Differential regulation of the Drosophila sleep homeostat by circadian and arousal inputs
Figures
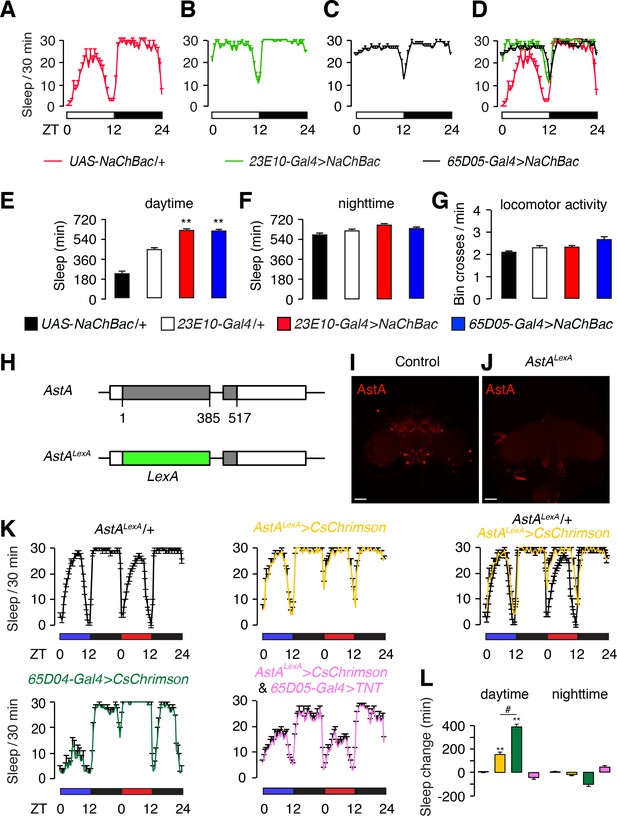
Effects on sleep resulting from hyperactivation of dFB (23E10) and AstA (65D05) neurons.
(A—D) Sleep profiles of the indicated flies during the day (ZT0—ZT12) and night (ZT12—ZT24). The white and black bars beneath the sleep profiles indicate the day and night cycles. The amounts of sleep (0—30 min) are plotted per 30 min bins. (A) Control flies (UAS-NaChBac/+). (B) Flies expressing NaChBac (UAS-NaChBac) using the 23E10-Gal4. Note that the 23E10-Gal4 > NaChBac annotation indicates transgenic flies that bear one copy of the 23E10-Gal4 transgene and one copy of the UAS-NaChBac transgene. Similar annotation applies elsewhere as all flies contain one copy each of the UAS and Gal4 transgenes. (C) 65D05-Gal4 > NaChBac. (D) Combined sleep profiles from A—C. (E,F) Quantification of daytime and nighttime sleep exhibited by the indicated flies. The genotypes are indicated below. (G) Bin crosses/min during the wake periods. The genotypes are indicated below. Error bars, SEMs. **p<0.01, one-way ANOVA with Dunnett’s test. n = 16 for UAS-NaChBac/+, n = 48 for 23E10-Gal4/+, n = 35 for 23E10-Gal4 > NaChBac, and n = 30 for 65D05-Gal4 > NaChBac. (H) Illustration of the AstALexA allele containing the LexA gene inserted at the position of the normal translation start codon for AstA. The numbers indicate the genomic nucleotide residues, with one defined as the first nucleotide of the start codon of the wild-type AstA gene. (I,J) Whole-mount AstA+ + AstALexA brains stained with anti-AstA. The scale bars represent 40 µm. (K) Sleep profiles of flies exposed to optogenetic stimulation with CsChrimson. Flies were entrained under 12 hr blue light/12 hr dark cycles for 3 days, and then shifted to a red light/dark cycle on the 4th day. Blue light does not activate CsChrimson. Shown are the sleep profiles under the blue and red light conditions as indicated by the blue and red bars. The total sleep time (0—30 min) is plotted in 30 min bins. The genotypes are indicated below. The flies include one copy of each of the indicated transgenes. LexOp-CsChrimson/+and UAS-CsChrimson/+were expressed under control of either the AstALexA/+or 65D04-Gal4/+, respectively. (L) Quantification of the change in daytime and nighttime sleep induced by CsChrimson activation. AstALexA/+serves as the control. n = 10–32. Error bars, SEMs. **p < 0.01, one-way ANOVA with Dunnett’s test. n = 32 for AstALexA/+, n = 10 for AstALexA >CsChrimson, n = 70 for 65D05-Gal4 > CsChrimson, and n = 23 for AstALexA >CsChrimson and 65D05-Gal4 > TNT..
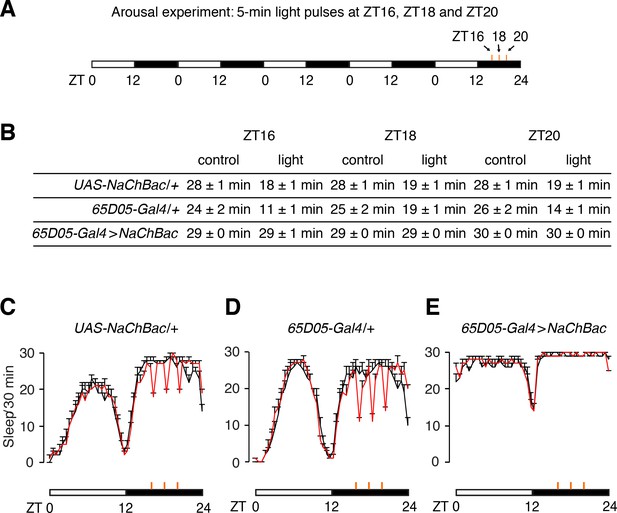
Arousal effects of 5 min light delivered at ZT16, ZT18, and ZT20.
(A) Paradigm for testing arousal. Flies were maintained for five days under 12 hr light/12 hr dark cycles. On the 5th night we exposed the flies to three 5 min light pulses delivered at ZT16, ZT18, and ZT20. The white and black horizontal bars indicate the day/night cycles and the small orange vertical bars indicate the light pulses. (B) Reductions in sleep during the 30 min period following the onset of the light stimuli. Each 30 min period includes the 5 min light pulse plus the following 25 min. Control, the sleep time during the same 30 min period of the 4th night. (C—E) Sleep profiles of the indicated flies during the 4th and 5th days. The profile during the 4th day (black traces) establishes baseline sleep (black line) and the profile during the 5th day (red) includes the effects on sleep (arousal) due to the three 5 min light pulses (orange traces). n = 24 for all flies. Error bars, SEMs.
