesters Apoer2 in intracellular compartments thereby reducing the ability of the postsynaptic neuron to phosphorylate (activate) NMDA receptors in response to Reelin (shown on the right), whereas ApoE2 or ApoE3 efficiently recycle back to the cell surface and thus deplete surface Apoer2 levels to a lesser extent (illustrated on the left for ApoE3). Aβ oligomers interfere with NMDAR tyrosine phosphorylation by activating tyrosine phosphatases. This panel has been reproduced from Chen et al. (2010). (B) When NHE6 (red) is functioning normally, early endosomal pH is maintained at ~6.5 by the action of the proton pump and proton leakage through NHE6. This pH level is close to the isoelectric point (pI) of ApoE4, which is particularly sensitive to structural unfolding into a molten globule state upon entering acidic compartments (Morrow et al., 2002). The resulting reduced solubility may hamper efficient release of ApoE4 from its receptors (such as Apoer2 in neurons). Upon NHE6 inhibition, endosomal pH is further reduced, resulting in the release of ApoE4 from its receptors through improved solubility and the repulsion forces proteins exert upon each other when the pH is below their pI. Thus, trafficking and recycling of Apoer2 and the neurotransmitter receptors it associates with is restored. It should be noted that this property of ApoE4 to undergo impaired trafficking through endosomal compartments is universal and not restricted to neurons. Therefore, NHE6 inhibition may also aid in the clearance of amyloid-β in the brain and LDL removal in the liver.
© 2010 PNAS. All rights reserved. Subpanel A in Figure 9 is reproduced from Chen et al. (2010), PNAS with permission.
Videos
Supporting material for Figure 1C. 3D View of Apoer2 co-localizes with Apoe in primary neurons.
N-terminal mCherry-labeled Apoer2 (red) and C-terminal GFP-labeled ApoE3 (green) co-localize intracellularly in primary neurons. Rat primary cortical neurons were infected with lentiviral mCherry-Apoer2 and subsequently exposed to ApoE3-GFP-conditioned media. Confocal microscopy was performed as described in the Materials and methods section.
Supporting material for Figure 1C.
3D View of Apoer2 co-localizes with Apoe in primary neurons. N-terminal mCherry-labeled Apoer2 (red) and C-terminal GFP-labeled ApoE3 (green) co-localize intracellularly in primary neurons. Rat primary cortical neurons were infected with lentiviral mCherry-Apoer2 and subsequently exposed to ApoE3-GFP-conditioned media. Confocal microscopy was performed as described in the Materials and methods section.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293 | Thermo Fisher | R70507, RRID: CVCL_0045 | Tested mycoplasma free annually, last test January 16,2018 |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216, RRID: CVCL_0063 | Tested mycoplasma free annually, last test January 16,2018 |
| Strain, strain background (Mus musculus) | Mouse/ApoE3ki (B6.129P2- Apoetm2 (APOE*3)Mae N8) | (Knouff et al., 1999; Sullivan et al., 1997) | Originally provided by Dr. Nobuyo Maeda | |
| Strain, strain background (Mus musculus) | Mouse/ApoE4ki (B6.129P2- Apoetm2 (APOE*4)Mae N8) | (Knouff et al., 1999;Sullivan et al., 1997) | Originally provided by Dr. Nobuyo Maeda | |
| Strain, strain background (Rattus norvegicus) | SD rat | Charles River | SC:400 | |