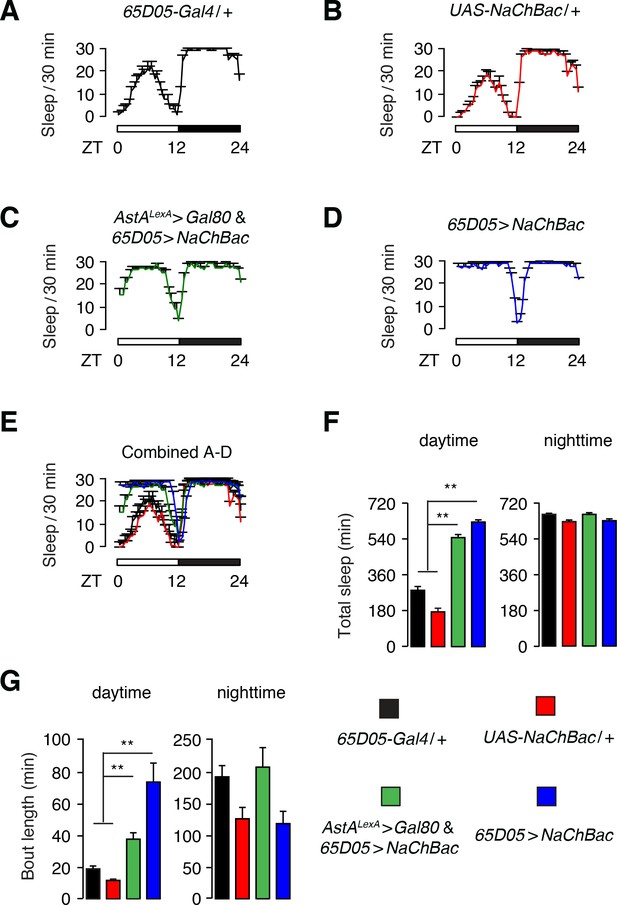
Sleep promoting effects resulting from hyperactivation of a subset of 65D05-Gal4 neurons that are not labeled by the AstAlexA reporter.
(A—E) Sleep profiles of the indicated flies during a single day (ZT0—ZT12) and night (ZT12—ZT24). The amount of sleep is plotted per 30 min bins. (F) Quantification of total daytime and nighttime sleep exhibited by the indicated flies (see color coding of the genotypes in A—D). (G) Quantification of daytime and nighttime sleep-bout length of the indicated flies (see color coding of the genotypes in A—D). Error bars, SEMs. **p<0.01, one way ANOVA with Dunnett’s test. n = 32 for 65D05-Gal4/+, n = 32 for UAS-NaChBac/+, n = 28 for AstALexA >Gal80 and 65D05 > NaChBac, and n = 23 for 65D05-Gal4 > NaChBac.
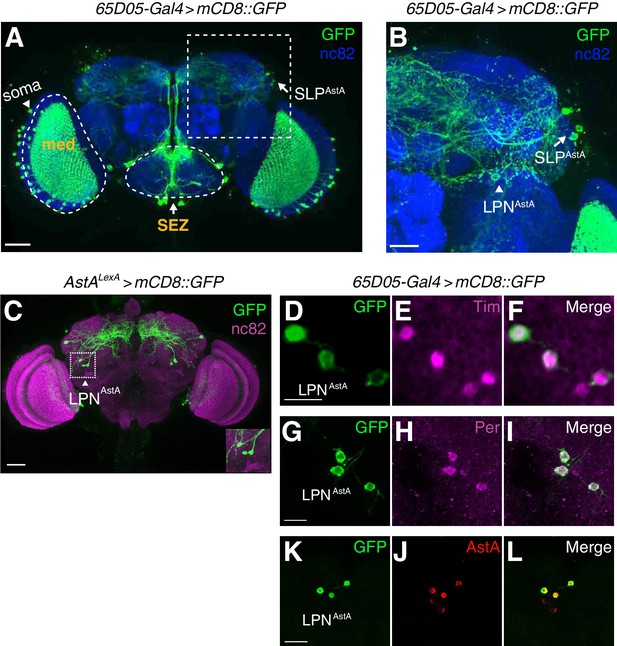
LPNAstA circadian pacemaker neurons are labeled by AstA reporters (65D05-Gal4 and AstALexA) and anti-AstA.
(A) Whole-mount of a brain expressing UAS-mCD8::GFP under the control of the 65D05-Gal4. Green, anti-GFP; blue: anti-nc82 (a pan-neuronal marker labeling active zones). The dashed box indicates the region shown at higher magnification in B. The scale bar represents 40 µm. (B) Zoomed-in view of boxed region in A. The arrow and arrowhead indicate SLPAstA and LPNAstA neurons, respectively. The scale bar represents 20 µm. (C) Immunostaining of a brain whole-mount (AstALexA >mCD8::GFP) with anti-GFP (green) and anti-nc82 (magenta) The scale bar represents 40 µm. The inset at the right bottom corner shows the three LPNAstA neurons. (D—L) Immunostaining of LPNAstA neurons in brain whole-mounts from 65D05-Gal4 > mCD8::GFP flies. (D—F) Co-staining with anti-GFP (green) and anti-Tim (magenta). The scale bar represents 10 µm. (G—I) Co-staining with anti-GFP (green) and anti-Per (magenta). The scale bar represents 10 µm. (J—K) Co-staining with anti-GFP (green) and anti-AstA (red), the scale bar represents 10 µm.
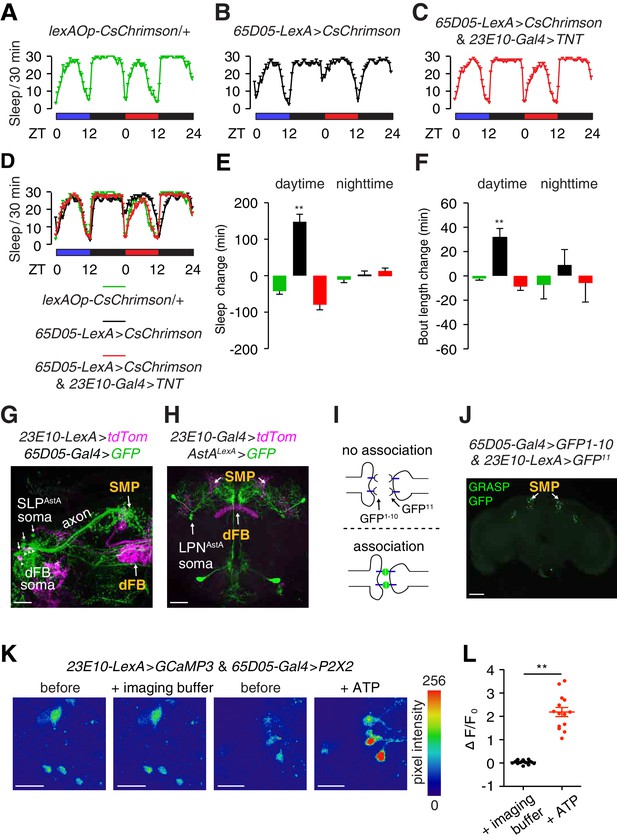
Anatomical and functional connectivity between LPNAstA/SLPAstA and dFB neurons.
(A—D) Sleep profiles of the indicated flies under 12 hr blue light/12 hr dark and 12 hr red light/12 hr dark cycles. Red but not blue lights activate CsChrimson. (E) Quantification of changes in daytime and nighttime sleep due to neuronal activation by red lights in 65D05 neurons expressing CsChrimson. (F) Quantification of changes in daytime and nighttime sleep bout length due to neuronal activation by red lights in 65D05 neurons expressing CsChrimson. The changes in total sleep time and the average bout lengths were calculated by subtracting these sleep parameter values obtained during the blue-light/dark cycles from those obtained during the red-light/dark cycles. Error bars, SEMs. **p < 0.01, one-way ANOVA with Dunnett’s test. n = 15 for lexAOp-CsChrimson/+, n = 24 for 65D05-LexA > CsChrimson, and n = 31 for 65D05-LexA > CsChrimson and 23E10-Gal4 > TNT. (G) Anti-tdTomato (magenta) and anti-GFP (green) staining of a whole-mount brain expressing tdTomato in dFB neurons (23E10-LexA > tdTom) and GFP in SLPAstA neurons (65D05-Gal4 > GFP). Both reporters showed innervation in the SMP. The scale bar represents 20 µm. (H) Anti-tdTomato (magenta) and anti-GFP (green) staining of a whole-mount brain expressing tdTomato in dFB neurons and GFP in LPNAstA neurons (AstALexA >GFP). Both reporters stained the SMP region. The scale bar represents 40 µm. (I) Cartoon illustrating the GRASP assay. Fluorescence is produced only when the two segments of GFP associate on the extracellular surfaces of adjacent cells. (J) Image of GRASP GFP fluorescence revealing close association between 65D05-Gal4 and AstAR1-LexA labeled neurons in the brain. The scale bar represents 60 µm. (K) Representative images of GCaMP3 fluorescence in dFB neurons upon activating the ATP-gated P2X2 cation channel in LPNAstA and SLPAstA neurons (65D05-Gal4 > P2X2) Shown are images before and during application of the imaging buffer (AHL) only, or ATP (2.5 mM final concentration) in imaging buffer. The scale bars represent 20 µm. (L) Quantification of the changes in GCaMP3 fluorescence in dFB neurons before and after adding the imaging buffer only or during exposure to 2.5 mM ATP to activate P2X2 expressing neurons. The effects of adding ATP versus the imaging buffer only were compared. Error bars, SEM. **p<0.01, unpaired Student’s t-test. n = 14 for ATP treatment and 16 for treatment with the imaging buffer from three independent imaging experiments.
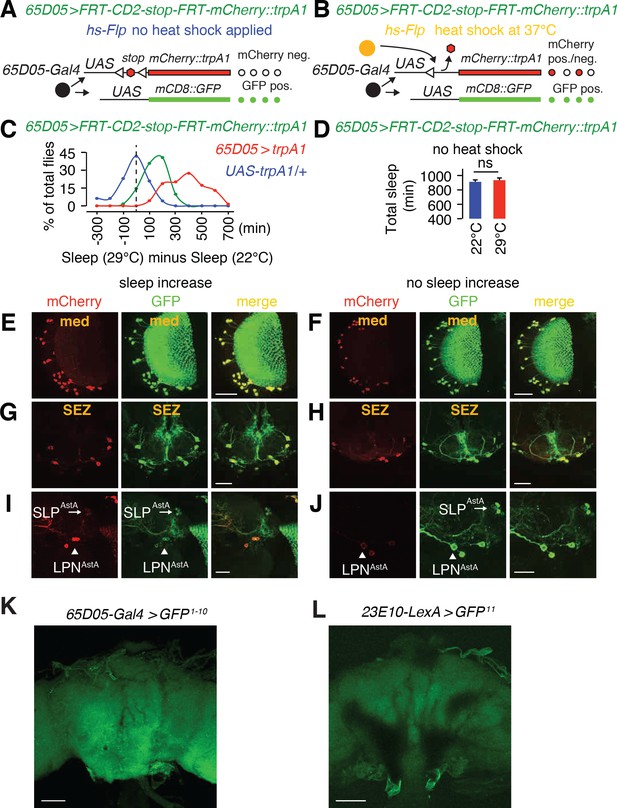
LPNAstA and SLPAstA are the sleep-promoting neurons labeled by the 65D05-Gal4.
(A, B) Diagrams of the FlpOut method used to express mCherry::TRPA1 in random subsets of 65D05-Gal4 neurons. The flies were then maintained under light/dark cycles at 22°C (no TRPA1 activation) and then switched to a light/dark cycle at 29°C to activate TRPA1. The sleep at the two temperatures was compared. The flies used in this analysis included the following four transgenes: 1) 65D05-Gal4, 2) UAS-FRT-CD2-stop-FRT-mCherry::trpA1, 3) hs-Flp, and 4) UAS-mCD8::GFP. However, for brevity, they are referred to as 65D05 > FRT-stop-FRT-mCherry::trpA1. Regardless of whether or not there was heat shock treatment to induce expression of the Flp, the UAS-mCD8::GFP was expressed in all 65D05-Gal4 neurons. (A) If the flies were not exposed to a heat shock treatment, the Flp would not be expressed and the stop cassette would not be excised, thereby precluding expression of mCherry::trpA1. (B) All flies used in this analysis were exposed to a heat shock treatment resulting in excision of the stop codon and expression of mCherry::trpA1 in random subsets of 65D05-Gal4 neurons. These flies are referred to as 65D05 > FlpOut-mCherry::trpA1. (C) Percentages of heat-shock treated flies (AstA >FlpOut-mCherry::trpA1) with the indicated sleep change due to thermal activation by TRPA1 at 29°C relative to the sleep level exhibited by the same animals at 22°C. The positive control was heat-shock treated flies expressing UAS-trpA1 in all 65D05-Gal4 neurons (65D05 > trpA1) and the negative control was heat-shock treated UAS-trpA1 flies. The vertical dashed line indicates no sleep change. n = 63 for 65D05 > FlpOut- mCherry::trpA1, n = 71 for 65D05-Gal4 > trpA1, and n = 49 for UAS-trpA1 only. (D) No increase of total sleep time in 65D05 > FRT-CD2-stop-FRT-mCherry::trpA1 flies, which do not express mCherry::trpA1 due to absence of a heat shock treatment. Error bar, SEMs. ns, not significant. Unpaired Student’s t-test. n = 16 for each temperature condition. (E—J) 65D05 > FlpOut-mCherry::trpA1 flies. Because the FlpOut is stochastic different subsets of 65D05 neurons express trpA1. However, all AstA1 positive neurons express mCD8::GPF. TRPA1 is activated at 29°C but not 22°C. Therefore, we analyzed sleep in flies first at 22°C and then at 29°C. Some flies showed an increase in sleep, and others did not. All 65D05-Gal4 neurons are labeled with anti-GFP (green), while mCherry::trpA1 expressing 65D05-Gal4 neurons are labeled with anti-mCherry. The scale bars represent 20 µm. (E, G and I) Immunostaining of different regions of a brain from a representative fly that exhibited an increase in sleep due thermoactivation of mCherry::TRPA1 at 29°C. (F, H and J) Immunostaining of different regions of a brain from a fly that did not show an increase in sleep due thermoactivation of mCherry::TRPA1 at 29°C. (K, L) Negative controls for GRASP analysis shown in Figure 3J. The scale bars represent 60 µm.