| Antibody | goat anti- ApoE, pAb | EMD Millipore | 178479, RRID: AB_10682965 | 1:1000 (WB) |
| Antibody | rabbit anti-Apoer2 | Herz Lab, #2561 | 1:1000 (WB) | |
| Antibody | rabbit anti-Dab1 | Herz Lab, #5091 | 1:1000 (WB) | |
| Antibody | mouse anti-FLAG M2 | Sigma-Aldrich | F3165, RRID: AB_259529 | 1:1000 (WB) |
| Antibody | rabbit anti-GluA1 | Abcam | ab31232, RRID: AB_2113447 | 1:1000 (WB) |
| Antibody | rabbit anti-GluA2/3 | EMD Millipore | 07–598, RRID: AB_310741 | 1:1000 (WB) |
| Antibody | rabbit anti-GluN2B | Cell Signaling Technology | 4207S, RRID: AB_1264223 | 1:1000 (WB) |
| Antibody | rabbit anti -Insulin Receptor B (4B8), mAb | Cell Signaling Technology | 3025, RRID: AB_ 2280448 | 1:1000 (WB) |
| Antibody | rabbit anti-Lrp1 | Herz Lab | 1:5000 (WB) | |
| Antibody | rabbit anti-Ldlr | Herz Lab | 1:1000 (WB) | |
| Antibody | rabbit anti -NHE6 (C-terminus) | Herz Lab | 1:1000 (WB) | |
| Antibody | mouse anti-phosphotyrosine (4G10) mAb | EMD Millipore | 05–321, RRID: AB_309678 | 1:1000 (WB) |
| Antibody | rabbit anti-Transferrin receptor | Abcam | Ab61134, RRID: AB_943620 | 1:1000 (WB) |
| Antibody | rabbit anti-B-Actin | Abcam | Ab8227, RRID: AB_2305186 | 1:3000 (WB) |
| Peptide, recombinant protein | 6-cyano-7- nitroquinoxaline- 2,3-dione, CNQX | Sigma-Aldrich | C127 | |
| Peptide, recombinant protein | ApoE3, human | Sigma-Aldrich | SRP4696 | |
| Chemical compound, drug | B-27 Supplement (50X), serum free | Thermo Fisher | 17504044 | |
| Chemical compound, drug | Bafilomycin A1 | Cayman Chemical | CAS88899-55-2 | |
| Chemical compound, drug | DMEM | Sigma-Aldrich | D6046 | |
| Chemical compound, drug | FuGENE | Promega | E2311 | |
| Chemical compound, drug | HBSS (1X) | Gibco | 14175 | |
| Chemical compound, drug | L-Glutamic acid (Glutamate) | Sigma-Aldrich | G1251 | |
| Chemical compound, drug | Neurobasal Medium (1X) Liquid without Phenol Red | Thermo Fisher | 12348017 | |
| Chemical compound, drug | NeutrAvidin Agarose | Thermo Fisher | 29201 | |
| Chemical compound, drug | Nimodipine | Sigma- Aldrich | N3764 | |
| Chemical compound, drug | NP-40 Alternative | EMD Millipore | 492016 | |
| Chemical compound, drug | 32% Paraformaldehyde AQ solution | Fisher Scientific | 15714S | |
| Chemical compound, drug | PBS (1X) | Sigma- Aldrich | D8537 | |
| Chemical compound, drug | Penicillin- Streptomycin Solution, 100X | Corning | 30–002 CI | |
| Chemical compound, drug | Phosphatase Inhibitor Cocktail | Thermo Fisher | 78420 | |
| Chemical compound, drug | Poly-D -Lysine Solution | Sigma-Aldrich | A-003-M | |
| Chemical compound, drug | Protein A- Sepharose 4B | Thermo Fisher | 101042 | |
| Chemical compound, drug | Protein G- Sepharose 4B | Thermo Fisher | 101142 | |
| Chemical compound, drug | Proteinase Inhibitor Cocktail | Sigma-Aldrich | P8340 | |
| Chemical compound, drug | Sodium- hydrogen exchanger inhibitor | Merck KGaA | EMD87580 | |
| Chemical compound, drug | Sulfo-NHS- SS-biotin | Pierce | 21331 | |
| Chemical compound, drug | Tetrodotoxin | Sigma-Aldrich | T8024 | |
| Chemical compound, drug | Triton X-100 | Sigma- Aldrich | CAS9002-93-1 | |
| Chemical compound, drug | Vectashield with DAPI | Vector Labs | H-1200 | |
| Recombinant DNA reagent | pcDNA3.1-Zeo | Invitrogen | V79020 | |