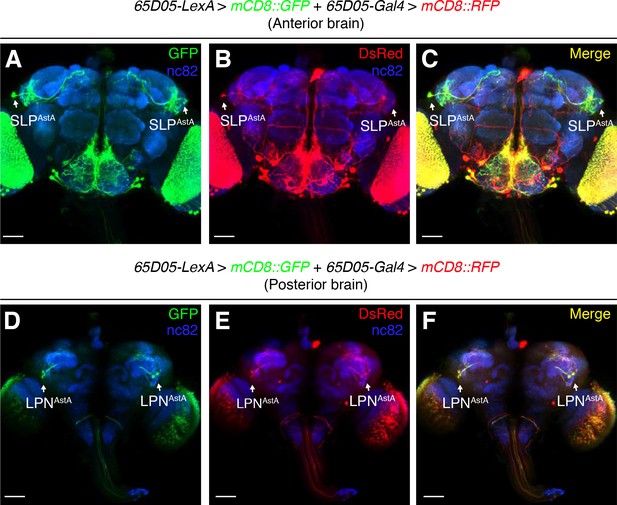
Expression pattern of 65D05-LexA and 65D05-Gal4.
A whole-mount fly brain expressing lexAOp-mCD8::GFP and UAS-mCD8::RFP under the control of the 65D05-LexA and 65D05-Gal4, respectively. The brain was stained with anti-GFP (green), anti-RFP (red), and anti-nc82 (blue). The anterior and posterior views of the brain are presented separately for better visualization of the SLPAstA and LPNAstA neurons. (A—C) Anterior brain. (D—F) Posterior brain.
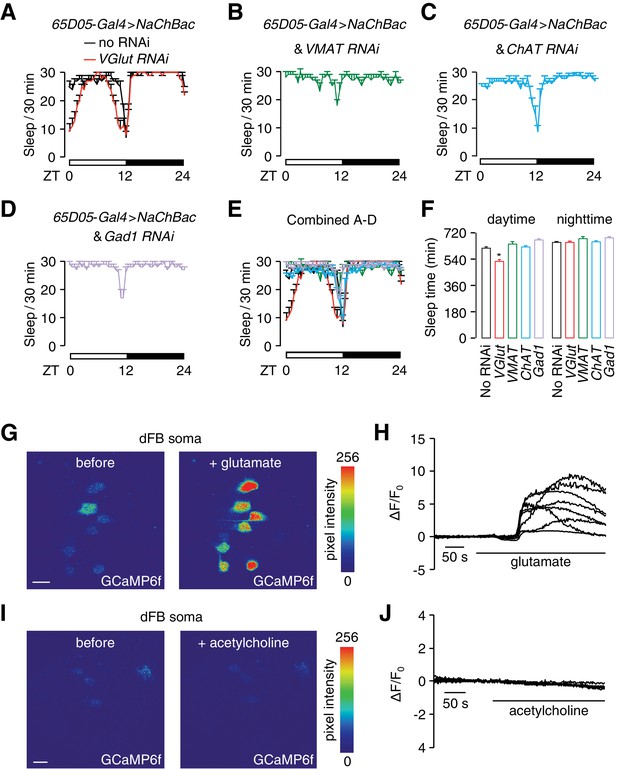
Glutamate is the excitatory neurotransmitter in 65D05-Gal4-positive neurons, which activates dFB neurons and promotes sleep.
(A) Sleep profile of 65D05-Gal4 > NaChBac flies with and without the VGlut RNAi (UAS-VGlut) expressed under control of the 65D05-Gal4. (B—D) Sleep profiles of 65D05-Gal4 > NaChBac flies expressing VMAT-RNAi, ChAT-RNAi or Gad1-RNAi transgenes under control of the 65D05-Gal4. (E) Combination of sleep profiles shown in A—D. (F) Quantification of daytime and nighttime sleep of 65D05-Gal4 > NaChBac flies expressing the indicated RNAi lines. Error bars, SEM. **p<0.01, one way ANOVA with Dunnett’s test. n = 85 for no RNAi, n = 51 for VGlut RNAi, n = 42 for VMAT RNAi, n = 46 for ChAT RNAi, and n = 40 for Gad1 RNAi. (G) Representative images of GCaMP6f fluorescence before and after bath application of 50 mM glutamate. (H) Representative traces of changes in GCaMP6f fluorescence (∆F/F0) upon bath application of 50 mM glutamate. (I) Representative images of GCaMP6f fluorescence before and after bath application of 50 mM acetylcholine. (J) Representative traces showing changes in GCaMP6f fluorescence (∆F/F0) upon bath application of 50 mM acetylcholine. The scale bars in panels G and I represent 20 µm.
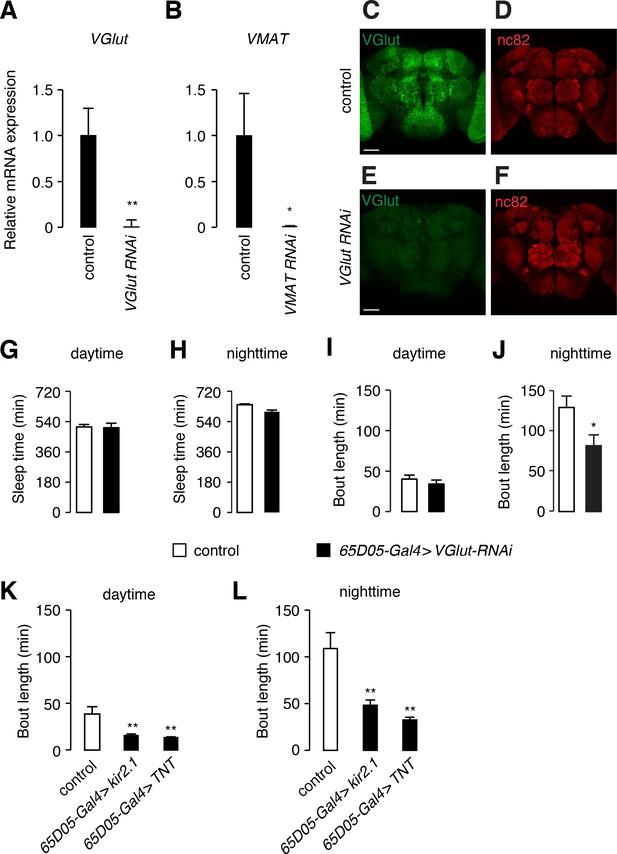
RNAi knock-down of VGlut and VMAT.