| Recombinant DNA reagent | psPAX2 | Addgene | 12260 | |
| Recombinant DNA reagent | pMD2.G | Addgene | 12259 | |
| Recombinant DNA reagent | pLKO.1 | Addgene | 10878 | |
| Recombinant DNA reagent | pLVXCMV100 | (Dean et al., 2017) | N/A | |
Transfected construct (Mus musculus) | pCrl, Reelin expression vector | (D'Arcangelo et al., 1997) | N/A | |
| Transfected construct (Mus musculus) | pcDNA3.1-Apoer2-Fc | (Hiesberger et al., 1999) | N/A | |
| Transfected construct (Homo sapiens) | pcDNA3.1-ApoE2 | (Chen et al., 2010) | N/A | progenitor pcDNA3.1-Zeo |
| Transfected construct (Homo sapiens) | pcDNA3.1-ApoE3 | (Chen et al., 2010) | N/A | progenitor pcDNA3.1-Zeo |
| Transfected construct (Homo sapiens) | pcDNA3.1-ApoE4 | (Chen et al., 2010) | N/A | progenitor pcDNA3.1-Zeo |
| Transfected construct (shRNA construct) | pLKO.1-shRNA scramble | this paper | N/A | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE1 | Open Biosystem | TRCN0000044651 | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE5 | this paper | N/A | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE6 a | Open Biosystem | TRC N000 0068828 | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE6 b | Open Biosystem | TRCN0000068830 | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE6 c | Open Biosystem | TRCN0000068832 | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE7 | Open Biosystem | TRCN0000068812 | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE8 | this paper | N/A | progenitor pLKO.1 |
| Transfected construct (shRNA construct) | pLKO.1-shNHE9 | Open Biosystem | TRCN0000068856 | progenitor pLKO.1 |
| Transfected construct (Mus musculus) | pLVX-mCherry- Apoer2 | this paper | N/A | progenitor pLVXCMV100 |
| Transfected construct (Homo sapiens) | pcDNA3.1- ApoE3-GFP | this paper | N/A | progenitor pcDNA3.1-Zeo |
| Sequence-based reagent (oligo) | Scramble shRNA forward | IDT Inegrated DNA Technologies | N/A | 5’-CCGGCCTAAGGTTAAGTCGCCCT CGCTC-3' |
| Sequence-based reagent (oligo) | Scramble shRNA reverse | IDT Inegrated DNA Technologies | N/A | 5'-GAGCGAGGGCGACTTAACCTTAGG TTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE1 (SLC9A1) forward | Open Biosystem | TRCN0000044651 | 5’-CCGCCATC GGATCTTCCCTTCCTTACTCG-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE1 (SLC9A1) reverse | Open Biosystem | TRCN0000044651 | 5'-AGTAAGGAAGGGAAGATCCGATGTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE5 (SLC9A5) forward | IDT Inegrated DNA Technologies | N/A | 5’-CCGGAAGGACCACACTCATCTTAG TCTCG-3' |
| Sequence based reagent (oligo) | shRNA anti NHE5 (SLC9A5) reverse | IDT Inegrated DNA Technologies | N/A | 5'-AGACTAAGATGAGTGTGGTCCTTT TTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6) -a forward | Open Biosystem | TRCN0000068828 | 5’-CCGGGCCGTTTATATGGCATAGGAACTC-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6)- a reverse | Open Biosystem | TRCN0000068828 | 5'-GAGTTCCTATGCCATATAAACGGCTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6)-b forward | Open Biosystem | TRCN0000068830 | 5’-CCGGCCCTTGTCTCTCTTACTTAATCTCG-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6)-b reverse | Open Biosystem | TRCN0000068830 | 5'-AGATTAAGTAAGAGAGACAAGGGTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6)-c forward | Open Biosystem | TRCN0000068832 | 5’-CCGGCCTTGGGTCTATCTTAGCATACTCG-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE6 (SLC9A6)-c reverse | Open Biosystem | TRCN0000068832 | 5'-AGTATGCTAAGATAGACCCAAGGTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE7 (SLC9A7) forward | Open Biosystem | TRCN0000068812 | 5’-CCGGCCATTGTACT ATCCTCGTCTACTCG-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE7 (SLC9A7) reverse | Open Biosystem | TRCN0000068812 | 5'-AGTAGACGAGGATAGTACAATGGTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE8 (SLC9A8) forward | IDT Inegrated DNA Technologies | N/A | 5’-CCGGAAGGCTTCATGTGGTTGGATGCTC-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE8 (SLC9A8) reverse | IDT Inegrated DNA Technologies | N/A | 5'-GAGCATCCAACCACAT GAAGCCTTTTTTTG-3’ |
| Sequence-based reagent (oligo) | shRNA anti NHE9 (SLC9A9) forward | Open Biosystem | TRCN0000068856 | 5’-CCGGCTGGGCAGAAA GCAGAAGATTCTC-3' |
| Sequence-based reagent (oligo) | shRNA anti NHE9 (SLC9A9) reverse | Open Biosystem | TRCN0000068856 | 5'-GAGAATCTTCTGCTTT CTGCCCAGTTTTTG-3’ |
| Sequence-based reagent (oligo) | Apoer2 NT cloning site (sdm) forward | Inegrated DNA Technologies | N/A | 5'-TACAAATCTAGAGATCCG CTGCCGGGCGGCCAAG-3' |
| Sequence-based reagent (oligo) | Apoer2 NT cloning site (sdm) reverese | Inegrated DNA Technologies | N/A | 5'-ACTCATGTCGACCGCTG CGGAGAGATG CTGAAGCTG-3' |
| Sequence-based reagent (oligo) | mCherry (for mCherry-Apoer2) forward | Inegrated DNA Technologies | N/A | 5'-AAATTCGTCGACATGGTG AGCAAGGGCGA GGAGGATAAC-3' |
| Sequence-based reagent (oligo) | mCherry (for mCherry-Apoer2) reverse | Inegrated DNA Technologies | N/A | 5'-GGGAACGTCTAGAG GACTTGTACAGCTC GTCCATG-3' |
| Sequence-based reagent (oligo) | Apoer2 forward | Inegrated DNA Technologies | N/A | 5'-TGGAGCGCTAGCGC CACCATGGGCCGCCC AGAACTGG-3' |
| Sequence-based reagent (oligo) | Apoer2 reverse | Inegrated DNA Technologies | N/A | 5'-AACCCGGAATTCTCA GGGCAGTCCAT CATCTTCAAGAC-3' |
| Sequence-based reagent (oligo) | NheI-site removal (sdm) forward | Inegrated DNA Technologies | N/A | 5'-GTTTACCGTCGA CCTCTAGCTAG-3' |
| Sequence-based reagent (oligo) | NheI-site removal (sdm) reverse | Inegrated DNA Technologies | N/A | 5'-AATGTCAAGGCCTCTCACTCTCTG-3' |
| Sequence-based reagent (oligo) | CMVfull forward | Inegrated DNA Technologies | N/A | 5'-CAGTTTATCGATG GCCAGATATACGCG TTGACATTG-3' |
| Sequence-based reagent (oligo) | CMVfull reverse | Inegrated DNA Technologies | N/A | 5'-TTTCCGCTAGCGGATCC CAGCTTGGGTCT CCCTATAGTGAGT-3' |
| Sequence-based reagent (oligo) | ApoE3 (ApoE3-GFP) forward( | Inegrated DNA Technologies | N/A | 5'-ATCAGGGAATTCAAC CATGAAGGTTCTG TGGGCTGCG-3' |
| Sequence-based reagent (oligo) | GFP (ApoE3-GFP) reverse | Inegrated DNA Technologies | N/A | 5'-ATTGGTGGATCCGCGT GATTGTCGCTG GGCACAG-3' |
| Software, algorithm | Adobe Creative Cloud | Adobe | RRID: SCR_010279 | |
| Software, algorithm | GraphPad Prism 7.0 | GraphPad Software | RRID: SCR_002798 | |
| Software, algorithm | Fiji/ImageJ | NIH | RRID: SCR_002285 | |
| Software, algorithm | LabView7.0 | National Instruments | RRID: SCR_014325 | |
| Software, algorithm | Odyssey Imaging System | LI-COR | RRID: SCR_014579 | |
| Software, algorithm | Clustal Omega | EMBL-EBI | RRID: SCR_001591 | |
| Leica TCS SPE | Leica | RRID: SCR_002140 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40048.021