(A, B) Quantification of VGlut and VMAT mRNA expression assessed by quantitative real-time (RT) PCR. Total RNAs were extracted from control flies (w1118) or flies expressing the indicated UAS-RNAi transgene in all neurons under control of the elav-Gal4, and used for reverse transcription. Expression of VGlut or VMAT RNAs was quantified by RT-PCR. Error bars, SEMs. *p < 0.05, **p < 0.01. For VGlut RT-PCR: n = 4 for control flies, and n = 5 for VGlut RNAi. For VMAT RT-PCR: n = 5 for control flies, and n = 5 for VMAT RNAi. (C, D) A whole-mount control brain immunostained with anti-VGlut (green) and anti-nc82 (red). (E, F) A whole-mount brain that expressed VGlut-RNAi in all neurons immunostained with anti-VGlut (green) and anti-nc82 (red). (G–L) Quantification of parameters of daytime and nighttime sleep of the indicated flies. 65D05-Gal4/+served as the control. (G, H) Total sleep. (I—L) Bout length. Error bars, SEMs. *p < 0.05, **p < 0.01. Unpaired Student’s t-test for G—J, and one way ANOVA with Dunnett’s test for K and L. In panels G—J, n = 27 for the control, and n = 12 for 65D05-Gal4 > VGlut RNAi. In panels K and L, n = 32 for the control, n = 27 for 65D05-Gal4 > kir2.1, and n = 29 for 65D05-Gal4 > TNT.
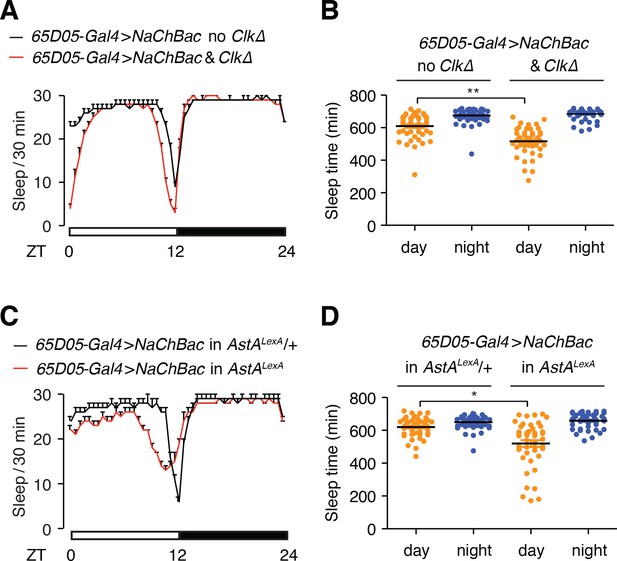
Testing whether Clk and AstA function in sleep in LPNAstA neurons.
(A) Sleep profiles of the indicated flies. (B) Quantification of daytime and nighttime sleep of the indicated flies. Error bars, SEMs. n = 48 for all flies. (C) Sleep profiles of the indicated flies. (D) Quantification of daytime and nighttime sleep of the indicated flies. Error bars, SEMs. n = 48 for 65D05-Gal4 > NaChBac in AstALexA/+. n = 46 for 65D05-Gal4 > NaChBac in AstALexA. *p < 0.05, **p < 0.01. Unpaired Student’s t-test of the indicated two groups.
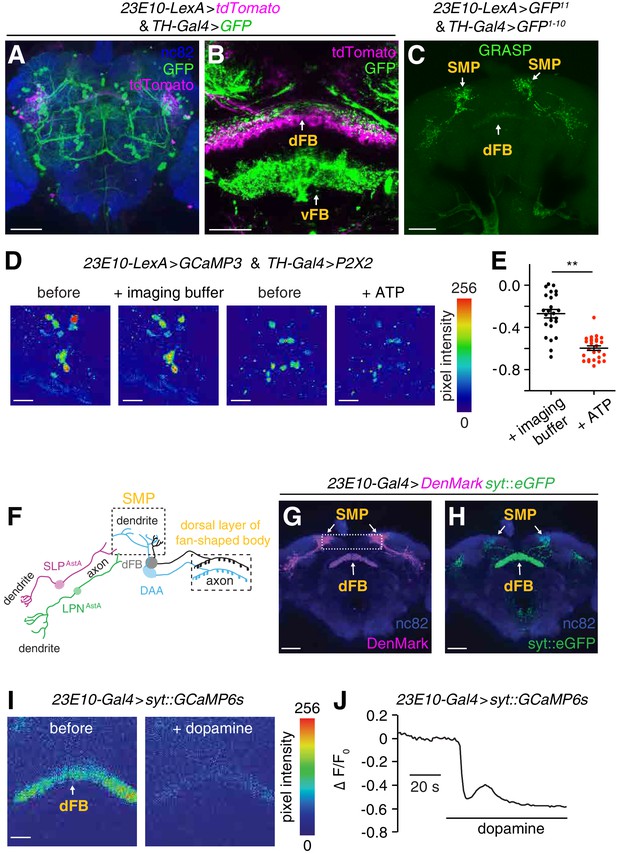
Dopaminergic arousal (DAA) neurons associate closely with dFB neurons and downregulate their activity.
(A, B) Brains expressing GFP in dopaminergic neurons (TH-Gal4 >GFP) and tdTomato in dFB neurons (23E10-LexA > tdTomato) stained with anti-GFP (green), anti-tdTomato (magenta) and anti-nc82. The dFB and vFB regions are indicated in B. The scale bars represent 60 and 20 µm in A and B, respectively. (C) GRASP GFP fluorescence revealing close associations between sleep-promoting dFB neurons and dopaminergic neurons in the brain. The dFB (23E10-LexA positive) and dopaminergic neurons (TH-Gal4 positive) expressed the GFP11 and GFP1-10 fragments, respectively. The scale bar represents 60 µm. (D) Representative images of GCaMP3 fluorescence in dFB neurons (23E10-LexA positive) upon activating the ATP-gated P2X2 cation channel (UAS-P2X2) expressed in DAA neurons under control of the TH-Gal4. Shown are images before and during application of the imaging buffer (AHL) only, or ATP (2.5 mM final concentration) in the imaging buffer. The scale bars represent 20 µm. (E) Quantification of the changes in GCaMP3 fluorescence in dFB neurons before and after adding the imaging buffer (AHL) only or during exposure to 2.5 mM ATP to activate P2X2 expressing DAA neurons. The genotype of the flies is as indicated in (D). **p < 0.01. Unpaired Student’s t-test. n = 24 for ATP treatment and n = 26 for treatment with the imaging buffer only from three independent imaging experiments. (F) Cartoon showing the positions of the dendrites and axons of LPNAstA/SLPAstA neurons and DAA neurons relative to the processes of the dFB neurons. (G, H) Whole-mount brain expressing dendritic (UAS-DenMark) and axonal (UAS-syt-eGFP) markers in dFB neurons under control of the 23E10-Gal4. The brain was stained with anti-dsRed and anti-GFP to detect DenMark and syt::eGFP, respectively, and with anti-nc82. The boxed region in (G) indicates the superior medial protocerebrum (SMP). The scale bars represent 40 µm. (I) Representative images showing syt::GCaMP6s fluorescence in axonal terminals of dFB neurons before the after bath application of 10 mM dopamine. UAS-syt::GCaMP6s was expressed in dFB neurons under control of the 23E10-Gal4. The scale bar represents 20 µm. (J) Representative trace showing the change in syt::GCaMP fluorescence (∆F/F0) in axons of dFB neurons (23E10-Gal4 > syt::GCaMP6s) upon bath application of 10 mM dopamine.
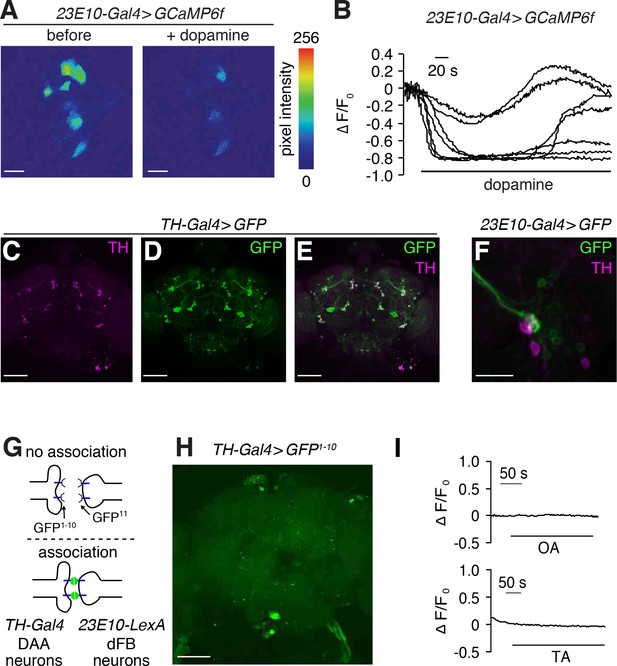
DAA arousal neurons regulate dFB neurons.
(A) Representative images of GCaMP6f fluorescence in the soma of dFB (23E10-Gal4) neurons before and during bath application of 10 mM dopamine. The scale bar represents 20 µm. (B) Representative traces showing changes in GCaMP6f fluorescence (∆F/F0) in the soma of 7 dFB neurons soma before and during bath application of 10 mM dopamine. (C—E) Whole-mount brains expressing UAS-GFP under control of the TH-Gal4 immunostained with anti-GFP and anti-TH. The scale bars represent 60 µm. (F) Immunostaining of whole-mount brain expressing GFP in dFB neurons. Anti-GFP (green). Anti-TH (red) labels dopaminergic neurons. The scale bar represents 60 µm. (G) Cartoon showing the GRASP analysis shown in Figure 5C. (H) Negative control for GRASP analysis shown in Figure 5C. The scale bar represents 60 µm. (I) Representative traces showing lack of changes in syt::GCaMP6s fluorescence (∆F/F0) in dFB (AstAR1) neurons (23E10-Gal4 > syt::GCaMP6s) upon bath application of 10 mM octopamine (OA) and 10 mM tyramine (TA). 3—four independent samples were imaged.
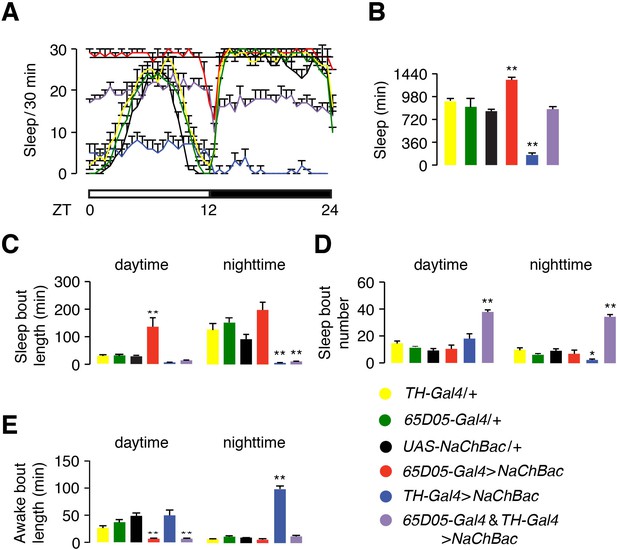
Hyperactivation of dopaminergic neurons antagonizes the sleep-promoting effect of hyperactivation of LPNAstA and SLPAstA (65D05-positive) neurons.
(A) Effects on sleep profiles due to hyperactivation of TH-positive (dopaminergic) and 65D05-positive (includes LPNAstA and SLPAstA neurons) with NaChBac. See the legend at the bottom right for the genotypes. (B) Quantification of total sleep time during a 24 hr light/dark cycle by the flies indicated in (A). (C—E) Quantification of daytime and nighttime sleep and awake parameters in flies of the indicated genotypes. (C) Sleep bout length. (D) Sleep bout number. (E) Awake bout length. n = 12—24. Error bars, SEMs. *p < 0.05, **p < 0.01. One-way ANOVA with Dunnett’s test. n = 16 for TH-Gal4/+, n = 47 for 65D05-Gal4/+, n = 16 for UAS-NaChBac/+, n = 12 for 65D05-Gal4 > NaChBac, n = 14 for TH-Gal4 >NaChBac, and n = 24 for 65D05-Gal4 and TH-Gal4 >NaChBac.
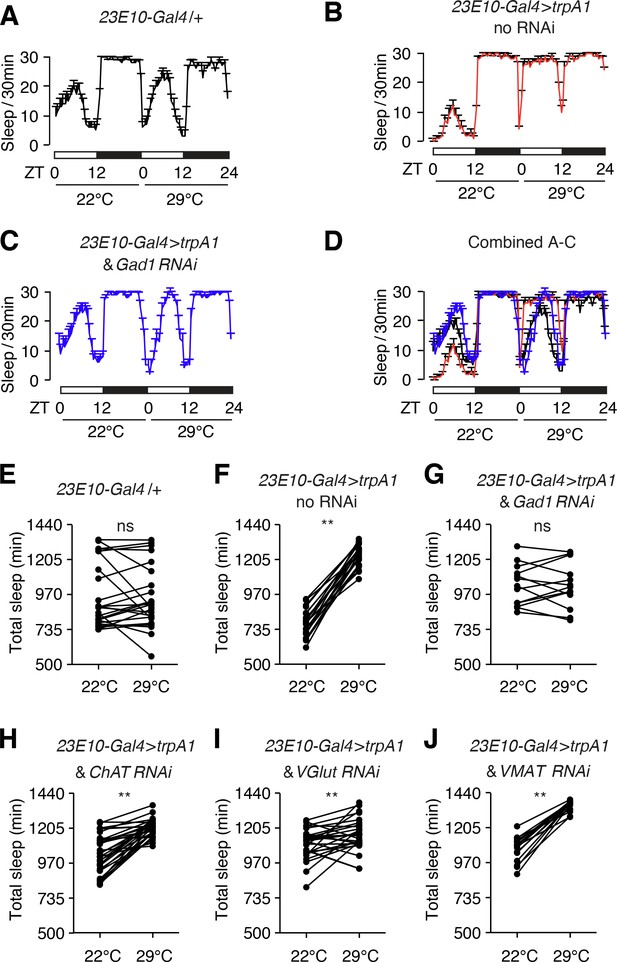
Sleep promotion due to hyperactivation of dFB neurons (23E10-Gal4 positive) depends on GABA produced by dFB neurons.
(A—D) Elevated sleep due to thermal hyperactivation of dFB neurons with TRPA1 (23E10-Gal4 > trpA1) is reduced by expression of the Gad1 RNAi in dFB neurons. TRPA1 is activated at 29°C but not 22°C. (E—J) Quantification of total sleep-time at 22°C and 29°C exhibited by 23E10-Gal4 > trpA1 flies with or without RNAi transgenes directed at genes required for neurotransmitter synthesis or packaging. **p < 0.01. Paired Student’s t-test. n = 24 for 23E10-Gal4/+, n = 24 for 23E10-Gal4 > trpA1 no RNAi, n = 13 for 23E10-Gal4 > trpA1 and Gad1 RNAi, n = 32 for 23E10-Gal4 > trpA1 and ChAT RNAi, n = 28 for 23E10-Gal4 > trpA1 and VGlut RNAi, and n = 9 for 23E10-Gal4 > trpA1 and VMAT RNAi.
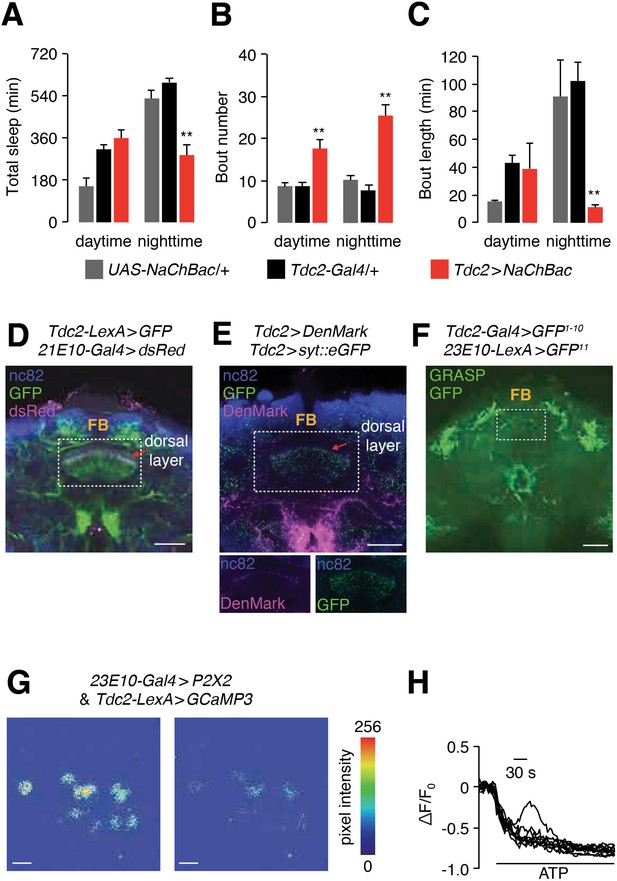
dFB neurons inhibit TDC2-positive arousal neurons.
(A—C) Quantification of daytime and nighttime sleep parameters in flies expressing UAS-NaChBac under control of the Tdc2-Gal4. (A) Total sleep time. (B) Bout number. (C) Bout length. n = 24 for UAS-NaChBac/+, n = 16 for Tdc2-Gal4/+, n = 16 for Tdc2 >NaChBac. Error bars, SEMs. Unpaired Student’s t-test. (D) A whole-mount brain co-expressing dFB and Tdc2 reporters (23E10-Gal4 > dsRed and Tdc2-LexA > GFP). The brain was co-stained with anti-dsRed and anti-GFP. The scale bar represents 40 µm. (E) Top, a whole-mount brain expressing UAS-DenMark and UAS-syt-eGFP under control of the Tdc-Gal4. The brain was co-stained with anti-dsRed (recognizes DenMark) and anti-GFP. The scale bar represents 40 µm. Bottom, images of the FB region showing the DenMark (anti-dsRed) and GFP signals separately. The boxed region contains the dFB. (F) Image of GRASP GFP fluorescence to test for close associations between Tdc2 and dFB neurons in the brain. The scale bar represents 60 µm. (G) Testing effect of GCaMP3 fluorescence in Tdc2-positive neurons after activating the P2X2 channel in dFB (23E10-Gal4 positive) neurons with ATP. Representative images showing GCaMP3 fluorescence before and after bath application of 2.5 mM ATP. The scale bars represent 20 µm. (H) Representative traces showing the changes in GCaMP3 fluorescence (∆F/F0) upon bath application of 2.5 mM ATP. The genotype is the same as in (G).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | UAS-NaChBac | Bloomington Drosophila Stock Center | stock # 9469; RRID:BDSC_9469 | |
| Genetic reagent (D. melanogaster) | UAS-TetxLC.tnt | Bloomington Drosophila Stock Center | stock# 28997; RRID:BDSC_28997 | |
| Genetic reagent (D. melanogaster) | UAS-TrpA1(B).K | Bloomington Drosophila Stock Center | stock# 26263; RRID:BDSC_26263 | |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6f | Bloomington Drosophila Stock Center | stock# 42747; RRID:BDSC_42747 | |
| Genetic reagent (D. melanogaster) | VGlut-Gal80 | Bloomington Drosophila Stock Center | stock# 58448; RRID:BDSC_58448 | |
| Genetic reagent (D. melanogaster) | Tdc2-Gal4 | Bloomington Drosophila Stock Center | stock# 9313; RRID:BDSC_9313 | |
| Genetic reagent (D. melanogaster) | AstAR1-Gal423E10 | Bloomington Drosophila Stock Center | stock# 49032; RRID:BDSC_49032 | |
| Genetic reagent (D. melanogaster) | UAS-Shits | Bloomington Drosophila Stock Center | stock# 44222; RRID:BDSC_44222 | |
| Genetic reagent (D. melanogaster) | Gad1-RNAi | Bloomington Drosophila Stock Center | stock# 28079; RRID:BDSC_28079 | |
| Genetic reagent (D. melanogaster) | VMAT-RNAi | Bloomington Drosophila Stock Center | stock# 31257; RRID:BDSC_31257 | |
| Genetic reagent (D. melanogaster) | hs-FLP,UAS-mCD8::GFP | Bloomington Drosophila Stock Center | stock# 28832; RRID:BDSC_28832 | |
| Genetic reagent (D. melanogaster) | lexAOp2-CsChrimson.mVenus | Bloomington Drosophila Stock Center | stock# 55139; RRID:BDSC_55139 | |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson.mVenus | Bloomington Drosophila Stock Center | stock# 55136; RRID:BDSC_55136 | |
| Genetic reagent (D. melanogaster) | UAS-DenMark,UAS-syt.eGFP | Bloomington Drosophila Stock Center | stock# 33064; RRID:BDSC_33064 | |
| Genetic reagent (D. melanogaster) | UAS-mCD8-GFP | Bloomington Drosophila Stock Center | stock# 5137; RRID:BDSC_5137 | |
| Genetic reagent (D. melanogaster) | lexAOp2-Gal80 | Bloomington Drosophila Stock Center | stock# 32217; RRID:BDSC_32217 | |
| Genetic reagent (D. melanogaster) | Tdc2-LexA | Bloomington Drosophila Stock Center | stock# 52242; RRID:BDSC_52242 | |
| Genetic reagent (D. melanogaster) | AstAR1-LexA23E10 | Bloomington Drosophila Stock Center | stock# 53618; RRID:BDSC_53618 | |
| Genetic reagent (D. melanogaster) | AstA-LexA65D05 | Bloomington Drosophila Stock Center | stock# 53625; RRID:BDSC_53625 | |
| Genetic reagent (D. melanogaster) | ChAT-RNAi | Bloomington Drosophila Stock Center | stock# 60028; RRID:BDSC_60028 | |
| Genetic reagent (D. melanogaster) | VGlut-RNAi | BloomingtonDrosophila Stock Center | stock# 40927; RRID:BDSC_40927 | |
| Genetic reagent (D. melanogaster) | UAS-Clk.Δ | Bloomington Drosophila Stock Center | stock# 36318; RRID:BDSC_36318 | |
| Genetic reagent (D. melanogaster) | AstA-Gal465D05 | Bloomington Drosophila Stock Center | stock# 39351; RRID:BDSC_39351 | |
| Genetic reagent (D. melanogaster) | UAS-mCD8-RFP, LexAOp2-mCD8-GFP | Bloomington Drosophila Stock Center | stock# 32229; RRID:BDSC_32229 | |
| Genetic reagent (D. melanogaster) | TH-Gal4 | Bloomington Drosophila Stock Center | stock# 8848; RRID:BDSC_8848 | |
| Genetic reagent (D. melanogaster) | UAS-FRT-CD2-stop-FRT-mCherry::trpA1 | PMID: 24860455 | ||
| Genetic reagent (D. melanogaster) | UAS-P2X2, lexOp-GCaMP3 | PMID: 22539819 | ||
| Genetic reagent (D. melanogaster) | UAS-cd4::spGFP1-10, lexAOp-cd4::spGFP11 | PMID: 19217375 | ||
| Genetic reagent (D. melanogaster) | AstALexA | This paper | ||
| Antibody | anti-dsRed (rabbit polyclonal) | Takara | Cat. 632496 | (1:1000) |
| Antibody | anti-GFP (rabbit polyclonal) | ThermoFisher Scientific | Cat. A11122 | (1:500) |
| Antibody | anti-GFP (chicken polyclonal) | ThermoFisher Scientific | Cat. A10262 | (1:1000) |
| Antibody | anti-GFP (mouse monoclonal) | Sigma-Aldrich | Cat. G6539 | (1:100) |
| Antibody | anti-Brp (termed as nc82, mouse monoclonal) | Developmental Studies Hybridoma Bank | DHSB: nc82 | (1:200) |
| Antibody | anti-Timeless (rat polyclonal) | other | Obtained from Amita Sehgal's lab; (1:1000) | |
| Antibody | anti-TH (rabbit polyclonal) | EMD Millipore | Cat. AB152 | (1:1000) |
| Antibody | anti-VGlut | other | Obtained from Aaron Dianonio's lab; (1:1000) | |
| Antibody | anti-AstA (mouse monoclonal) | Developmental Studies Hybridoma Bank | DHSB: 5F10 | (1: 50) |
| Antibody | Goat anti-mouse IgG1 | ThermoFisher Scientific | Alexa Fluor 488, 568, 647; (1:1000) | |
| Antibody | Goat anti-rat IgG | ThermoFisher Scientific | Alexa Fluor 555; (1:1000) | |
| Antibody | Goat anti-rabbit IgG | ThermoFisher Scientific | Alexa Fluor 488, 568; (1:1000) | |
| Antibody | Goat anti-chicken | ThermoFisher Scientific | Alexa Fluor 488; (1:1000) | |
| Recombinant DNA reagent | pBPLexA::p65Uw | Addgene | Cat. 26231 | pBPLexA::p65Uw was a gift from Gerald Rubin (Addgene plasmid # 26231; http://n2t.net/addgene:26231; RRID:Addgene_26231) |
| Commercial assay or kit | In-Fusion cloning kit | Takara | Cat. 121416 | |
| Chemical compound, drug | Mounting media | Vector Laboratories | Cat. H-1000 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | Cat. T8787 | |
| Chemical compound, drug | L-glutamic Acid | Sigma-Aldrich | Cat. G1251 | |
| Chemical compound, drug | ATP | Sigma-Aldrich | Cat. 10519987001 | |
| Chemical compound, drug | Octopamine | Sigma-Aldrich | Cat. O0250 | |
| Chemical compound, drug | Tyramine | Sigma-Aldrich | Cat. T2879 | |
| Chemical compound, drug | Dopamine | Sigma-Aldrich | Cat. H8502 | |
| Software, algorithm | Sleeplab | other | shared by Dr. William Joiner | |
| Other | Drosophila Embryo Injection Services | BestGene Inc. |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.40487.017





